Abstract
Proteases help plants maintain protein quality and break down protein subsets in reply to environmental, developmental, biotic, and abiotic stressors. Filamentation temperature-sensitive H (FtsH) is an ATP-dependent metalloprotease detected in both prokaryotes and eukaryotes cells. The present research, the FtsH gene in the bean, which has an important place in the legume family and is an important agricultural product, was characterized for the first time using various bioinformatic tools, and qRT-PCR measured its expression level. In the study, effectively identified and characterized 17 FtsH genes present in genome the of Phaseolus vulgaris. The MW of FtsH proteins varied from 71.16 to 147.07 kDa, their amino acid lengths ranged from 642 to 1284, and their pI values varied from 5.39 to 9.60. Interestingly, the distribution of these 17 distinct PvFtsH genes across the 8 chromosomes was not uniform, exhibiting an uneven pattern throughout the genome. A pair of segmental duplication fragments were found, revealing probable processes of gene expansion and evolution. Collinearity with related genes in Arabidopsis and rice was thoroughly examined to determine the evolutionary conservation and differentiation of PvFtsH genes. Additionally, we used RNAseq and qRT-PCR to investigate the expression patterns of PvFtsH in leaf tissue under salt and drought conditions. Our data showed unique expression patterns, suggesting PvFtsH may respond to environmental and physiological stressors. Overall, this work makes major contributions to our understanding of PvFtsH genes and their roles in the context of gene evolution, chromosomal distribution, and expression patterns under various environmental situations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants encounter a diverse array of abiotic and biotic stressors throughout each stage of their development (Gull et al. 2019). Drought, salinity, and heat are the most widely studied abiotic stressors in the literature (Turhan et al. 2021; Zandalinas et al. 2022). Due to growing global warming, high heat stress has already resulted in billions of dollars’ worth of crop losses throughout the world (Perry et al. 2020). The detrimental impacts on agricultural growth, development, and productivity are evident in both drought and salt stress conditions, posing significant challenges to crop yield and overall agricultural sustainability (Yu et al. 2022; Wu et al. 2022). Furthermore, it induces substantial damage to the physiological and biochemical processes of the plant, often leading to the denaturation of proteins and the generation of reactive oxygen species (ROS) (Aleem et al. 2022; Zhao et al. 2022). Hence, to endure and mitigate detrimental consequences, plants make use of a vast variety of physiological and chemical defense mechanisms (Praveen et al. 2023). Biotic and abiotic stress factors often cause protein dysfunction, and abnormal proteins disrupt cell viability (Ali and Baek 2020). Unwanted or damaged proteins are removed by a tightly regulated proteolytic process (Dougan et al. 2012).
Proteolytic enzymes are commonly referred to as proteases, proteinases, or peptidases (Rawlings et al. 2020). Although proteases play essential roles in various critical plant cell processes, including senescence, cell death, and responses to environmental changes, there exists limited knowledge regarding the substrate specificity and physiological functions of numerous plant proteases. Even for ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco), the most prevailing plant protein, it is unknown which proteases are involved in the process of cellular localization or destruction (García-Lorenzo et al. 2006). Plants depend on proteases to maintain rigorous protein quality control and to break down certain protein subsets in response to various environmental and developmental cues. The similarities and variances among the proteases produced in various animals may provide important clues about their physiological functions and evolutionary history (Kato and Sakamoto 2019).
Filamentation temperature-sensitive H (FtsH) protease is known as a membrane-bound ATP-dependent Zn2+ metalloproteinase, belongs to the protein family AAA, and was initial found in E. coli (Guo et al. 2021; Pu et al. 2022). FtsH proteins are present in both prokaryotic and eukaryotic organisms. Especially, endosymbiotic organelles, including chloroplasts and mitochondria, contain them (Wan and Ling 2022). FtsH metalloproteases are widely known for playing a crucial part in the upkeep and proteolysis of membrane proteins in plants, animals, and eubacteria (Wagner et al. 2012). FtsH genes have been detected in an extensive variability of plant species, including the potato (Hajibarat and Saidi 2023), pear (Guo et al. 2021), soybean (Wang et al. 2023), rice (Wu et al. 2021), pepper (Xiao et al. 2021), tobacco (Pu et al. 2022), maize (Yue et al. 2010), Arabidopsis (Mielke et al. 2021).
Specifically, completing the genome sequencing of plants such as Arabidopsis and rice with next-generation sequencing technologies makes it possible to identify all FtsH genes in both genomes of these plants. There are 17 and 19 FtsH proteases that are membrane-bound in the model organism Arabidopsis and rice respectively (Zhang and Sun 2009; Mishra and Funk 2021).
One of the most popular legumes consumed worldwide is the Phaseolus vulgaris L., which offers the human diet with proteins, vitamins, and minerals (Ilhan et al. 2018). It is a necessary food in many nations due to its nutritional value as well as its affordability and adaptability for long-term storage. Because this legume is grown in peripheral parts of Brazil, where unfavorable environmental conditions like drought and salt regularly occur, productivity there is thought to be low (Inal et al. 2017; Ito et al. 2017). To improve bean breeding for abiotic stress tolerance, it is necessary to develop novel, effective ways. Abiotic stress causes changes in gene expression, which are crucial for plants to adapt and persist in extreme environments (Baloglu et al. 2014). Research on the growth, development, and stress tolerance of legumes is therefore practically important for agricultural output. Despite the entire sequencing of the legume genomes, a comprehensive assessment of the metalloprotease FtsH gene family members in beans have not been done.
Applying plant hormones and/or growth regulators to plants have been demonstrated an effective method for decreasing the harmful effects of environmental stress throughout their growth period (Amiri et al. 2024). Melatonin, also known as N-acetyl-5-methoxytryptamine, is a compound derivative from the essential amino acid tryptophan and is found in the leaves, stems, shoots, fruits, seeds, and roots of many different plant species, serving a variety of functions in vascular plants (Malabadi et al. 2021; Kasapoglu et al. 2023; Yusuf et al. 2024). Melatonin not only exhibits antioxidant properties but also safeguards against abiotic stress impacts through the regulation of plant hormones, activation of endoplasmic reticulum stress- responsive genes, and enhancement of protein homeostasis, heat shock proteins, and transcription factors (Colombage et al. 2023; Tiwari et al. 2023). Furthermore, melatonin's role under stressful circumstances is to eliminate reactive oxygen species (ROS) and reactive nitrogen species (RNS) (Wang et al. 2024). According to reports, melatonin is more efficient than α-tocopherol and glutathione in removing ROS and RNS (Sun et al. 2021a, b). Consequently, it essentially minimizes the oxidative damage to proteins, nucleic acids sustain and lipids (Tiwari et al. 2024). Due to its amphiphilic nature, melatonin has a significant ability to penetrate all cellular compartments (Choi et al. 2024).
Bioinformatics methods were employed in the current study to find new proteins and genes. Although omics methods like transcriptomics, metabolomics, and genomics are widely used, there has been no previous study that comprehensively identifies and analyzes the expression of the metalloprotease FtsH gene family in common beans at a genome-wide level. Owing to the significance of the FtsH gene family in stress and plant growth and development, the results of this investigation will deliver essential data for Phaseolus vulgaris basic breeding research’s that seeks to identify genes responding to drought and salt stress.
Material and methods
Identification of FtsH protein in common bean genome
Protein sequences for the FtsH gene family in the Phaseolus vulgaris genome were acquired from the Phytozome database (v12.1) using the Pfam entry IDs structural domains AAA (PF00004), FtsH extracellular structural domain (PF06480) and peptidase M41 (PF01434). Both blastp from the Phytozome database (v12.1) and the hidden Markov model (HMM) database were used to identify all probable FtsH proteins in the bean genome (Schmutz et al. 2014). Unrelated sequences were extracted using the “decrease redundancy tool”. Using the HMMER database, the presence of the FtsH domain in the associated sequences was investigated. The “ProtParam” tool was utilized to ascertain the molecular weight (mw) and putative isoelectric point (pI) of the FtsH proteins that were acquired.
Identification of structure, physical locations, gene duplications and motifs of PvFtsH genes
The Gene Structure Display Server (GSDS) v2.0 was utilized to get data on the exon–intron sections of PvFtsH proteins (Guo et al. 2007). The location information of the PvFtsH genes was determined using the coding DNA (CDS) and genome sequences. The chromosomal locations and sizes of the PvFtsH genes were determined using the Phytozome database (v12.1).
The PvFtsH genes were first assigned to specific locations on each of the P. vulgaris chromosomes and observed using the TBtools program (Chen et al. 2023). In addition, gene duplication was identified using the TBtools software. It was calculated using the TBtools basic Ka/Ks calculator algorithm to calculate the exchange rates for non-homologous (Ka), homologous (Ks) and non-homologous to homologous (Ka/Ks) among duplicate pairs of PvFtsH genes (Chen et al. 2023). T = Ks/2 (= 6.56E-9) formula was used to calculate each PvFtsH gene's divergence and duplication period (measured in Million Years Ago, MYA) (Yang and Nielsen 2000; Muslu et al. 2023).
