Abstract
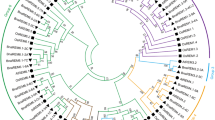
Auxin plays key roles in a wide variety of plant activities, including embryo development, leaf formation, phototropism, fruit development and root initiation and development. Auxin/indoleacetic acid (Aux/IAA) genes, encoding short-lived nuclear proteins, are key regulators in the auxin transduction pathway. But how they work is still unknown. In order to conduct a systematic analysis of this gene family in Solanaceae species, a genome-wide search for the homologues of auxin response genes was carried out. Here, 26 and 27 non redundant AUX/IAAs were identified in tomato and potato, respectively. Using tomato as a model, a comprehensive overview of SlIAA gene family is presented, including the gene structures, phylogeny, chromosome locations, conserved motifs and cis-elements in promoter sequences. A phylogenetic tree generated from alignments of the predicted protein sequences of 31 OsIAAs, 29 AtIAAs, 31 ZmIAAs, and 26 SlIAAs revealed that these IAAs were clustered into three major groups and ten subgroups. Among them, seven subgroups were present in both monocot and dicot species, which indicated that the major functional diversification within the IAA family predated the monocot/dicot divergence. In contrast, group C and some other subgroups seemed to be species-specific. Quantitative real-time PCR (qRT-PCR) analysis showed that 19 of the 26 SlIAA genes could be detected in all tomato organs/tissues, however, seven of them were specifically expressed in some of tomato tissues. The transcript abundance of 17 SlIAA genes were increased within a few hours when the seedlings were treated with exogenous IAA. However, those of other six SlIAAs were decreased. The results of stress treatments showed that most SIIAA family genes responded to at least one of the three stress treatments, however, they exhibited diverse expression levels under different abiotic stress conditions in tomato seedlings. SlIAA20, SlIAA21 and SlIAA22 were not significantly influenced by stress treatments even though at least one stress-related cis-element was identified in their promoter regions. In conclusion, our comparative analysis provides an insight into the evolution and expression patterns in various tissues and in response to auxin or stresses of the Aux/IAA family members in tomato, which will provide a very useful reference for cloning and functional analysis of each member of AUX/IAA gene family in Solanaceae crops.







Similar content being viewed by others
Abbreviations
- ARF:
-
Auxin response factor
- Aux/IAA:
-
Auxin/indoleacetic acid
- AuxRE:
-
Auxin responsive elements
- NLS:
-
Nuclear localization signal
- qRT-PCR:
-
Quantitative real-time PCR
References
Abel S, Theologis A (1995) A polymorphic bipartite motif signals nuclear targeting of early auxin-inducible proteins related to PS-IAA4 from pea (Pisum sativum). Plant J 8:87–96
Bailey TL, Boden M, Buske FA, Frith M, Grant E, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:202–208
Berleth T, Krogan NT, Scarpella E (2004) Auxin signals—turning genes on and turning cells around. Curr Opin Plant Biol 7:553–563
Brukhin V, Hernould M, Gonzalez N, Chevalier C, Mouras A (2003) Flower development schedule in tomato Lycopersicon esculentum cv. Sweet Cherry. Sex Plant Reprod 15:311–320
Chaabouni S, Jones B, Delalande C, Wang H, Li Z, Mila I, Frasse P, Latche A, Pech JC, Bouzayen M (2009) Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. J Exp Bot 60(4):1349–1362
Cle’ment B, Pollmann S, Wielder E, Urbanczyk-Wochniak E, Otter L (2006) The Agrobacterium vitis T-6b oncoprotein induces auxin-independent cell expansion in tobacco. Plant J 45:1017–1027
Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435:441–445
Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29:153–168
Ghanashyam C, Jain M (2009) Role of auxin-responsive genes in biotic stress responses. Plant Signal Behav 4:846–848
Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S (2004) Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134:1555–1573
