Abstract
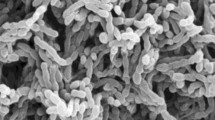
A novel, Gram-positive, spore-forming actinomycete, designated strain 7K107T, was isolated from a soil sample collected from the Karakum Desert, Turkmenistan. Strain 7K107T forms extensively branched substrate mycelia and aerial mycelia which differentiate into short chains of spores. The novel strain contains meso-diaminopimelic acid as the diagnostic wall amino acid and glucose, galactose, madurose and ribose as whole cell sugars. The predominant menaquinones were identified as MK-10(H4), MK-9(H4), MK-10(H6) and MK-9(H6). The polar lipids were found to be diphosphatidylglycerol, phosphatidylethanolamine, glycolipids, phospholipids, unidentified lipids and an aminolipid. Major fatty acids were identified as C17:0 10-methyl and C14:0. The G + C content of the genomic DNA was determined to be 70.8%. Phylogenetic analysis based on 16S rRNA gene sequences showed that the strain is a member of the family Streptosporangiaceae. The strain shares high 16S rRNA gene sequence similarity (96.2%) with Sphaerisporangium album YIM 48782T followed by Sphaerisporangium corydalis NEAU-YHS15T (96.0%) and Nonomuraea candida HMC10T (95.9%). However, phylogenetic analyses based on 16S rRNA and gyrB genes, as well as whole genome comparison, confirmed the distinctiveness of the strain from closely related type strains of the genera Sphaerisporangium, Nonomuraea and Thermostaphylospora. On the basis of morphological, chemotaxonomic and phylogenetic as well as genomic analyses, strain 7K107T is concluded to represent a new genus within the family Streptosporangiaceae, for which the name Desertiactinospora gelatinilytica gen. nov., sp. nov is proposed. The type strain of D. gelatinilytica is 7K107T (= DSM 107423T = JCM 32585T = KCTC 49108T).




Similar content being viewed by others
References
Ara I, Kudo T (2007) Sphaerosporangium gen. nov., a new member of the family Streptosporangiaceae, with descriptions of three new species as Sphaerosporangium melleum sp. nov., Sphaerosporangium rubeum sp. nov. and Sphaerosporangium cinnabarinum sp. nov., and transfer of Streptosporangium viridialbum Nonomura and Ohara 1960 to Sphaerosporangium viridialbum comb. nov. Actinomycetologica 21:11–21
Aziz R, Bartels D, Best A, DeJongh M, Disz T, Edwards R, Formsma K, Gerdes S, Glass E, Kubal M, Meyer F, Olsen G, Olson R, Osterman A, Overbeek R, McNeil L, Paarmann D, Paczian T, Parrello B, Pusch G, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genom 9(1):75
Bertels F, Silander OK, Pachkov M, Rainey PB, van Nimwegen E (2014) Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol Biol Evol 31:1077–1088
Brosius J, Palmer ML, Kennedy PJ, Noller HF (1978) Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA 75:4801–4805
Bull AT (2011) Actinobacteria of the extremobiosphere. In: Horikoshi K (ed) Extremophiles handbook. Springer, Tokyo, pp 1203–1240
Carro L, Nouioui I, Sangal V, Meier-Kolthoff JP, Trujillo ME, Montero-Calasanz MC, Sahin N, Smith DL, Kim KE, Peluso P, Deshpande S, Woyke T, Shapiro N, Kyrpides NC, Klenk H-P, Göker M, Goodfellow M (2018) Genomebased classification of micromonosporae with a focus on their biotechnological and ecological potential. Sci Rep 8:525
Castejon M, Menéndez MC, Comas I, Vicente A, Garcia MJ (2018) Whole-genome sequence analysis of the Mycobacterium avium complex and proposal of the transfer of Mycobacterium yongonense to Mycobacterium intracellulare subsp. yongonense subsp. nov. Int J Syst Evol Microbiol 68:1998–2005
Chun J, Goodfellow M (1995) A phylogenetic analysis of the genus Nocardia with 16S rRNA gene sequences. Int J Syst Evol Microbiol 45:240–245
Couch JN (1955) A new genus and family of the Actinomycetales, with a revision of the genus Actinoplanes. J Elisha Mitchell Sci Soc 71:148–155