Conserved motifs of PvFtsH proteins were examined using the "Multiple EM for Motif Elimination (MEME)" methods (Bailey et al. 2006). The maximum number of motifs and minimum/maximum width are limited to 10, 50, and 2, respectively. The motif region is between 2 and 300. ny number of repetitions can be chosen for the region distribution. The InterPro database's default parameters were used to scan the identified motifs (Jones et al. 2014).
Phylogenetic analysis and sequence alignment
Neighbor-joining (NJ) approach was utilized for phylogenetic analyses, and 1000 repeated bootstrap values were used. Using ClustalW, PvFtsH protein sequences were aligned (Sinha and Meller 2007). The phylogenetic tree was drawn by Molecular Evolutionary Genetic Analysis (MEGA) v11 program (Tamura et al. 2011). The Interactive Tree of Life (iTOL) interface was used to shape the tree (Letunic and Bork 2011). In addition, the FtsH proteins were subjected to sequence logo analysis utilizing the WEBLOGO online web program, as part of the multiple sequence alignment study (Crooks et al. 2004).
Comparative mapping between common beans and other species
Comparative genomics aims to identify synteny, which refers to gene blocks or other evolutionarily conserved markers and quantify the evolutionary relationship between genomes based on chromosomal rearrangements (Sinha and Meller 2007). With the use of the TBtools, the synteny map of the FtsH genes in Oryza sativa, Arabidopsis thaliana, and Phaseolus vulgaris was created.
Promotor analysis and intracellular localization of the PvFtsH gene family
The cis-acting elements present in the 5' upstream regions of the bean FtsH gene family were obtained using the PlantCARE database. These areas consist of about 2 kb of DNA fragments from each gene (Oner et al. 2022). Their intracellular localization was estimated using WoLFPSORT (Horton et al. 2007).
Three-dimensional homology modeling of FtsH proteins in beans
Using the found FtsH protein sequences, three-dimensional (3D) structures were produced using the Phyre2 library for protein homology modeling (Kelley et al. 2015). The finest 3D image was chosen after examining the protein models' dependability rates. During the final validation phase, the models with the highest level of confidence have been used and visualizations were created using the modeling technique that provided the most achievable match.
Construction of protein–protein interactions network and gene ontology analysis
Protein–protein interactions (PPI) are concerned with various biological processes, such as cellular regulation, metabolic growth, and intercellular communication (Braun and Gingras 2012). To ascertain the physical and functional interactions of proteins, the STRING database was utilized. Obtained data were divided and integrated with the score for entire PPIs on the STRING website. Functional genomics is a crucial component in plant biotechnology research methodologies as it enables the functional characterization of novel sequence data. The Gene Ontology (GO) database was utilized to collect protein ontology data (Gene Ontology Consortium et al. 2023). The GO analysis was conducted utilizing the TBtools software, and the results were presented using an identical application.
Analyzing in silico gene expression in various tissues
By utilizing the Phytozome database (v12.1), we evaluated the expression levels of the PvFtsH genes in different plant tissues throughout different developmental stages. The levels of in-silico expression were measured using a metric called FPKM, which stands for the anticipated number of transcript fragments per kilobase per million base pairs sequenced. The CIMMiner algorithm was utilized to create a heat map using the log2-transformed FPKM values.
The expression profiles of the bean FtsH genes were analyzed using Illumina RNA-seq data acquired from the Sequence Read Archive (SRA) data library. The accession numbers for salt [SRR957668 (salt stress treated leaf), SRR958469 (leaf salt control)] and drought stress [SRR8284481 (drought stress treated leaf), and SRR8284480 (leaf drought control)] were ascertained to locate pertinent RNA-seq data (Hiz et al. 2014; Kasapoglu et al. 2023) were used. The reads per kilobase (RPKM) algorithm of the exon model per million mapped reads was employed to regularize gene expression levels (Mortazavi et al. 2008). After converting RPKM data to log2, with the CIMMiner algorithm produced a heatmap.
Prediction miRNAs that target the PvFtsH genes
MiRBase v21.0 (www.mirbase.org) was utilized to obtain entirely known miRNA plant sequences. The psRNATarget Server was used, which may be found at www.plantgrn.noble.org/psRNATarget, and the defaulting miRNA estimation parameters were applied (Zhang 2005). BLASTX was employed with a threshold of 1e-10 to assess in silico indicated miRNA targets with Phaseolus vulgaris expressed sequence tags (EST) in the NCBI.
Plant material, stress induction, and melatonin treatments.
In this study, the plant material consisted of Serra and Elkoca-05 Phaseolus vulgaris cultivars. All experiments and applications were achieved by using the laboratories facilities in Erzurum Technical University, Faculty of Science, Department of Molecular Biology and Genetic. To impose drought stress, Polyethylene glycol 6000 (PEG 6000) was applied at concentrations of 0% (control) and 20%. For inducing salt stress, Sodium chloride (NaCl) was used at concentrations of 0 mM (control) and 150 mM. The cultivation of plants, as well as the application of stress agents and melatonin, strictly adhered to the protocol outlined in reference (Aygören et al. 2023). Melatonin application was performed by foliar spraying of the leaves, using concentrations of 0 mM (control) and 200 mM. This application was carried out 24 h before subjecting the plants to the stressors. The hydroponic system, including 1/10 Hoagland solution, served as the medium for melatonin application and stress induction. The plants underwent 24 h of drought stress, and 9 days in salt stress to evaluate the impact of ions under salt stress conditions. Upon completing the stress periods, leaf tissues of the bean genotypes were promptly harvested and freeze up in liquid nitrogen. These tissue samples were thereafter held at − 80 °C to maintain their molecular integrity. The study used a completely randomized design with three replicated. For every treatment, five plants were carefully selected from each replicate and combined using the bulking technique. This approach allowed for efficient molecular analysis in the study.
RNA extraction and cDNA synthesis
The RNA extraction method was performed with TRIzol® reagent (Invitrogen Life Technologies, USA) based on the manufacturer's instructions. RNA concentrations were quantified in Nanodrop (Multiskan GO instrument (Thermo Fisher Scientific, Massachusetts, USA), and A260/280 ratio data were acquired. The 1.2% agarose gel was prepared to visualize the RNA. 3 µl of 6X loading dye, and 3 µl of RNA sample were loaded into the wells after mixing and run at 80–100 V for 40 min. The gel was observed in the UV Transilluminator. The process described in the high-quality cDNA synthesis kit (Roche, USA) was utilized to produce complementary DNA. The primers were constructed using Primer3 software, utilizing the sequences of 4 PvFtsH genes.
qRT-PCR analysis.
The RT-qPCR experiments employed the iTaq Universal SYBR Green Supermix (Bio-Rad, USA), and the reaction requirements outlined by Inal et al. (2017) were followed. Utilizing the LightCycler® Nano Device, RT-qPCR reactions have been carried out (Roche). A total of three technical and biological replicates were utilized. The β-Actin gene was utilized to normalize the qRT-PCR data by the 2CT method. (Livak and Schmittgen 2001). The data were analyzed using GraphPad Prism 9 with a two-way ANOVA approach. Fisher's least significant difference (LSD) test was used to test the differences between means at the 0.05 significance level.
Results and discussion
Identification, chromosomal location, duplication, and synteny analysis of PvFtsH genes
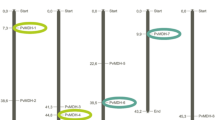
Lately, with the advance of genome-wide research and the spread of bioinformatics databases; the development of stress-resistant plant lines, the development of plant breeding methodologies, and a better understanding of the genes and proteins of defense against stress have been provided. Plants protect themselves under stress by altering gene-genome expression for a variety of physiological activities. RNA-seq analysis has been successfully used in the analysis of gene expression and identification of gene homologues in plants, under different treatment conditions, in different organs (Muñoz-Bodnar et al. 2014). Plant biology has a limited understanding of proteolysis. Proteases are essential players in the proteolysis process during a variety of critical processes, involving those involving senescence, cell death, and environmental changes. In current study, as protease, FtsH gene family was bioinformatically characterized and carried out the relative expression of chosen genes in conditions drought and salt stress and it was found to be their relation to the plant defense mechanism. As a result of in silico analysis, 17 candidate PvFtsH genes were detected by using the bean (Phaseolus vulgaris L.) genome. They were sorted as PvFtsH1 − PvFtsH17 (Table 1). The chromosomal location, amino acid (aa) length, molecular weight (mw), pI: isoelectric point; instability index, classify and aliphatic index information of PvFtsH genes were also shown (Table 1). The proteins ranged in aa length from 642 (PvFtsH5) to 1284 (PvFtsH17). The proteins’ pI ranged from 5.39 (PvFtsH12) to 9.60 (PvFtsH5). Also, their mw varied from 71.16 kDa (PvFtsH5) to 147.07 kDa (PvFtsH17) (Table 1). According to the WoLFPSORT database, PvFtsH-like genes were mostly located in the chloroplast but also in other subcellular components (Table 1). Currently, genome-wide analyses of the FtsH gene family are being conducted using a variety of organisms with different number of FtsH genes, including plants like Arabidopsis (12), pear (19), rice (9), corn (2) and tobacco (20) ( Adam et al. 2001; Sakamoto et al. 2003; Zhang and Sun 2009; Yue et al. 2010; Guo et al. 2021; Pu et al. 2022), as well as microorganisms like Acidovorax citrulli, Bacillus subtilis, cyanobacteria, Escherichia coli, and Lactobacillus plantarum (Ji et al. 2019).