Gray WM, Kepinski S, Rouse D, Leyser D, Estelle M (2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 15:414
Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49:373–385
Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen G, Guilfoyle TJ, Berleth T (2004) Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131:1089–1100
Hayashi K, Tan X, Zheng N, Hatate T, Kimura Y, Kepinski S, Nozaki H (2008) Small-molecule agonists and antagonists of F-box protein–substrate interactions in auxin perception and signaling. Proc Natl Acad Sci USA 105:5632–5637
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Hompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Jain M, Khurana JP (2009) Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J 276:3148–3162
Jain M, Kaur N, Garg R, Jitendra K, Thakur, Akhilesh K, Tyagi, Jitendra P, Khurana (2006) Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6:47–59
Kalluri UC, DiFazio SP, Brunner AM, Tuskan GA (2007) Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol 7:59
Kim J, Harter K, Theologis A (1997) Protein–protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA 94:11786–11791
Kloosterman B, Visser RGF, Bachem CWB (2006) Isolation and characterization of a novel potato Auxin/indole-3-acetic Acid family member (StIAA2) that is involved in petiole hyponasty and shoot morphogenesis. Plant Physiol Biochem 44:766–775
Ku HM, Vision T, Liu JP, Tanksley SD (2000) Comparing sequenced segments of the tomato and Arabidopsis genomes: large-scale duplication followed by selective gene loss creates a network of synteny. Proc Natl Acad Sci USA 97:9121–9126
Kumar R, Tyagi AK, Sharma AK (2011) Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol Genet Genomics 285:245–260
Leyser O (2002) Molecular genetics of auxin signalling. Ann Rev Plant Biol 53:377–398
Li H, Tiwari SB, Hagen G, Guilfoyle TJ (2011) Identical amino acid substitutions in the repression domain of auxin/indole-3-acetic acid proteins have contrasting effects on auxin signalling. Plant Physiol 155:1252–1263
Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49:387–400
Nakamura A, Umemura I, Gomi K, Hasegawa Y, Kitano H, Sazuka T, Matsuoka M (2006) Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J 46:297–306
Nebenführ A, White TJ, Lomax TL (2000) The diageotropica mutation alters auxin induction of a subset of the Aux/IAA gene family in tomato. Plant Mol Biol 44:73–84
Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2(9):e258
Ouellet F, Overvoorde PJ, Theologis A (2001) IAA17/AXR3: biochemical insight into an auxin mutant phenotype. Plant Cell 13:829–841
Park JY, Kim HJ, Kim J (2002) Mutation in domain II of IAA1 confers diverse auxin-related phenotypes and represses auxin-activated expression of Aux/IAA genes in steroid regulator-inducible system. Plant J 32:669–683
Park HC, Kim ML, Kang YH, Jeon JM, Yoo JH, Kim MC, Park CY, Jeong JC, Moon BC, Lee JH, Yoon HW, Lee S, Chung WS, Lim CO, Lee SY, Hong JC, Cho MJ (2004) Pathogen- and NaCl-induced expression of the scam-4 promoter is mediated in part by a gt-1 box that interacts with a gt-1-like transcription factor. Plant Physiol 135:2150–2161
Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park CM (2007) GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem 282:10036–10046
Quint M, Gray WM (2006) Auxin signaling. Curr Opin Plant Biol 9:448–453
Raikhel N (1992) Nuclear targeting in plants. Plant Physiol 100:1627–1632
Reed JW (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6(9):420–425
Remington DL, Vision TJ, Guilfoyle TJ, Reed WJ (2004) Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135:1738–1752
Rieping M, Sehfffl F (1992) Synergistic effect of upstream sequences, CCAAT box elements, and HSE sequences for enhanced expression of chimaeric heat shock genes in transgenic tobacco. Mol Gen Genet 231(2):226–232
Rogg LE, Lasswell J, Bartel B (2001) A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13:465–480
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sato A, Yamamoto KT (2008) Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol Plant 2(133):397–405
