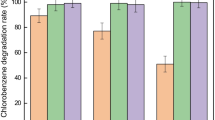
Abstract
In this study, a strain was isolated from a sewage treatment plant in Jiangsu Province, China. The strain was identified as Brevundimonas sp. KX-1. After 5 days, 50.2% 3-chlorocarbazole (3-CCZ) was degraded under the optimum condition as follows: 1 g/L starch, 30 °C, pH 6.5 and 50 mg/L 3-CCZ. The degradation of 3-CCZ by KX-1 conformed to the first-order kinetic model under different initial concentrations in this experiment. The intermediate product of 3-CCZ degradation was identified as (2E,4Z)-6-(2-amino-5-chlorophenyl)-2-hydroxy-6-oxohexa-2,4-dienoic acid. The activities of the meta-cleavage enzymes for biphenyl-2,3-diol (the analogs of intermediate product 2'-amino-5'-chloro-[1,1'-biphenyl]-2,3-diol) were measured with the crude extracts of cells grown in the presence of 3-CCZ. The complete genome of KX-1 was sequenced and compared with the Brevundimonas diminuta BZC3. BZC3 and KX-1 belonged to the same species, displaying the genetic similarity of 99%. But BZC3 could efficiently degrade gentamicin for the potential microbial function analysis. Compared with BZC3, KX-1 possessed the primary function annotations about transportation and metabolism of amino acids (6.65%) and the transportation and metabolism of carbohydrates (5.96%). In addition, KX-1 was rich in sucrose and starch metabolism pathways (ko00500) compared with the genome of BZC3, indicating the high efficiency of KX-1 for starch utilization during degradation. This article reveals the difference between strain KX-1 and bacteria of the same genus in terms of the whole genome sequence, demonstrating that KX-1 is a novel strain Brevundimonas with the ability to degrade 3-CCZ.
Graphical abstract







Similar content being viewed by others
Data availability statement
All data generated or analysed during this study are included in this published article [and its supplementary information files].
References
Agteren MHV, Keuning S, Janssen DB (1998) Polyaromatic hydrocarbons (PAHs) [M]
Ahmed N, Fariha H, Shah AA (2015) Degradation of poly(ε-caprolactone) (PCL) by a newly isolated Brevundimonas sp. strain MRL-AN1 from soil. FEMS Microbiol Lett, pp 1–7.
Bindra SK, Narang RS (1995) Combustion of flame retardants. Chemosphere 31:4413–4425
Borodovsky M, Mcininch J (1993) GENMARK: parallel gene recognition for both DNA strands. Comput Chem 17:123–133
Bradford MM (1976) A rapid and sensitive method for the quantization of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem 72:248–254
Buchanan RE, Gibbons NE (1984) Berger bacterial identification manual. Springer-Verlag, America
Cao Y, Jia L, Xu L, Xie J (2010) Analysis of different PCBs degradation abilities of biphenyl dioxygenase derived from Enterobacter sp. LY402 by molecular simulation. J Biotechnol 150:230–231
Chiang BY, Chen TC, Pai CH, Chou CC, Chen HH, Ko TP, Hsu WH, Chang CY, Wu WF,Wang, AHJ, Lin CH (2010) Protein S-thiolation by Glutathionylspermidine (Gsp): the role of Escherichia coli Gsp synthetASE/amidase in redox regulation. J Biol Chem, p 285.
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679
Dsouza M, Taylor MW, Turner SJ, Aislabie J (2014) Genome-based comparative analyses of Antarctic and temperate species of Paenibacillus. PLoS One, p 9.
Dumur F (2015) Carbazole-based polymers as hosts for solution-processed organic light-emitting diodes: Simplicity, efficacy. Org Electron 25:345–361
Fa Rajzadeh Z, Karbalaei-Heidari HR (2012) Isolation and characterization of a new Achromobacter sp. strain CAR1389 as a carbazole-degrading bacterium. World J Microbiol Biotechnol 28:3075–3080
Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L, Land ML, Pelletier DA, Beatty JT, Lang AS, Tabita FR, Gibson JL, Hanson TE, Bobst C, Torres JLTY, Peres C, Harrison FH, Gibson J, Harwood CS (2003) Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotechnol 22:55–61
Gieg LM, Otter A, Fedorak PM (1996) Carbazole degradation by Pseudomonas sp. LD2: metabolic characteristics and the identification of some metabolites. Environ Sci Technol 30:575–585
Gribble GW (2012) Occurrence of halogenated alkaloids-Chapter 1. Alkaloids Chem Biol 71:1–165.
Grifoll M, Selifonov SA, Chapman PJ (1995) Transformation of substituted fluorenes and fluorene analogs by Pseudomonas sp. Strain F274. Appl Environ Microbiol 61:3490–3493
Grigoriadou A, Schwarzbauer J (2011) Non-target screening of organic contaminants in sediments from the industrial Coastal Area of Kavala City (NE Greece). Water Air Soil Pollut 214:623–643
Haryono M, Cho ST, Fang MJ, Chen AP, Chou SJ, Lai EM, Kuo CH (2019) Differentiations in gene content and expression response to virulence induction between two agrobacterium strains. Front Microbiol, p 10.