P. vulgaris was found to have PvFtsH genes on 8 of its 11 chromosomes, with none detected on chromosomes 3, 7, and 10. (Fig. 1). Moreover, only one gene (PvFtsH17) could be mapped to any of the chromosomes because it was found on the unassembled scaffolds (Table 1). It was found that a PvFtsH gene is present on chromosomes 4, 5, and 6, while two genes are found on chromosomes 1, 2, and 8. Additionally, three genes are located on chromosome 11, and four genes are found on chromosome 9 (Fig. 1). Consistent with the above results, and only 13 out of the 24 chromosomes have NtFtsH genes in Nicotiana tabacum L. (Pu et al. 2022). The density of genes on chromosomes and the distribution of the FtsH gene did not correlate. This shows that gene loss may have affected the FtsH gene family during evolution. As a result, certain FtsH genes may have developed through duplication processes, indicating that these processes were crucial to the rapid expansion of the FtsH family in plants.
Duplications are important genomic occurrences that aid in the enlargement of gene families. In protein evolution, gene duplication actions have facilitated the acquisition of novel functional traits by proteins (Kondrashov et al. 2002). Tandem duplication refers to the presence of duplications on the same chromosome, whereas segmental duplication refers to the occurrence of the same genes on distinct chromosomes (Fu et al. 2020; Qu et al. 2023a, b). These types of duplication have a significant role in fostering species variety hence, it may be easier for plants to adjust to changing environmental circumstances (Qin et al. 2023).
In this research, it was identified one segmentally duplicated gene (PvFtsH3 and PvFtsH13) pair among 17 PvFtsH genes (Table S1). Furthermore, to assess selection pressure and the timing of PvFtsH gene duplications, the measures of non-synonymous divergence (Ka), synonymous divergence (Ks), and Ka/Ks were computed. The duplicated PvFtsHs exhibited Ka/Ks ratios ranging was found as 0,0951 (Table S1). Values larger than 1 in the Ka/Ks ratio often indicate positive selection, values smaller than 1 imply purifying selection, and a value of 1 indicates neutral selection (Nekrutenko et al. 2002). According to this hypothesis, it was estimated that the duplicated PvFtsH genes were subjected to purifying selection pressure. The divergence of the PvFtsH3 and PvFtsH13 genes occurred approximately 42.620 million years ago. Gene duplication, especially segmental and tandem duplication results, plays an imperative function in the evolutionary process and the enlargement of gene families (Liu et al. 2023). Similarly, it was observed that the NtftsH gene had four pairs of segmental duplications. Contrary to a prior study conducted on pear trees (Guo et al. 2021), no instances of tandem duplication were identified in the tobacco genome for the NtFtsH gene. These findings indicate that segmental duplication might have a greater impact on the expansion of the NtFtsH family compared to tandem duplication. This finding aligns with previous research conducted on other plant species, suggesting that segmental duplication events had a role in the creation and evolution of specific NtFtsH genes (Pu et al. 2022).
In phylogenetics, orthologous genes, which are genes with similar functions that are present in different species, are frequently investigated (Bengoa Luoni et al. 2021). In study, we indicated the orthologous relationships of PvFtsH genes among the genomes of P. vulgaris, A. thaliana, and O. sativa. The correlation enables a comparative analysis of the variations in the presence and organization of FtsH genes among the three-plant species. The analysis revealed that the genomes of P. vulgaris and A. thaliana shared seven orthologous gene pairs (PvFtsH1-AT2G2640.1, PvFtsH4-AtFtsH7, PvFtsH6-AT3G16290.1, PvFtsH7-AtFtsH12, PvFtsH9-AtFtsH9, PvFtsH10-AtFtsH5, and PvFtsH16-AtFtsH4) while P. vulgaris L. and O. sativa exhibited one orthologous gene pair (PvFtsH3-OsFtsH2) (Fig. 2). One gene pair revealed no collinearity with A. thaliana or O. sativa proposing that the homologous gene pairs formed after the species diverged (Dey et al. 2023). Previous research has indicated that FtsH genes show orthology among plants such as Arabidopsis thaliana, Glycine max, Medicago truncatula, Medicago sativa, and Oryza sativa (Zhu et al. 2023).
The gene architecture, phylogenetic analysis, cis‑acting elements, motif analysis, and multiple sequence analysis of FtsH members in P. vulgaris.
Homology modeling, often referred to as comparative modeling or template-based modeling, operates under the assumption that the 3D structures of proteins are more conserved than their amino acid sequences. Consequently, it posits that similar amino acid sequences should exhibit similar 3D structures (Yang et al. 2023). The FtsH proteins available in the Protein Data Bank (PDB) were subjected to Blastp analysis, and their 3D homology models were generated using Phyre2, a database renowned for predicting the structure and function of proteins (Kelley et al. 2015). Homology modeling and 3D structure prediction were performed on a set of 17 PvFtsH proteins (Fig. S1). Using homology modeling, the 17 PvFtsH proteins' 3D structures were predicted with a 100% modeling confidence level. The results indicated that when the secondary structures of PvFtsH proteins were looked at, most of them were made up of disordered structures. It is recognized that certain amino acids disrupt hydrogen bonding during the folding process, leading to a conflict among the conformational energies of the side chain and the maximal hydrogen bond formation (Hooft et al. 1996). Specifically, a distinctive structure known as a twisted β planar layer (twisted β sheet) and α-helix structure were observed in PvFtsH proteins (Fig. S1).
The analysis of multiple sequence alignment between PvFtsH protein sequences revealed highly conserved regions with specific amino acid sequences. To understand the evolutionary relationship, FtsH protein sequences from Arabidopsis (dicotyledonous), Oryza (monocotyledonous), and common bean were aligned, and a phylogenetic tree was constructed (Fig. 3). Phylogenetic tree analysis of 38 PvFtsH proteins from three plant species (Fig. 3) was conducted using the NJ method in MEGA v11 software. The 38 FtsH genes were categorized into three groups based on the percentage of homology among them. Group A consisted of only 1 PvFtsH gene, group B 15 common bean, 11 Arabidopsis, and 9 rice genes, and group C 1 common bean and 1 Arabidopsis gene (Fig. 3).
Phylogenetic tree of Phaseolus vulgaris, Arabidopsis thaliana, and Oryza sativa gene family members using the Neighbor-joining method. The sativa genes of P. vulgaris are represented by the genes starting in “Pv”, those of A. thaliana by “At”, and those of O. sativa by “Os”. The tree diverged into 3 classes with 38 proteins, containing Group A, Group B, and Group C
Comparing bean with rice, it was observed that the homology between Arabidopsis and common bean FtsH genes was higher. This difference in homology could potentially be attributed to variations in leaf structure among common bean, Arabidopsis, and rice, resulting in divergent levels of homology. Subcellular localization study indicated a greater degree of similarity between genes expressed in the same cellular location, whether in common bean or Arabidopsis. This finding implies that these genes may have comparable functions. Therefore, it can be inferred that PvFtsH and AtFtsH proteins likely perform similar biological functions within the cell. Consistent with our findings, other plants also possess an equivalent amount of FtsH genes, despite variations in genome size across different species. For example, rice contains 9 and Arabidopsis 12 FtsH genes (Pu et al. 2022). This condition suggests that the quantity of FtsH family members is generally constant and does not have a direct relationship with the extent of the genome. Shan et al. (2023) demonstrate that the phylogenetic tree categorization of Glycine max identified 80 GmFtsH genes, which were divided into 8 groups together with Arabidopsis thaliana, Nicotiana tabacum, and Oryza sativa.