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Goodfellow M (1971) Numerical taxonomy of some nocardioform bacteria. J Gen Microbiol 69:33–90
Goodfellow M, Quintana ET (2012) Family 1. Streptosporangiaceae Goodfellow, Stanton, Simpson and Minnikin 1990a 321VP. In: Goodfellow M, Kampfer P, Busse H-J, Trujillo M, Suzuki KE, Ludwig W, Whitman WB (eds) Bergey’s manual of systematic bacteriology, vol. 5, vol 2. Springer, New York
Goodfellow M, Stanton LJ, Simpson KE, Minnikin DE (1990) Numerical and chemical classification of ActinopIanes and some related actinomycetes. J Gen Microbiol 136:19–36
Goodfellow M, Maldonado LA, Quintana ET et al (2005) Reclassification of Nonomuraea flexuosa (Meyer 1989) Zhang et al. 1998 as Thermopolyspora flexuosa gen. nov., comb. nov., nom. rev. Int J Syst Evol Microbiol 55:1979–1983
Gordon RE, Barnett DA, Handerhan JE, Pang C (1974) Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int J Syst Bacteriol 24:54–63
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Jensen A, Scholz CF, Kilian M (2016) Re-evaluation of the taxonomy of the Mitis group of the genus Streptococcus based on whole genome phylogenetic analyses, and proposed reclassification of Streptococcus dentisani as Streptococcus oralis subsp. dentisani comb. nov., Streptococcus tigurinus as Streptococcus oralis subsp. tigurinus comb. nov., and Streptococcus oligofermentans as a later synonym of Streptococcus cristatus. Int J Syst Bacteriol 66(11):4803–4820
Jones KL (1949) Fresh isolates of actinomycetes in which the presence of sporogenous aerial mycelia is a fluctuating characteristic. J Bacteriol 57:141–145
Kelly KL (1964) Inter-Society Color Council-National Bureau of Standards color name charts illustrated with centroid colors. US Government Printing Office, Washington
Kroppenstedt RM (1985) Fatty acid and menaquinone analysis of actinomycetes and related organisms. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 173–199
Kudo T, Itoh T, Miyadoh S, Shomura T, Seino A (1993) Herbidospora gen. nov., a new genus of the family Streptosporangiaceae Goodfellow et al. 1990. Int J Syst Bacteriol 43:319–328
Kumar S, Stecher G, Tamura K (2016) Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870
Kuykendall LD, Roy MA, O’Neill JJ, Devine TE (1988) Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int J Syst Bacteriol 38:358–361
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Lechevalier MP, Lechevalier HA (1970a) Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol 20:435–443
Lechevalier HA, Lechevalier MP (1970b) A critical evaluation of the genera of aerobic actinomycetes. In: Prauser H (ed) The actinomycetales. Gustav Fischer Verlag, Jena, pp 393–405
Lechevalier MP, De Bièvre C, Lechevalier HA (1977) Chemotaxonomy of aerobic actinomycetes: phospholipid composition. Biochem Syst Ecol 5:249–260
Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60
Meyers PR (2014) Gyrase subunit B amino acid signatures for the actinobacterial family Streptosporangiaceae. Syst Appl Microbiol 37:252–260
Meyers PR (2015) Analysis of recombinase A (recA/RecA) in the actinobacterial family Streptosporangiaceae and identification of molecular signatures. Syst Appl Microbiol 38:567–577
Meyers PR (2017) Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxyl acids. J Clin Microbiol 16:584–586
Miller LT (1982) Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxyl acids. J Clin Microbiol 16:584–586
Mingma R, Duangmal K, Take A, Inahashi Y, Omura S et al (2016) Proposal of Sphaerimonospora cavernae gen. nov., sp. nov. and transfer of Microbispora mesophila (Zhang et al. 1998) to Sphaerimonospora mesophila comb. nov. and Microbispora thailandensis (Duangmal et al. 2012) to Sphaerimonospora thailandensis comb. nov. Int J Syst Evol Microbiol 66:1735–1744