Kilbane JJ, Daram A, Abbasian J, Kayser KJ (2002) Isolation and characterization of Sphingomonas sp. GTIN11 capable of carbazole metabolism in petroleum. Biochem Biophys Res Commun 297:242–248
Inoue K, Habe H, Yamane H, Omori T, Nojiri H (2010) Diversity of carbazole-degrading bacteria having the car gene cluster: isolation of a novel gram-positive carbazole-degrading bacterium. FEMS Microbiol Lett 245:145–153
Inoue K, Habe H, Yamane H, Nojiri M (2006) Characterization of novel carbazole catabolism genes from Gram-positive carbazole degrader nocardioides aromaticivorans IC177. Appl Environ Microbiol 72:3321–3329
Ji CY, Yan L, Chen YC, Yue SQ, Dong QX, Chen JF, Zhao MR (2019) Evaluation of the developmental toxicity of 2,7-dibromocarbazole to Zebrafish based on transcriptomics assay. J Hazard Mater 368:514–522
Karon K, Lapkowski M, Juozas G (2014) Electrochemical and UV-Vis/ESR spectroelectrochemical properties of polymers obtained from isomeric 2,7- and 3,6-linked carbazole trimers; influence of the linking topology on polymers properties. Electrochim Acta 123:176–182
Ii JJK, Daram A, Abbasian J et al (2002) Isolation and characterization of Sphingomonas sp. GTIN11 capable of carbazole metabolism in petroleum [J]. Biochem Biophys Res Commun 297(2):242–248
Inoue K, Habe H, Yamane H, Omori T, Nojiri H (2005) Diversity of carbazole-degrading bacteria having the car gene cluster: Isolation of a novel gram-positive carbazole-degrading bacterium. FEMS Microbiol Lett 245:145–153
Kronimus A, Schwarzbauer J, Dsikowitzky L, Heim S, Littke R (2004) Anthropogenic organic contaminants in sediments of the Lippe river. Germany Water Res 38:3473–3484
Kuehl DW, Durhan E, Butterworth BC, Linn D (1984) Tetrachloro-9H-Carbazole, a previously unrecognized contaminant in sediments of the Buffalo River. J Great Lakes Res 10:210–214
Leipe DD, Landsman D (1997) Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Res 25:3693–3697
Li RR, Gu PF, Fan XY, Shen JY, Wu YL, Huang LX, Li Q (2018) Isolation and characterization of PHA-producing bacteria from propylene oxide saponification wastewater residual sludge. Appl Biochem Biotechnol 186:233–244
Lin KD, Chen YQ, Yuan DX (2016) Environmental behaviors and ecotoxicology of the emerging contaminants polyhalogenated carbazoles. Huanjing Kexue 37:1576
Liu Y, Chang HQ, Li ZJ, Feng Y, Cheng DM, Xue JM (2017) Biodegradation of gentamicin by bacterial consortia AMQD4 in synthetic medium and raw gentamicin sewage. Sci Rep, p 7.
Lobastova TG, Sukhodolskaya GV, Nikolayeva VM, Baskunov BP, Turchin KF, Donova MV (2004) Hydroxylation of carbazoles by Aspergillus flavus VKM F-1024. FEMS Microbiol Lett 235:51–56
Ma Y, Li ZW, Yuan M, Chen LH, Zhou SS (2017) Isolation and identification of 3-bromocarbazole-degrading bacteria. J Environ Health b 52:796–801
Ouchiyama N, Miyachi S, Omori T (1998) Cloning and nucleotide sequence of carbazole catabolic genes from Pseudomonas stutzeri strain OM1, isolated from activated sludge. J Gen Appl Microbiol 44:57–63
Nojiri H (2012) Structural and molecular genetic analyses of the bacterial carbazole degradation system. Biosci Biotechnol Biochem 76:1–18
Omori T, Sugimura K, Ishigooka H et al (1986) Purification and Some Properties of a 2-Hydroxy-6-oxo-6-phenylhexa-2,4-dienoic Acid Hydrolyzing Enzyme from Pseudomonas cruciviae S93 B1 involved in the Degradation of Biphenyl [J]. Agri Biol Chem
Pan XX, Li CG, Chen J, Liu JQ, Ge JL, Yao JY, Wang SY, Wang ZY, Qu RJ, Li A (2019) The photodegradation of 1,3,6,8-tetrabromocarbazole in n-hexane and in solid-mediated aqueous system: Kinetics and transformation mechanisms. Chem Eng J, p 375.