The presence of PvFtsH genes was observed in 17 cellular sites, with a particular abundance in the chloroplast. Typically, the localization of FtsH protein in a plant's mitochondria or chloroplast membrane system plays a crucial role in facilitating membrane protein breakdown and repair. This serves as an essential mechanism for maintaining the quality control of membrane-bound proteins in plants (Sun et al. 2021a, b). Additionally, to controlling photosynthetic processes, FtsH genes regulate metabolic growth, plastid differentiation, and other root-shoot critical functions (Van Der Hoom 2008). These regulations and controls are provided through cis regulators in the promoter region. Cis-regulatory sequences are non-coding DNA linear nucleotide segments with varying locations and orientations regarding genes and activity (Biłas et al. 2016). The 2000 bp upstream region of the PvFtsH gene promoter was investigated for cis-acting elements, with a focus on 17 PvFtsH genes (Fig. 4A). Understanding the role of cis-acting elements is crucial for the regulation of a wide range of biological processes, such as hormone production and responses to abiotic stress (Duraisamy et al. 2016). Thus, present study was conducted to examine the cis-acting elements present in PvFtsH genes. Cis-acting elements are categorized in a prior study according to their roles, which included growth, hormone, environmental stress, light, promoter, site binding, and others (Akbulut et al. 2022). Promoter regions of FtsH genes could be separated into four main categories, including 36 hormone-related elements [ABRE (ABA responsive element), TCA-element (salicylic acid responsiveness), TGA-element (auxin-related responsiveness) and TATC-box (gibberellin-responsive elements)] and 61 promoter-related elements (TATA and CAAT box), 90 abiotic stress responsive elements [(DRE (drought responsive elements), MYB and MYC (drought-related regulatory), STRE (stress responsive element) and TC-rich repeats (defense and stress-responsive element)] and 1 biotic stress-responsive element (box S) (Fig. 4A). According to the analysis results, there were still cis-acting elements with unknown functions. Among the FtsH genes, compared to other metalloproteases, the FtsH3 has the highest number of elements with 37 cis-acting elements, and FtsH5 has the lowest number of elements with 19 cis-acting elements.
A The upstream cis-acting elements of the PvFtsH gene, specifically focusing on a 2000 bp upstream region. The cis-acting elements were classified into different types, and a distinct color represented each. B Gene structures of PvFtsH gene family members. Introns are represented with lines. UTR and CDS are denoted by filled purple and red boxes, respectively. C Potential conserved motif arrangement among members of the FtsH gene family in beans. Each motif type and potential sequence information related to colored box symbols are represented by the various colored blocks
MYB (46) and MYC (24) had the highest number of cis-acting elements associated with abiotic stress when all PvFtsH genes were evaluated. It has been declared that most GmFtsH genes incorporate MYB (239) and MYC binding sites (139) (Shan et al. 2023). Research has indicated that MYB, MYC, DRE, and STRE transcription mechanisms (TFs) can directly or indirectly engage in the drought signal response pathway through multiple factors (Ebeed 2022; Feng et al. 2023; Kodackattumannil et al. 2023; Zhou et al. 2023). These findings suggest that PvFtsHs may play a role in developmental processes as well as in response to biotic and abiotic stresses. FtsH has been implicated in the response to a variety of abiotic stressors, containing light, drought, high temperature, and salinity (Pu et al. 2022; Hajibarat and Saidi 2023).
The PvFtsH genes' gene structure analysis showed that each one has multiple exons and introns. Interestingly, genes clustered within the same branch of the evolutionary tree exhibited similar numbers and lengths of introns/exons. The exon numbers of PvFtsH genes range from 4 to 19, while the intron counts range from 3 to 18 (Fig. 4B). The highest intron–exon number belongs to PvFtsH16 and 17, while the lowest intron–exon number belongs to PvFtsH3, 5 and 13. Out of the 17 PvFtsH genes, 2 of them had NO untranslated regions (UTRs) (Fig. 4B). All 17 PvFtsH genes exhibited conserved domains related to FtsH proteins, which are characteristic features of the FtsH protein family (Fig. 4B). In addition to being highly conserved in mitochondria and chloroplasts, the FtsH gene is also homologous with the prokaryotic membrane-bound ATP-dependent protease (Pu et al. 2022). Additionally, the preservation of the FtsH domain ensures the gene family's fundamental functions, as diversity within the family reduces the selection pressure during evolutionary processes (William Roy and Gilbert 2006). The genetic structure of the PvFtsH gene resembles that of Nicotiana, Arabidopsis, pear, and other plant species, indicating a high level of conservation across different organisms (Pu et al. 2022). Certain genes within this group possess extensive introns, suggesting they are relatively recent additions to the PvFtsH gene family. In addition, PvFtsH genes that clustered together on the phylogenetic tree shared similar patterns of intron–exon distribution and conserved domains, advising they play comparable roles in plant function.
Further analysis was performed by the MEME software to determine the structure and number of conservative motifs in PvFtsH genes. Ten conservative motifs were found and labeled Motif 1–10 (Fig. 4C). The amino acid numbers of conserved motifs are between 21 and 50. Motifs 1 and 3 were conserved in all common bean PvFtsH within the gene family, indicating that these motifs may function as identifiers for distinguishing the PvFtsH gene (Fig. 4C). Additionally, while motif1 contains the METALLOPROTEASE M41 FTSH domain, motif2 contains the ATPase_AAA_core domain (Table S2). The genes with the most motifs are PvFtsH1, 2, 3, 8, 10, 11, 13, 14 and 15 and contain all conserved motifs. The gene containing the fewest motifs is the PvFtsH17 gene. Additionally, it was observed that genes within the same branch generally possessed the same number and type of motifs. The sequences of closely related individuals had a high degree of similarity, and their motifs also demonstrated commonality. The shared existence of identical conserved motifs suggests that members of the PvFtsH gene family have comparable roles and confirms the reliable creation of the evolutionary tree. Significantly, PvFtsH exhibits structural similarities and conserved domains when compared to FtsH genes found in soybean, pear, M. truncatula and M. sativa (Guo et al. 2021; Shan et al. 2023; Zhu et al. 2023; Li et al. 2024).
The WEBLOGO database was utilized to find conserved domain sequences and motif logos of P. vulgaris FtsH gene families. The multiple sequence alignment (Fig. S2) shows conserved Walker A (GX1X2X3PX4GX5LLX6GX7PGTGKT) and Walker B (PX1X2X3FIDEIDA) motifs in the ATP binding domain, where X is a variable residue. We have also identified a conserved motif known as second region of homology (SHR), which is a unique domain found in FtsH. The conserved sequences of this motif are TNX1X2X3X4LDX5X6X7X8RX9GRX10DR. The SHR distinguishes FtsH proteins from other ATP-dependent proteases. The zinc-binding motif, HEX1X2H, is the proteases' catalytic center in the M41 peptidase domain. In P. vulgaris, the residue X2 is conserved as ‘G’ (Fig. S2). These conserved sequences served as a criterion for isolating and identifying FtsH genes from other plants (Guo et al. 2021).
Gene ontology analysis and protein–protein interactions
The rate of participation of PvFtsH genes in molecular function, cellular components, and biological processes was determined (Fig. 5). The FtsH enzyme is primarily involved in proteolysis processes as well as protein, organonitrogen compound, and macromolecule metabolic process. The investigation of the molecular functions of the FtsH enzyme revealed that its primary metabolic activities include endopeptidase activity, peptidase activity, metallopeptidase activity, and metalloendopeptidase activity (Fig. 5). The role of the FtsH gene as a cellular component appears to be solely involved in membrane integrity.
PPIs play a vital role in some biological processes, containing DNA replication, transcription, metabolic cycles, and signal transmission (Zhang et al. 2016). Because proteins interact with one another to achieve their functions, it's critical to understand the nature of these connections and how intricate cellular processes work. An average plant's proteome is estimated to comprise 36,795 proteins, of which 75,000–150,000 are projected to interact with one another (Struk et al. 2019). PPIs can often be divided into two categories: constitutive and regulative. Constitutive PPIs are found almost everywhere and have a high affinity while regulative PPIs, on the other hand, are only found in certain cell or developmental situations or are activated by certain stimuli (Zhang et al. 2022). The PvFtsH protein sequences were utilized to identify PPIs of PvFtsHs using the STRING interface. Analysis results show that all PvFtsH proteins interact with each other (Fig. S3). PvFtsH2 and PvFtsH6 are the higher interacting level with the number 15, which means these proteins interacts 15 other PvFtsH proteins except PvFtsH11 and PvFtsH10 respectively. Whereas PvFtsH3 was found to least interacting FtsH protein of bean FtsH proteins.