Shibasaki K, Uemura M, Tsurumi S, Rahman A (2009) Auxin response in Arabidopsis under cold stress: underlying molecular mechanisms. Plant Cell 21:3823–3838
Song Y, Wang L, Xiong LZ (2009) Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta 229:577–591
Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319:1384
Tan X, Luz IA, Villalobos C, Sharon M, Zheng CX, Robinson CV, Estelle M, Zheng N (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446:640–645
Terrile MC, Fiol DF, Casalongue CA (2010) Solanum tuberosum Aux/IAA family: new members and characterization of StIAA1 interacting proteins. Plant Growth Regul 62:93–99
Thakur JK, Tyagi AK, Khurana JP (2001) OsIAA1: an Aux/IAA cDNA from rice, and changes in its expression as influenced by auxin and light. DNA Res 8:193–203
Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13:2809–2822
Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15:533–543
Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16:533–543
Ulmasov T, Hagen G, Guilfoyle TJ (1997a) ARF1, a transcription factor that binds to auxin response elements. Science 276:1865–1868
Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997b) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9:1963–1971
Walker JC, Key JL (1982) Isolation of cloned cDNAs to auxinresponsive poly (A)+RNAs of elongating soybean hypocotyls. Proc Natl Acad Sci USA 79(23):7185–7189
Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latche A, Pech JC, Bouzayena M (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17:2676–2692
Wang DK, Pei KM, Fu YP, Sun ZX, Li SJ, Liu H, Tang K, Han B, Tao YZ (2007) Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394:13–14
Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, Latche A, Pech JC, Fernie AR, Bouzayen M (2009) Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21:1428–1452
Wang SK, Bai YH, Shen CJ, Wu YR, Zhang SN, Jiang D, Guilfoyle TJ, Chen M, Qi YH (2010a) Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct Integr Genomics 10:533–546
Wang Y, Deng D, Bian Y, Lv Y, Xie Q (2010b) Genome-wide analysis of primary auxin-responsive Aux/IAA gene family in maize (Zea mays L.). Mol Biol Rep 37:3991–4001
Winter D, Vinegar B, Nahal H, Ammar R, Wilson G, Provart N (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2:e718
Wu J, Wang F, Cheng L, Kong F, Peng Z, Liu S, Yu X, Lu G (2011) Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Rep 30:2059–2073
Xu X, Pan S, Cheng S, Zhang B, Mu D, Ni P, Zhang G, Yang S, Li R, Wang J et al (2011) Genome sequence and analysis of the tuber crop potato. Nature 475(7355):189–195
Yamamoto KT (1994) Further characterization of auxin-regulated mRNAs in hypocotyl sections of mung bean [Vigna radiata (L.) Wilczek]: sequence homology to genes for fatty-acid desaturases and atypical late-embryogenesis-abundant protein, and the mode of expression of the mRNAs. Planta 192:359–364
Yamamoto KT, Mori H, Imaseki H (1992) cDNA cloning of indole-3-acetic acid regulated genes: Aux22 and SAUR from mung bean (Vigna radiata) hypocotyls tissue. Plant Cell Physiol 33:93–97
Yang XP, Lee SS, So J, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle1 M (2004) The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. Plant J 40:772–782
Zanetti ME, Terrile MC, Godoy AV, San Segundo B, Casalongue CA (2003) Molecular cloning and characterization of a potato cDNA encoding a stress regulated Aux/IAA protein. Plant Physiol Biochem 41:755–760
Acknowledgments
This work was supported by the State Key Basic Research and Development Plan of China (2009CB119000), the National Natural Science Foundation of China (30771470) and Zhejiang Provincial Natural Science Foundation of China (R3110209 and 2009C32025). Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: JN043331(SlIAA12), JN043332(SlIAA13), JN043333(SlIAA16).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
438_2012_675_MOESM6_ESM.xls
Table S6. The expression analysis of AtIAA family genes in various Arabidopsis tissues. The expression data were searched against Arabidopsis eFP Browser using the AtIAA gene accession number (XLS 31 kb)