Parette R, McCrindle R, McMahon KS, Pena-Abaurrea M, Reiner E, Chittim B, Riddell N, Voss G, Dorman FL, Pearson WN (2015) Halogenated indigo dyes: a likely source of 1,3,6,8-tetrabromocarbazole and some other halogenated carbazoles in the environment. Chemosphere 127:18–26
Pena-Abaurrea M, Jobst KJ, Ruffolo R, Shen L, McCrindle R, Helm PA, Reiner EJ (2014) Identification of potential novel bioaccumulative and persistent chemicals in sediments from Ontario (Canada) using scripting approaches with GC×GC-TOF MS analysis. Environ Sci Technol 48:9591–9599
Pereira WE, Rostad CE, Taylor HE (1980) Mount St. Helens, Washington, 1980 volcanic eruption: characterization of organic compounds in ash samples. Geophys Res Lett 7:954–954
Qiu Y, Zheng MG, Wang L, Zhao QS, Lou YH, Shi L, Qu LY (2019) Sorption of polyhalogenated carbazoles (PHCs) to microplastics. Mar Pollut Bull 146:718–728
Radula-Janik K, Kupka T, Ejsmont K, Daszkiewicz Z, Sauer SPA (2015) Molecular modeling and experimental studies on structure and NMR parameters of 9-benzyl-3,6-diiodo-9H-carbazole. Struct Chem 26:997–1006
Salam LB, Ilori MO, Amund OO, Numata M, Horisaki T, Nojiri H (2014) Carbazole angular dioxygenation and mineralization by bacteria isolated from hydrocarbon-contaminated tropical African soil. Environ Sci Pollut Res 21:9311–9324
Santos SCC, Alviano DS, Alviano CS, Padula M, Leitao AC, Martins OB, Ribeiro CMS, Sassaki MYM, Matta CPS, Bevilaqua J, Sebastian GV, Seldin L (2006) Characterization of Gordonia sp. strain F.5.25.8 capable of dibenzothiophene desulfurization and carbazole utilization. Appl Microbiol Biotechnol 71:355–362
Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM (2017) Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736
Zhou SS, Pan XX, Tang QZ, Zhu HB, Zhou JY, Zhao L, Guo JH, Wang ZY, Liu WP, Li A (2019) Photochemical degradation of polyhalogenated carbazoles in hexane by sunlight. Sci Total Environ 671:622–631
Shotbolt-Brown J, Hunter DWF, Aislabie J (1996) Isolation and description of carbazole-degrading bacteria. Can J Microbiol 42:79–82
Suvorova IA, Gelfand MS (2019) Comparative genomic analysis of the regulation of aromatic metabolism in betaproteobacteria. Front Microbiol, p. 10.
Takase I, Omori T, Minoda Y (1986) Microbial degradation products from biphenyl-related compounds. Agric Biol Chem 50:681–686
Trobs L, Henkelmann B, Lenoir D, Reischl A, Schramm KW (2011) Degradative fate of 3-chlorocarbazole and 3,6-dichlorocarbazole in soil. Environ Sci Pollut R 18:547–555
Wang Y, Yang JK, Lee OO, Dash S, Lau SCK, Al-Suwailem A, Wong TYH, Danchin A, Qian PY (2011) Hydrothermally generated aromatic compounds are consumed by bacteria colonizing in Atlantis II Deep of the Red Sea. Isme J 5:1652–1659
Woo SJ, Kim NY, Kim SH, Ahn SJ, Seo JS, Jung SH, Cho MY, Chung JK (2018) Toxicological effects of trichlorfon on hematological and biochemical parameters in Cyprinus carpio L. following thermal stress. Comp Biochem Physiol C-Toxicol Pharmacol 209:18–27
Zhang XJ, Zheng MG, Yin XC, Wang L, Lou YH, Qu LY, Liu XW, Zhu HH, Qiu Y (2019) Sorption of 3,6-dibromocarbazole and 1,3,6,8-tetrabromocarbazole by microplastics. Mar Pollut Bull 138:458–463
Zhu L, Hites RA (2005) Identification of brominated carbazoles in sediment cores from Lake Michigan. Environ Sci Technol 39:9446–9451
Acknowledgements
The authors acknowledge the financial support from the National Key Research and Development Project (Grant No.2019YFC0408604); Research project of ecological environment protection and restoration of Yangtze River in Zhoushan (SZGXZS2020068).
Funding
This study was funded by the National Key Research and Development Project (Grant No.2019YFC0408604) and Research project of ecological environment protection and restoration of Yangtze River in Zhoushan (SZGXZS2020068).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by XK. The first draft of the manuscript was written by XK and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript. Conceptualization: XK; Methodology: XK; Formal analysis and investigation: XK; Writing—original draft preparation: XK; Writing—review and editing: XK and XX Z; Funding acquisition: XX Z and XS S; Resources: XX Z; Supervision: XX Z and XS S.
Corresponding author
Ethics declarations
Conflict of interest
Xin Kang declares that he/she has no conflict of interest. Xiaoxiang Zhao declares that he/she has no conflict of interest. Xinshan Song declares that he/she has no conflict of interest. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to Publish
Not applicable.
Human or animal participants
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kang, X., Zhao, X. & Song, X. Analysis of a novel strain Brevundimonas KX-1 capable of degrading 3-chlorocarbazole based on the whole genome sequence. Antonie van Leeuwenhoek 116, 577–593 (2023). https://doi.org/10.1007/s10482-023-01831-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-023-01831-2