miRNAs analysis of PvFtsH genes
Through the analysis of projected miRNA targets, valuable knowledge regarding the regulatory interactions between miRNAs and FtsH genes was obtained. MicroRNAs (miRNAs) correspond to a group of endogenous, small RNAs, typically 20–22 nucleotides in length (Bajczyk et al. 2023). Each miRNA can reduce gene expression by targeting multiple messenger RNAs (mRNAs) containing the cognate miRNA binding site, thereby causing cleavage or inhibition of translation (Zhao et al. 2021; Huang et al. 2022; Shah et al. 2023). The discovery of miRNAs related with FtsH genes will help elucidate possible regulatory mechanisms affecting the expression and function of FtsH. The miRNAs considered to be related to FtsHs were acquired from the psRNATarget database, and the findings are shown in Table S3. In present study identified 32 miRNAs related to the PvFtsH genes. miR477 and miR2673 targets 2 genes including PvFtsH14 and PvFtsH15. The PvFtsH13 gene was revealed to be targeted by miR7121. The miRNAs mostly targeted the PvFtsH17 gene. Recent years have seen a growing body of evidence suggesting that the miR477 family is a group of microRNAs that are generated by stress and play a significant role in responding to both abiotic and biotic stressors (Wang et al. 2020). miR477 families were observed to be upregulated under salt stress in P. cathayana and cold stress in Populus (Lu et al. 2008; Zhou et al. 2012). Another study demonstrated that miR2673 was down-regulated in potatoes during drought stress (Yang et al. 2013). Small-miR169s in maize and their targets in Zea mays leaves are responsive to salt treatment, ABA, and osmotic stress (Luan et al. 2015). It has been found that under normal conditions, the Osa-miR172d family mediates the transmission of flowering-related characteristics, but this miRNA is downregulated during drought stress (Kumar et al. 2024).
Expression patterns of PvFtsH genes in different tissues
A heat map of 17 FtsH genes represented by FPKM values in various tissues and organs was created using RNA-seq data (Fig. S4). Eleven various tissues and organs were contained in the analysis. The most of PvFtsH genes were expressed in at least one tissue except for PvFtsH8, which were scarcely expressed in any of the tissue or organ. Furthermore, certain gene expressions exhibited specificity to particular tissues. PvFtsH3 and PvFtsH10 displayed relatively similar high expression levels in leaves, flowers, and young trifoliates. Consequently, a more increased expression level of the PvFtsH13 genes was determined in the flower than in the other tissues and organs (Fig. S4). These three genes are potentially associated with the process of flower formation. Previous research has shown that FtsH genes are predominantly expressed in a range of organs, including leaves, floral organs, and seeds. (Yue et al. 2010; Wang et al. 2023). According to Mishra et al. (2019), FtsHi enzymes evolved alongside FtsH12 in flowering plants, particularly eudicots. Pu et al. (2022) examined the calyx, fruits, leaves, petals, roots, stems, and stigma of N. tabacum plants to determined tissue-specific expression of the FtsH gene family. Several genes have been determined as exhibiting high levels of expression in different tissues. All tissues and organs were found to express nine chosen genes, with the leaf expressing four of these genes more frequently than other organs. These genes that are expressed at high levels may have distinct housekeeping functions (Wang et al. 2021).
In silico expression analysis and qRT-PCR analysis of PvFtsH genes in drought and salt stress conditions
The world population has expanded dramatically over the last few decades and is anticipated to get 10 billion by 2050 (Qu et al. 2023a, b). Urbanization and land degradation are reducing arable land, making it impossible to meet global food demand (Raza et al. 2023). Thus, considerable efforts and realistic techniques are needed to boost crop productivity, particularly in marginal areas, due to climate change and other biotic and abiotic pressures (Alotaibi et al. 2023). Recently, identifying genes' role in stress using bioinformatics tools and using them in breeding programs and agricultural biotechnology may be a viable strategy (Roychowdhury et al. 2023).
The Illumina RNAseq dataset was taken from the NCBI Sequence Read Archive (SRA) database to perform this investigation. PvFtsH gene expression patterns in P. vulgaris leaf tissue were evaluated. Different patterns of gene regulation in response to salt stress and drought are shown by the expression analysis. Looking at drought stress, there was rise in the expression levels of PvFtsH2, PvFtsH6, and PvFtsH11 genes, while the expression levels of PvFtsH1, PvFtsH8, PvFtsH9, PvFtsH12, PvFtsH14, PvFtsH15, PvFtsH16, and PvFtsH17 genes decreased (Fig. 6). The FtsH3 mutant exhibited a drought-tolerant response in its above-ground tissues as a result of the drought-responsiveness of its root-dependent bacterial clone (Mishra and Funk 2021). As a result of salt stress, there was an increase in the expression levels of PvFtsH2, PvFtsH4, PvFtsH5, PvFtsH6, PvFtsH8, PvFtsH11, PvFtsH12, PvFtsH16, and PvFtsH17 genes, while the expression levels of PvFtsH14 and PvFtsH15 genes decreased (Fig. 6). The increase in PvFtsH3, PvFtsH10, and PvFtsH13 gene expressions in salt treatment was highly significant compared to the salt control. These results indicate that PvFtsH3, PvFtsH10, and PvFtsH13 genes have the potential to respond to salt stress situations (Fig. 6). Li et al. (2024) showed that overexpression of MsFtsH8 ensures resistance to oxidative and salt stress, which is accompanied by decreased ROS levels and increased expression and activity of antioxidant enzymes. PvFtsH8 showed a decrease the most prominent in control samples. The results of the in-silico analysis conducted for this study are encouraging; nevertheless, qPCR laboratory experiments will serve to further validate the conclusions.
Heatmap-based expression analysis of genes in leaf tissues of bean plant under different stress conditions. The heatmap's color gradients, where red denotes higher expression and blue denotes lower expression, show the different gene expression levels. Yellow or near-yellow regions denote insignificant changes in expression levels
RNAseq and CDS data were utilized to design primers for qRT-PCR reactions. Expression levels of PvFtsH genes were determined by performing qRT-PCR using leaf tissues from bean cultivars Elkoca-05 and Serra. The investigation focused on the expression levels of the genes PvFtsH3, PvFtsH6, PvFtsH10, and PvFtsH15 (Figs. 7 and 8). Primary sequence information of the genes to be investigated in the study is shown in Table S4. This study examined how the expression levels of these four genes changed when plants were stressed by drought (a 20% concentration of PEG6000), salt stress (a 150 mM concentration of NaCl), and melatonin (a 200 µM concentration). The effect of melatonin, drought, and salinity, on the expression of PvFtsH genes was investigated through the utilization of in silico and in vitro gene expression analysis techniques.
The expression profile of 5 PvFtsH genes was investigated using qRT-PCR analysis on leaf tissues of Elkoca-05 and Serra cultivars in drought stress. The 2−ΔΔCt method was utilized to calculate the expression levels. The presented data are depicted as bars, representing the mean ± SE (n = 3). Melatonin was included as a treatment factor. Statistical analysis was conducted using the Dunnett test to determine significant differences between the control and treatment groups. The results were denoted as follows: *p < 0.05, **p < 0.01, and ns (non-significant) for cases where there was no significant difference between the control and treatment conditions
The expression profile of 5 PvFtsH genes was investigated using qRT-PCR analysis on leaf tissues of Elkoca-05 and Serra cultivars in salt stress. The 2−ΔΔCt method was utilized to calculate the expression levels. The presented data are depicted as bars, representing the mean ± SE (n = 3). Melatonin was included as a treatment factor. Statistical analysis was conducted using the Dunnett test to determine significant differences between the control and treatment groups. The results were denoted as follows: *p < 0.05, **p < 0.01, and ns (non-significant) for cases where there was no significant difference between the control and treatment conditions
In drought stress, 4 PvFtsH genes in Elkoca-05 genotypes and PvFtsH3 and PvFtsH15 in Serra genotypes exhibited a considerable decrease in expression level (Fig. 7). Following melatonin administration, significant up-regulation was observed in the expression levels of all genes in Serra, and only the PvFtsH15 gene in Elkoca-05. In addition, as a result of melatonin application alone, the expression of the PvFtsH6 gene in the Elkoca-05 cultivar was down-regulated, while this gene was up-regulated in the Serra cultivar. Upon combining both treatments, the expression levels of the PvFtsH3 and PvFtsH6 genes significant decreased, while no significant alteration was detected in the expression of the other genes in Elkoca-05. In Serra, it was shown that the expression levels of the PvFtsH3 and PvFtsH15 genes were significantly reduced with combination treatments, whereas the PvFtsH6 gene was found to be increased.
In the Serra cultivar, there was a boost in the expression of the FtsH3 gene under salt stress (Fig. 8). In melatonin applying, an important rise was seen in the expression levels of three genes excluding PvFtsH10 gene in Elkoca-05. The treatment of melatonin against salt stress significantly raised the expression level of PvFtsH6 and reduced the expression level of PvFtsH10, but did not affect a significant difference in the expression levels of PvFtsH3 and PvFtsH15 in Elkoca-05. However, with the exception of PvFtsH3, the treatment of melatonin against salt stress resulted in a considerable increase in the expression of the remaining genes in Serra cultivars. The findings were attributed to the presence of cis-acting elements, namely ABRE, MYB, MYC, and STRE in this gene.