438_2012_675_MOESM7_ESM.xls
Table S7. The expression analysis of AtIAA family genes in response to auxin and abiotic treatments. The expression data were serached against Arabidopsis eFP Browser using each AtIAA gene accession number. (XLS 23 kb)
438_2012_675_MOESM10_ESM.jpg
Supplemental Figure 10. Alignment of potato Aux/IAA proteins obtained with the ClustalX program. The height of the bars indicates the number of identical residues per position. b: Multiple alignments of the domains I to IV of the tomato Aux/IAA proteins obtained with ClustalX and manual correction. Black and light gray shading indicate identical and conversed amino acid residues, respectively. Conserved domains are also underlined and correspond to part a. (JPEG 1417 kb)
438_2012_675_MOESM11_ESM.jpg
Supplemental Figure 11 a. Alignment of tobacco Aux/IAA proteins obtained with the ClustalX program. The height of the bars indicates the number of identical residues per position. b: Multiple alignments of the domains I to IV of the tobacco Aux/IAA proteins obtained with ClustalX and manual correction. Black and light gray shading indicate identical and conversed amino acid residues, respectively. Conserved domains are also underlined and correspond to part a. (JPEG 936 kb)
438_2012_675_MOESM12_ESM.jpg
Supplemental Figure 12. The five conserved consensus motifs in AUX/IAA of tomato, rice, maize and Arabidopsis IAA proteins were found by MEME. The symbol heights represent the relative frequency of each residue. The numbers of sites and e-value for each motif are also shown. (JPEG 432 kb)
438_2012_675_MOESM14_ESM.jpg
Supplemental Figure 14. Expression profiles of all the 26 SlIAA genes in different tomato flower tissues. qRT-PCR analyses using RNA generated from sepal (Se), petal (P), stamen (St), and ovary (O) were performed. For other details see Figure 5. (JPEG 557 kb)
438_2012_675_MOESM15_ESM.jpg
Supplemental Figure 15. Expression profiles of all the 26 SlIAA genes at different tomato flower developmental stages. QRT-PCR analyses were performed using RNA generated from tomato flower buds at three stages of early floral development, roughly defined by the length of flower buds as follows: stage I: 3-4 mm, stage II: 5-6 mm, and stage III at 7-8 mm. (JPEG 485 kb)
438_2012_675_MOESM16_ESM.jpg
Supplemental Figure 16. Expression profiles of all the 26 SlIAA genes in response to environmental stress. QRT-PCR analyses were used to assess SlIAA transcript levels in tomato seedlings exposed to drought (D), salt (S), heat (H) stresses when compared to control treatment. (JPEG 604 kb)
438_2012_675_MOESM17_ESM.jpg
Supplemental Figure 17. Phylogenetic relationships among the potato, tomato and tobacco IAA proteins. The unrooted tree was generated using MEGA4.1 program by the neighbor-joining method. Bootstrap support from 1000 replicates are indicated at each branch. (JPEG 90 kb)
438_2012_675_MOESM18_ESM.jpg
Supplemental Figure 18. Phylogenetic relationships among the Arabidopsis IAA proteins. The unrooted tree was generated using MEGA4.1 program by the neighbor-joining method. Bootstrap support from 1000 replicates are indicated at each branch. (JPEG 37 kb)
438_2012_675_MOESM19_ESM.jpg
Supplemental Figure 19. Phylogenetic relationships among the maize IAA proteins. The unrooted tree was generated using MEGA4.1 program by the neighbor-joining method. Bootstrap support from 1000 replicates are indicated at each branch. (JPEG 34 kb)
438_2012_675_MOESM20_ESM.jpg
Supplemental Figure 20. Phylogenetic relationships among the rice IAA proteins. The unrooted tree was generated using MEGA4.1 program by the neighbor-joining method. Bootstrap supports from 1000 replicates are indicated at each branch. (JPEG 38 kb)
Rights and permissions
About this article
Cite this article
Wu, J., Peng, Z., Liu, S. et al. Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Mol Genet Genomics 287, 295–311 (2012). https://doi.org/10.1007/s00438-012-0675-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-012-0675-y