Minnikin D, O’donnell A, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett J (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Montero-Calasanz JP, Meier-Kolthoff MDC, Zhang D-F, Yaramis A, Rohde M, Woyke T, Kyrpides NC, Schumann P, Li W-J, Göker M (2017) Genome-scale data call for a taxonomic rearrangement of Geodermatophilaceae. Front Microbiol 8:2501
Nash P, Krent MM (1991) Culture media. In: Ballows A, Hauser WJ, Herrmann KL, Isenberg HD, Shadomy HJ (eds) Manual of clinical microbiology, 5th edn. American Society for Microbiology, Washington DC, pp 1268–1270
Nicholson AC, Gulvik CA, Whitney AM, Humrighouse BW, Graziano J, Emery B, Bizet C (2018) Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov. Antonie Van Leeuwenhoek 111(1):55–72
Nonomura H, Ohara Y (1957) Distribution of actinomycetes in soil. II. Microbispora, a new genus of the Streptomycetaceae. J Ferment Technol 35:307–311
Nouioui I, Carro L, García-López M, Meier-Kolthoff J, Woyke T, Kyrpides N, Klenk H-P, Goodfellow M, Göker M (2018) Genome-based taxonomic classification of the phylum Actinobacteria. Front Microbiol 9:2007
Otoguro M, Yamamura H, Quintana ET (2014) The family Streptosporangiaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes. Springer, Berlin, pp 1011–1045
Rosselló-Móra R, Trujillo ME, Sutcliffe IC (2017) Introducing a digital protologue: a timely move towards a database-driven systematics of archaea and bacteria. Antonie Van Leeuwenhoek 110(4):455–456
Runmao H, Guizhen W, Junying L (1993) A new genus of actinomycetes, Planotetraspora gen. nov. Int J Syst Bacteriol 43:468–470
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sangal V, Goodfellow M, Jones AL, Schwalbe EC, Blom J, Hoskisson PA, Sutcliffe IC (2016) Next-generation systematic: an innovative approach to resolve the structure of complex prokaryotic taxa. Sci Rep 6:38392
Sanglier JJ, Whitehead D, Saddler GS, Ferguson EV, Goodfellow M (1992) Pyrolisis mass spectrometry as a method for the classification, identification and selection of actinomycetes. Gene 115:235–242
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. MIDI Inc., Newark
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tamura T, Suzuki S, Hatano K (2000) Acrocarpospora gen. nov., a new genus of the order Actinomycetales. Int J Syst Evol Microbiol 50:1163–1171
Tang B, Xie F, Zhao W, Wang J, Dai S, Zheng H, Ding X, Cen X, Liu H, Yu Y, Zhou H, Zhou Y, Zhang L, Goodfellow M, Zhao G-P (2016) A systematic study of the whole genome sequence of Amycolatopsis methanolica strain 239T provides an insight into its physiological and taxonomic properties which correlate with its position in the genus. Synth Syst Biotechnol 1:169–186
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624
Thiemann JE, Beretta G (1968) A new genus of the Actinoplanaceae: Planobispora, gen. nov. Arch Microbiol 62:157–166
Thiemann JE, Pagani H, Beretta G (1967) A new genus of the Actinoplanaceae: Planomonospora, gen. nov. Giorn Microbiol 15:27–38
Thiemann JE, Pagani H, Beretta G (1968) A new genus of the Actinomycetales: Microtetraspora gen. nov. J Gen Microbiol 50:295–303
Tindall BJ (1990) Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol Lett 66(1–3):199–202
Waksman SA (1961) The Actinomycetes, vol. 2, Classification, identification and descriptions of genera and species. Williams and Wilkins, Baltimore
Waksman SA (1967) The actinomycetes a summary of current knowledge. Ronald Press, New York
Weber T, Blin K, Duddela S, Krug D, Kim H-U, Bruccoleri R, Lee SY, Fischbach M-A, Müller R, Wohlleben W, Breitling R (2015) antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:W237–W243