FtsH has been implicated in the reply to a variety of abiotic stressors, including cold, drought, heat, light, and salt (Yue et al. 2010; Rollins et al. 2013; Hajibarat and Saidi 2023; Zhu et al. 2023). Pu et al. (2022) investigated the expression profiles of NtFtsH genes in response to abiotic stresses in tobacco plants. They found that under high-temperature stress, the NtFtsH3 gene showed up-regulation in stems, consistent with our findings. Moreover, the Arabidopsis mitochondrial protease AtFtsH4 enhances the plants' ability to withstand prolonged mild heat stress by reducing the excessive buildup of HSP23.6 in the mitochondria (Maziak et al. 2021). Chen et al. (2006). proposed that FtsH11, one of the twelve FtsHs that have been discovered, directly contributes to thermotolerance in Arabidopsis plants. The MsFtsH8 gene in alfalfa that induced by ABA and hydrogen peroxide (H2O2) treatment, and it plays a role in the plant's response and ability to tolerate salt stress (Qianwen et al. 2021). Hajibarat and Saidi (2023) revealed that StFtsH10 gene was expressed under conditions of high light, heat, and Potato virus S and Y stresses. The expression level of ZmFtsH2B was upregulated in the leaves of maize seedlings exposed to ABA treatment and water deficiency stress induced by 18% PEG (Yue et al. 2010). A previous study in pepper found that the CaFtsH6 gene plays an important role in stress tolerance. It was also detected that the CaFtsH6 gene was activated in response to drought, heat, and salinity stress. Silencing CaFtsH6 has been shown to reduce the stress tolerance of the pepper plant. In addition, the experimental results suggest that the CaFtsH6 gene plays a responsibility in stress tolerance by increasing the transcription levels of defense-related genes by inhibiting hydrogen peroxide accumulation (Xiao et al. 2021). With reduced stomatal conductance, increased intrinsic water-use efficiency, and delayed stress acclimation, plants carrying AtFtsHi3 also exhibit superior drought tolerance compared to wild-type plants (Mishra et al. 2021). This prior research provides valuable support to our current findings, strengthening the comprehension of the functions of FtsH genes in the plant's response to abiotic stress and their participation in complex regulatory networks.
Upon seeing a disparity in melatonin levels among plants of the same species cultivated in various stresses, it was deduced that melatonin plays a crucial role in responding to changes in the environment (Colombage et al. 2023). Endogenous melatonin inhibits or promotes the expression of genes that encode critical defense-related enzymes and transcription factors (Weeda et al. 2014). Due to the significant shifts in climate circumstances, the endogen systems in plants that promote stress tolerance are insufficient to safeguard crop yield. It has been reported that exogenous melatonin applications are protective against various factors such as drought stress (Iqbal et al. 2024), low-temperature (Bao et al. 2024), heavy metals (Saqip et al. 2024) and salinity (Kang et al. 2024). Yan et al. (2021) demonstrated that the levels of endogen melatonin in rice were elevated when exposed to salt stress. Furthermore, the application of external melatonin was found to enhance the regulation of ion balance in rice plants under salt stress by rising the levels of endogen melatonin. Zahedi et al. (2020) declared that 100 and 200 μM melatonin applications in strawberries alleviated salt stress by enhancing leaf antioxidant enzymes and abscisic acid. Exogenous melatonin applied to cucumber at a 200 µM dose lessened the deleterious effects of the chilling stress condition (Zhao et al. 2017). M. oleifera plants grew and yielded best with 100 mM melatonin foliar application under regular irrigation or drought stress (Sadak et al. 2020).
Conclusion
The current study conducted a genome-wide investigation of the FtsH gene family in beans. Based on comparable exon–intron architectures and motif patterns within the same groups and subgroups, seventeen PvFtsH genes were found and classified into three primary categories. The appearance of PvFtsH genes on chromosomes was shown in a sharp status. Phylogenetic comparison and synteny analysis of the PvFtsH genes from Arabidopsis thaliana and Oryza sativa plant species, analysis supplied important tips to the evolutionary process of bean PvFtsH genes. Based on subcellular localization analysis it was determined the PvFtsH genes can be found in the mitochondrion or chloroplast organelle. It was found also PvFtsH genes have significant roles in process of bean growth and development mechanisms. These results supply an important data for better explain the molecular biological roles of individual PvFtsH genes in bean.
References
Adam Z, Adamska I, Nakabayashi K, Ostersetzer O, Haussuhl K, Manuell A, Zheng B, Vallon O, Rodermel SR, Shinozaki K (2001) Chloroplast and mitochondrial proteases in Arabidopsis. A Proposed Nomenclature Plant Physiol 125(4):1912–1918
Akbulut SE, Okay A, Aksoy T, Aras ES, Büyük İ (2022) The genome-wide characterization of WOX gene family in Phaseolus vulgaris L. during salt stress. Physiol Mol Biol Plants 28(6):1297–1309
Aleem M, Riaz A, Raza Q, Aleem M, Aslam M, Kong K, Atif RM, Kashif M, Bhat JA, Zhao TJG (2022) Genome-wide characterization and functional analysis of class III peroxidase gene family in soybean reveal regulatory roles of GsPOD40 in drought tolerance. Genomics 114(1):45–60
Ali MS, Baek KH (2020) Protective roles of cytosolic and plastidal proteasomes on abiotic stress and pathogen invasion. Plants 9(7):832
Alotaibi M (2023) Climate change, its impact on crop production, challenges, and possible solutions. Not Bot Horti Agrobo 51(1):13020–13020
Amiri H, Zamani Z, Arnao MB, Ismaili A, Gavyar PHH, Khodayari H (2024) Optimal concentration of melatonin enhances drought stress tolerance in fenugreek. Acta Physiol Plant 2:15
Aygören AS, Güneş E, Muslu S, Kasapoğlu AG, Yiğider E, Aydın M, Büyük İ, İlhan E (2023) Genome-wide analysis and characterization of SABATH gene family in Phaseolus vulgaris genotypes subject to melatonin under drought and salinity stresses. Plant Mol Biol Rep 41(2):242–259
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34(2):W369–W373
Bajczyk M, Jarmolowski A, Jozwiak M, Pacak A, Pietrykowska H, Sierocka I, Swida-Barteczka A, Szewc L, Szweykowska-Kulinska Z (2023) Recent insights into plant miRNA biogenesis: Multiple layers of miRNA level regulation. Plants 12(2):342
Baloglu MC, Inal B, Kavas M, Unver T (2014) Diverse expression pattern of wheat transcription factors against abiotic stresses in wheat species. Gene 550(1):117–122
Bao Z, Zhou Q, Yu Y, Chen W, Yang Z, Cao S, Shi L (2024) Melatonin treatment induces DNA methylation to alleviate chilling induced-browning in cold stored peach fruit. Postharvest Biol Tec 208:112686
Bengoa Luoni SA, Cenci A, Moschen S, Nicosia S, Radonic LM, Sabio J, Langlade NB, Vile D, Rovere CV, Fernandez P (2021) Genome-wide and comparative phylogenetic analysis of senescence-associated NAC transcription factors in sunflower (Helianthus annuus). BMC Genomics 22:1–19
Biłas R, Szafran K, Hnatuszko-Konka K, Kononowicz AK (2016) Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Cult 127:269–287
Braun P, Gingras AC (2012) History of protein–protein interactions: From egg-white to complex networks. Proteomics 12(10):1478–1498
Chen J, Burke JJ, Velten J, Xin Z (2006) FtsH11 protease plays a critical role in Arabidopsis thermotolerance. Plant J 48(1):73–84
Chen C, Wu Y, Li J, Wang X, Zeng Z, Xu J, Liu Y, Feng J, Chen H, He Y, Xia R (2023) TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol Plant 16(11):1733–1742
Choi JW, Kim SW, Kim HS, Kang MJ, Kim SA, Han JY, Kim H, Ku SY (2024) Effects of melatonin, GM-CSF, IGF-1, and LIF in culture media on embryonic development: potential benefits of individualization. Int J Mol Sci 25(2):751
Colombage R, Singh MB, Bhalla PL (2023) Melatonin and abiotic stress tolerance in crop plants. Int J Mol Sci 24(8):7447
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14(6):1188–1190
Dey S, Malviya R, Pandey A, Banavath HN, Muthamilarasan M, Gayen D (2023) Identification and expression analysis of the FtsH protein family in chickpea in response to drought stress.
