Abstract
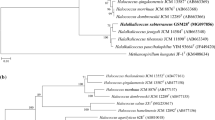
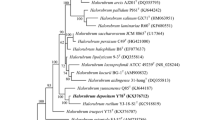
Two extremely halophilic archaea, designated YIM 93701T and YIM 93664, were isolated from Lop Nur region in Xinjiang Province, northwest of China. The cells of the two strains were observed to be cocci, non-motile and Gram-negative. The organisms were determined to be aerobic and required at least 6 % NaCl for growth (optimum 20–25 % and maximum 35 %). Growth was found to occur in the ranges of 16–50 °C (optimum 37 °C) and pH 6.0–8.5 (optimum 6.5–7.5). Cells did not lyse in distilled water. Phylogenetic analysis based on 16S rRNA gene sequences indicated that the two strains belongs to the genus Halalkalicoccus and possessed 99.3 and 99.5 % similarities with their closest phylogenetic relative Halalkalicoccus tibetensis JCM 11890T. Major polar lipids of the two strains were determined to be phosphatidylglycerol(PG),phosphatidylglycerol phosphate methyl ester (PGP-Me), phosphatidylglycerol sulfate (PGS) and three unidentified glycolipids. The DNA G+C contents were determined to be 60.0–60.4 mol%. The DNA hybridization between the two strains was 92.0 %. In addition, the hybridizations of both strains to H. tibetensis were 49 and 52 %, respectively, and to Halalkalicoccus jeotali were 38 and 33 %, respectively. On the basis of physiological, biochemical tests and phylogenetic differentiations, strains YIM 93701T and YIM 93664 were classified as the same species which represent a novel species in the genus Halalkalicoccus, for which the name Halalkalicoccus paucihalophilus sp. nov. is proposed. The type strain is YIM 93701T (=JCM 17505T = CCTCC 2012803T).

Similar content being viewed by others
References
Baker GC, Smith JJ, Cowan DA (2003) Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55:541–555
Barrow GI, Feltham RKA (1993) Cowan and steel’s manual for the identification of medical bacteria, 3rd edn. Cambridge University Press, Cambridge
Christensen H, Angen O, Mutters R, Olsen JE, Bisgaard M (2000) DNA–DNA hybridization determined in micro-wells using covalent attachment of DNA. Int J Syst Evol Microbiol 50:1095–1102
Cui HL, Lin ZY, Dong Y, Zhou PJ, Liu SJ (2007) Halorubrum litoreum sp. nov., an extremely halophilic archaeon from a solar saltern. Int J Syst Evol Microbiol 57:2204–2206
Cui HL, Li XY, Gao X, Xu XW, Zhou YG, Liu HC, Oren A, Zhou PJ (2010a) Halopelagius inordinatus gen. nov., sp. nov., a new member of the family Halobacteriaceae isolated from a marine solar saltern. Int J Syst Evol Microbiol 60:2089–2093
Cui HL, Gao X, Yang X, Xu XW (2010b) Halorussus rarus gen. nov., sp. nov., a new member of the family Halobacteriaceae isolated from a marine solar saltern. Extremophiles 14:493–499
Cui HL, Sun FF, Gao X, Dong Y, Xu XW, Zhou YG, Liu HC, Oren A, Zhou PJ (2010c) Haladaptatus litoreus sp. nov., an extremely halophilic archaeon from a marinesolar saltern, and emended description of the genus Haladaptatus. Int J Syst EvolMicrobiol 60:1085–1089
Dussault HP (1955) An improved technique for staining red halophilic bacteria. J Bacteriol 70:484–485
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid–deoxyribonucleic acid hybridization in micro dilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–789
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Gutiérrez MC, Castillo AM, Kamekura M, Ventosa A (2008) Haloterrigena salina sp. nov., an extremely halophilic archaeon isolated from a salt lake. Int J Syst Evol Microbiol 58:2880–2884
Kates M (1986). Techniques of lipidology, 2nd edn. Elsevier, Amsterdam, pp 106–107, 241–246
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Leifson E (1960) Atlas of Bacterial Flagellation. London: Academic Press Mesbah, M., Premachandran, U. & Whitman, W. B. (1989). Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol 47:87–95
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Ng WL, Yang CF, Halladay JT, Arora A, DasSarma S (1995) Protocol 25. Isolation of genomic and plasmid DNAs from Halobacterium halobium. In: S. DasSarma & E. M. Fleischmann (ed) Archaea a Laboratory Manual: Halophiles, pp. 179-180. Cold Spring Harbor Laboratory, New York
Oren A (2000). The order Halobacteriales. In: M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer & E. Stackebrandt (ed) The Prokaryotes: an Evolving Electronic Resource for the Microbiological Community, release 3.2. Springer, New York http://141.150.157.117:8080/prokPUB/index.htm
Oren A, Ventosa A, Grant WD (1997) Proposed minimal standards for description of new taxa in the order Halobacteriales. Int J Syst Bacteriol 47:233–238
Roh SW, Nam YD, Chang HW, Sung Y, Kim KH, Oh HM, Bae JW (2007) Halalkalicoccus jeotgali sp. nov., a halophilic archaeon from shrimp jeotgal, a traditional Korean fermented seafood. Int J Syst Evol Microbiol 57:2296–2298
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Savage KN, Krumholz LR, Oren A, Elshahed MS (2007) Haladaptatus paucihalophilus gen. nov., sp. nov., a halophilic archaeon isolated from a low-salt, sulfide-rich spring. Int J Syst Evol Microbiol 57:19–24
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterialsystematics. Int J Syst Bacteriol 37:463–464
Williams ST, Goodfellow M, Alderson G (1989) Genus Streptomyces Waksman and Henrici 1943, 339AL. In Bergey’s Manual Syst Bacteriol 4:2463–2468
Xue Y, Fan H, Ventosa A, Grant WD, Jones BE, Cowan DA, Ma Y (2005) Halalkalicoccus tibetensis gen. nov., sp. nov., representing a novel genus of haloalkaliphilic archaea. Int J Syst Evol Microbiol 55:2501–2505
Acknowledgments
We are grateful to Dr. Heng-Lin Cui for his kindly providing the type strains and valuable comments on our experiments. This research was supported by National Natural Science Foundation of China (No. 31200008), National New Drug Research and Development Project (No. 2010ZX09401-403) and 863 projects (No. 2012AA021705). Y.-G. Zhang wishes to thank the West Light Foundation of The Chinese Academy of Sciences. W.-J. Li was also supported by ‘Hundred Talents Program’ of the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Additional information
Bing-Bing Liu and Shu-Kun Tang contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, BB., Tang, SK., Zhang, YG. et al. Halalkalicoccus paucihalophilus sp. nov., a halophilic archaeon from Lop Nur region in Xinjiang, northwest of China. Antonie van Leeuwenhoek 103, 1007–1014 (2013). https://doi.org/10.1007/s10482-013-9880-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-013-9880-x