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA for phylogenetic study. J Bacteriol 173:697–703
Williams ST, Goodfellow M, Alderson G, Wellington EMH, Sneath PHA, Sackin MJ (1983) Numerical classification of Streptomyces and related genera. J Gen Microbiol 129:1743–1813
Wu H, Liu B, Shao Y, Ou X, Huang F (2018) Thermostaphylospora grisealba gen. nov., sp. nov., isolated from mushroom compost and transfer of Thermomonospora chromogena Zhang et al. 1998 to Thermostaphylospora chromogena comb. nov. Int J Syst Evol Microbiol 68:602–608
Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Zhang Z, Wang Y, Ruan J (1998) Reclassification of Thermomonospora and Microtetraspora. Int J Syst Bacteriol 48:411–422
Zhang Y-Q, Liu H-Y, Yu L-Y, Lee J-C, Park D-J, Kim C-J, Xu L-H, Jiang C-L, Li W-J (2011) Sinosporangium album gen. nov., sp. nov., a new member of the suborder Streptosporangineae. Int J Syst Evol Microbiol 61:592–597
Zhi X-Y, Li W-J, Stackebrandt E (2009) An update on the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int J Syst Evol Microbiol 59:589–608
Zhou EM, Tang SK, Sjoholm C, Song ZQ, Yu TT, Yang LL, Ming H, Nie GX, Li WJ (2012a) Thermoactinospora rubra gen. nov., sp. nov., a thermophilic actinomycete isolated from Tengchong, Yunnan province, south-west China. Antonie Van Leeuwenhoek 102:177–185
Zhou EM, Yang LL, Song ZQ, Yu TT, Nie GX et al (2012b) Thermocatellispora tengchongensis gen. nov., sp. nov., a new member of the family Streptosporangiaceae. Int J Syst Evol Microbiol 62:2417–2423
Zonn IS, Esenov PE (2012) The Karakum Desert. In: Zonn IS, Kostianoy AG (eds) The Turkmen Lake Altyn Asyr and water resources in Turkmenistan. Springer, Berlin, pp 23–37
Acknowledgements
This research was supported by Ondokuz Mayis University (OMU), project no. PYO.FEN.1901.16.001.
Author information
Authors and Affiliations
Contributions
NS designed study. HS, HA, KG and DC performed research. HA, NS and HS analysed data and wrote the manuscript. All of the authors assisted in writing the manuscript, discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants and/or animals performed by any of the authors. The formal consent is not required in this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S2
GyrB variable amino acid positions for strain 7K107T, Sphaerisporangium album DSM 45172T, Thermostaphylospora chromogena DSM 43794T and Sinosporangium album CPCC 201354T. Amino acid positions were numbered based on the Streptosporangium roseum DSM 43021T GyrB protein (DOCX 17 kb)
Fig. S1
Polar lipid profile of strain 7K107T. Key: DPG, Diphosphatidylglycerol; PE, Phosphatidylethanolamine; PL, Phospholipid; AL, Aminolipid; GL, Glycolipid; L, Lipid (DOCX 90 kb)
Fig. S2
Maximum-Likelihood tree based on almost complete 16S rRNA gene sequences showing relatyynships between strain 7K107T and closely related type strains of the family Streptosporangiaceae. The tree was inferred under the Tamura and Nei model (Tamura and Nei 1993) and the branches are scaled in terms of the expected number of substitutions per site. The numbers above the branches are support values when larger than 50% bootstrapping. Streptomyces ambofaciens ATCC 23877T was used as outgroup. The analysis involved 41 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1267 positions in the final dataset. Bar, 0.05 substitutions per nucleotide site (DOCX 507 kb)
Rights and permissions
About this article
Cite this article
Saygin, H., Ay, H., Guven, K. et al. Desertiactinospora gelatinilytica gen. nov., sp. nov., a new member of the family Streptosporangiaceae isolated from the Karakum Desert. Antonie van Leeuwenhoek 112, 409–423 (2019). https://doi.org/10.1007/s10482-018-1169-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-018-1169-7