Dougan DA, Micevski D (2012) The N-end rule pathway: from recognition by N-recognins, to destruction by AAA+ proteases. Biochim Biophys Acta Mol Cell Res 1:83–91
Duraisamy GS, Mishra AK, Kocabek T, Matoušek J (2016) Identification and characterization of promoters and cis-regulatory elements of genes involved in secondary metabolites production in hop (Humulus lupulus L.). Comput Biol Chem 64:346–352
Ebeed HT (2022) Genome-wide analysis of polyamine biosynthesis genes in wheat reveals gene expression specificity and involvement of STRE and MYB-elements in regulating polyamines under drought. BMC Genomics 23(1):734
Feng Y, Zeng S, Yan J, Li K, Xu H (2023) Genome-wide analysis and expression of MYC family genes in tomato and the functional identification of slmyc1 in response to salt and drought stress. Agron 13(3):757
Fu Y, Cheng M, Li M, Guo X, Wu Y, Wang J (2020) Identification and characterization of PLATZ transcription factors in wheat. Int J Mol Sci 21(23):8934
García-Lorenzo M, Sjödin A, Jansson S, Funk C (2006) Protease gene families in Populus and Arabidopsis. BMC Plant Biol 6:1–24
Gull A, Lone AA, Wani NUI (2019) Biotic and abiotic stresses in plants. Abiotic and biotic stress in plants, 1–19
Guo AY, Zhu QH, Chen X, Luo JC (2007) GSDS: a gene structure display server. Yi Chuan HEREDITAS 29(8):1023–1026
Guo Z, Gao X, Cai H, Yu L, Gu C, Zhang SL (2021) Genome-wide identification, evolution and expression analysis of the FtsH gene during fruit development in pear (Pyrus bretschneideri). Plant Biotechnol Rep 15(4):537–550
Hajibarat Z, Saidi A (2023) Filamentation temperature-sensitive (FtsH); key player in response to multiple environmental stress conditions and developmental stages in potato. J Plant Growth Regul 42(7):4223–4239
Hiz MC, Canher B, Niron H, Turet M (2014) Transcriptome analysis of salt tolerant common bean (Phaseolus vulgaris L.) under saline conditions. PLoS ONE 9:e92598
Hooft RW, Sander C, Vriend G (1996) Positioning hydrogen atoms by optimizing hydrogen-bond networks in protein structures. Proteins: Struct, Funct, Bioinform 26(4):363–376
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLFPSORT: protein localization predictor. Nucleic Acids Res 35(2):W585–W587
Huang T, He WJ, Li C, Zhang JB, Liao YC, Song B, Yang P (2022) Transcriptome-wide analyses of RNA m6A methylation in hexaploid wheat reveal its roles in mRNA translation regulation. Front Plant Sci 13:917335
İlhan E, Büyük İ, İnal B (2018) Transcriptome-Scale characterization of salt responsive bean TCP transcription factors. Gene 642:64–73
Inal B, Büyük İ, Ilhan E, Aras S (2017) Genome-wide analysis of Phaseolus vulgaris C2C2-YABBY transcription factors under salt stress conditions. 3 Biotech 7:1–10
Iqbal S, Hayat F, Hussain M, Mushtaq N, Rehman M, Asif A, Khan U, Shahid MA (2024) Melatonin supplementation alleviates drought stress in peach (Prunus persica) seedlings by improving photosynthesis, root morphological traits, and antioxidant defense system. Acta Physiol Plant 46(2):18
Ito TM, Trevizan CB, dos Santos TB, de Souza SGH (2017) Genome-wide identification and characterization of the dof transcription factor gene family in Phaseolus vulgaris L. Am J Plant Sci 8(12):3233–3257
Ji W, Yan J, Bai X, Qiao P, Li Z, Yang Y, Guan W, Zhao T (2019) Functional analysis of the gene ftsH in Acidovorax citrulli. Acta Phytopathol Sin 49(4):488–499
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240
Kang SM, Shaffique S, Injamum-Ul-Hoque M, Alomrani SO, Park YS, Lee IJ (2024) Foliar treatment with melatonin modulates photosynthetic and antioxidant responses in Silybum marianum L. under salt stress. Sci Hortic 325:112664
Kasapoglu AG, Ilhan E, Aydin M, Yigider E, Inal B, Buyuk I, Taspinar MS, Ciltas A, Agar G (2023) Characterization of Two-Component System gene (TCS) in melatonin-treated common bean under salt and drought stress. Physiol Mol Biol Plants 29:1–22
Kato Y, Sakamoto W (2019) Phosphorylation of the chloroplastic metalloprotease FtsH in Arabidopsis characterized by Phos-Tag SDS-PAGE. Front Plant Sci 10:1080
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6):845–858
Kodackattumannil P, Whitley K, Sasi S, Lekshmi G, Krishnan S, Al Senaani S, Kottackal M, Amiri KM (2023) Novel inducible promoter DREB1G cloned from date palm exhibits high fold expression over AtRD29 to drought and salinity stress. Plant Cell Tissue Organ Cult 154(2):367–380
Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV (2002) Selection in the evolution of gene duplications. Genome Biol 3:1–9
Kumar D, Mulani E, Singh BK, Dutta B, Singh A, Solanke AU, Sevanthi AM (2024) Understanding the role of miRNAs in governing the drought sensitive response of a rice mega variety. Swarna at Reproductive Stage Plant Stress 11:100302
Letunic I, Bork P (2011) Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39(2):W475–W478
Li M, Zhu X, Yu Q, Yu A, Chen L, Kang J, Wang X, Yang T, Yang Q, Long R (2024) FtsH proteases confer protection against salt and oxidative stress in Medicago sativa L. Plant Sci 338:111915
Liu X, Zhou G, Chen S, Jia Z, Zhang S, Ren M, He F (2023) Genome-wide analysis of the AP2/ERF gene family in Tritipyrum and the response of TtERF_B2-50 in salt-tolerance. BMC Genom 24:541
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4):402–408
Lu S, Sun YH, Chiang VL (2008) Stress-responsive microRNAs in Populus. Plant J 55(1):131–151
Luan M, Xu M, Lu Y, Zhang L, Fan Y, Wang L (2015) Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves. Gene 555(2):178–185
Malabadi RB, Kolkar KP, Meti NT, Chalannavar RK (2021) Melatonin: One molecule one-medicine for many diseases, coronavirus (SARS-CoV-2) disease (Covid-19); Function in plants. Int J Innov Sci 8(3):155–181
Maziak A, Heidorn-Czarna M, Weremczuk A, Janska H (2021) FTSH4 and OMA1 mitochondrial proteases reduce moderate heat stress-induced protein aggregation. Plant Physiol 187(2):769–786
Mielke K, Wagner R, Mishra LS, Demir F, Perrar A, Huesgen PF, Funk C (2021) Abundance of metalloprotease FtsH12 modulates chloroplast development in Arabidopsis thaliana. J Exp Bot 72(9):3455–3473
Mishra LS, Funk C (2021) The FtsHi enzymes of Arabidopsis thaliana: Pseudo-proteases with an Important function. Int J Mol Sci 22(11):5917
Mishra LS, Mielke K, Wagner R, Funk C (2019) Reduced expression of the proteolytically inactive FtsH members has impacts on the Darwinian fitness of Arabidopsis thaliana. J Exp Bot 70(7):2173–2184
Mishra LS, Mishra S, Caddell DF, Coleman-Derr D, Funk C (2021) The plastid-localized AtFtsHi3 pseudo-protease of Arabidopsis thaliana has an impact on plant growth and drought tolerance. Front Plant Sci 12:694727
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628
Muñoz-Bodnar A, Perez-Quintero AL, Gomez-Cano F, Gil J, Michelmore R, Bernal A, Szurek B, Lopez C (2014) RNAseq analysis of cassava reveals similar plant responses upon infection with pathogenic and non-pathogenic strains of Xanthomonas axonopodis pv. manihotis. Plant Cell Rep 33:1901–1912
Muslu S, Kasapoğlu AG, Güneş E, Aygören AS, Yiğider E, İlhan E, Aydın M (2023) Genome-wide analysis of glutathione S-transferase gene family in P vulgaris under drought and salinity stress. Plant Mol Biol Report 42:1–20
Nekrutenko A, Makova KD, Li WH (2002) The KA/KS ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res 12(1):198–202
Oner BM, Ilhan E, Kasapoglu AG, Muslu S, Aygoren AS, Ucar S, Yaprak E, Isıyel M, Aydinyurt R, Aydin M (2022) Genome wide analysis and characterization of npr-like gene family of Phaseolus vulgaris L. Nat pro Biotech 2(1):23–41
Perry ED, Yu J, Tack J (2020) Using insurance data to quantify the multidimensional impacts of warming temperatures on yield risk. Nat Commun 11(1):4542
Praveen A, Dubey S, Singh S, Sharma VK (2023) Abiotic stress tolerance in plants: a fascinating action of defense mechanisms. Biotechnology 13(3):102
Pu T, Mo Z, Su L, Yang J, Wan K, Wang L, Liu R, Liu Y (2022) Genome-wide identification and expression analysis of the ftsH protein family and its response to abiotic stress in Nicotiana tabacum L. BMC Genomics 23(1):1–15
Qin S, Wei F, Liang Y, Tang D, Lin Q, Miao J, Wei K (2023) Genome-wide analysis of the R2R3-MYB gene family in Spatholobus suberectus and identification of its function in flavonoid biosynthesis. Front Plant Sci 14:1219019
Qu J, Liu L, Guo Z, Li X, Pan F, Sun D, Yin L (2023a) The ubiquitous position effect, synergistic effect of recent generated tandem duplicated genes in grapevine, and their co-response and overactivity to biotic stress. Fruit Res 3(1):16
Qu Y, Mueller-Cajar O, Yamori W (2023b) Improving plant heat tolerance through modification of Rubisco activase in C3 plants to secure crop yield and food security in a future warming world. J Exp Bot 74(2):591–599
Rawlings ND (2020) Twenty-five years of nomenclature and classification of proteolytic enzymes. Biochim Biophys Acta Proteins Proteomics 1868(2):140345
Raza A, Tabassum J, Fakhar AZ, Sharif R, Chen H, Zhang C, Ju L, Fotopoulos V, Siddique KHM, Sing RK, Zhuang W, Varshney RK (2023) Smart reprograming of plants against salinity stress using modern biotechnological tools. Crit Rev Biotechnol 43(7):1035–1062
Rollins JA, Habte E, Templer SE, Colby T, Schmidt J, Von Korff M (2013) Leaf proteome alterations in the context of physiological and morphological responses to drought and heat stress in barley (Hordeum vulgare L.). J Exp Bot 64(11):3201–3212
Roychowdhury R, Das SP, Gupta A, Parihar P, Chandrasekhar K, Sarker U, Kumar A, Ramrao DP, Sudhakar C (2023) Multi-omics pipeline and omics-integration approach to decipher plant’s abiotic stress tolerance responses. Genes 14(6):1281
Sadak MS, Abdalla AM, Abd Elhamid EM, Ezzo MI (2020) Role of melatonin in improving growth, yield quantity and quality of Moringa oleifera L. plant under drought stress. Bull Natl Res Cent 44:1–13
Sakamoto W, Zaltsman A, Adam Z, Takahashi Y (2003) Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15(12):2843–2855
Saqib M, Khalofah A (2024) Alleviating effects of exogenous melatonin on nickel toxicity in two pepper genotypes. Sci Hortic 325:112635
Schmutz J, McClean PE, Mamidi S, Wu GA, Cannon SB, Grimwood J, Jackson SA (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46(7):707–713
Shah H, Manzoor A, Ashraf T, Maqbool R, Dar A (2023) MicroRNAs in Plants and Animals: Converging and Diverging Insights. Plant MicroRNAs and Stress Response 15–49
Shan Q, Zhou B, Wang Y, Hao F, Zhu L, Liu Y, Wang N, Wang F, Li X, Dong Y, Xu K, Zhou Y, Li H, Liu W, Gao H (2023) Genome-wide identification and comprehensive analysis of the Ftsh gene family in soybean (Glycine max). Int J Mol Sci 24(23):16996
Sinha AU, Meller J (2007) Cinteny: flexible analysis and visualization of synteny and genome rearrangements in multiple organisms. BMC Bioinform 8:1–9
Struk S, Jacobs A, Sánchez Martín-Fontecha E, Gevaert K, Cubas P, Goormachtig S (2019) Exploring the protein–protein interaction landscape in plants. Plant Cell Environ 42(2):387–409
Sun C, Liu L, Wang L, Li B, Jin C, Lin X (2021a) Melatonin: A master regulator of plant development and stress responses. J Integr Plant Biol 63(1):126–145
Sun JL, Li JY, Wang MJ, Song ZT, Liu JX (2021b) Protein quality control in plant organelles: current progress and future perspectives. Mol Plant 14(1):95–114
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
The Gene Ontology Consortium (2023) The gene ontology knowledgebase in 2023. Genetics 224:iyad031
Tiwari RK, Kumar R, Lal MK, Kumar A, Altaf MA, Devi R, Mangal V, Naz S, Altaf MM, Dey A, Aftab T (2023) Melatonin-polyamine interplay in the regulation of stress responses in plants. J Plant Growth Regul 42(8):4834–4850
Tiwari RK, Altaf MA, Kumar R, Lal MK (2024) Evolution of Melatonin as an Oxidative Stress Mitigator in Plant. In Melatonin in Plants: A Pleiotropic Molecule for Abiotic Stresses and Pathogen Infection (pp. 1–20). Singapore: Springer Nature Singapore.
Turhan S, Taspinar MS, Yigider E, Aydin M, Agar G (2021) The role of long terminal repeat (LTR) responses to drought in selenium-treated wheat. Environ Eng Manag J 20(6):917–925
Van Der Hoorn RA (2008) Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol 59:191–223
Wagner R, Aigner H, Funk C (2012) FtsH proteases located in the plant chloroplast. Physiol Plant 145(1):203–214
Wan C, Ling Q (2022) Functions of autophagy in chloroplast protein degradation and homeostasis. Front Plant Sci 13:993215
Wang S, Liu S, Liu L, Li R, Guo R, Xia X, Wei C (2020) miR477 targets the phenylalanine ammonia-lyase gene and enhances the susceptibility of the tea plant (Camellia sinensis) to disease during Pseudopestalotiopsis species infection. Planta 251:1–12
Wang Y, Cao W, Merritt J, Xie Z, Liu H (2021) Characterization of FtsH essentiality in Streptococcus mutans via genetic suppression. Front Genet 12:659220
Wang L, Yang Y, Yang Z, Li W, Hu D, Yu H, Li X, Cheng H, Kan G, Che Z, Zhang D, Zhang H, Wang H, Huang F, Yu D (2023) GmFtsH25 overexpression increases soybean seed yield by enhancing photosynthesis and photosynthates. J Integr Plant Biol 65(4):1026–1040
Wang L, Tanveer M, Wang H, Arnao MB (2024) Melatonin as a key regulator in seed germination under abiotic stress. J Pineal Res 76(1):e12937
Weeda S, Zhang N, Zhao X, Ndip G, Guo Y, Buck GA, Fu C, Ren S (2014) Arabidopsis transcriptome analysis reveals key roles of melatonin in plant defense systems. PLoS ONE 9(3):e93462
William Roy S, Gilbert W (2006) The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet 7(3):211–221
Wu Q, Han T, Yang L, Wang Q, Zhao Y, Jiang D, Ruan X (2021) The essential roles of OsFtsH2 in developing the chloroplast of rice. BMC Plant Biol 21:1–14
Wu F, Muvunyi BP, Yan Q, Kanzana G, Ma T, Zhang Z, Wang Y, Zhang J (2022) Comprehensive genome-wide analysis of polyamine and ethylene pathway genes in Cleistogenes songorica and CsSAMDC2 function in response to abiotic stress. Environ Exp Bot 202:105029
Xiao JJ, Zhang RX, Khan A, Ul Haq S, Gai WX, Gong ZH (2021) CaFtsH06, a novel filamentous thermosensitive protease gene, is involved in heat, salt, and drought stress tolerance of pepper (Capsicum annuum L.). Int J Mol Sci 22(13):6953
Yan F, Zhang J, Li W, Ding Y, Zhong Q, Xu X, Wei H, Li G (2021) Exogenous melatonin alleviates salt stress by improving leaf photosynthesis in rice seedlings. Plant Physiol Biochem 163:367–375
Yang Z, Nielsen R, Goldman N, Pedersen AMK (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genet 155(1):431–449
Yang J, Zhang N, Ma C, Qu Y, Si H, Wang D (2013) Prediction and verification of microRNAs related to proline accumulation under drought stress in potato. Comput Biol Chem 46:48–54
Yang Z, Zeng X, Zhao Y, Chen R (2023) AlphaFold2 and its applications in the fields of biology and medicine. Signal Transduct Target Ther 8(1):115
Yu Y, Xu J, Wang C, Pang Y, Li L, Tang X, Li B, Sun QJP (2022) Genome-wide analysis of the strigolactone biosynthetic and signaling genes in grapevine and their response to salt and drought stresses. PeerJ 10:e13551
Yue G, Hu X, He Y, Yang A, Zhang J (2010) Identification and characterization of two members of the FtsH gene family in maize (Zea mays L.). Mol Biol Rep 37:855–863
Yusuf M, Saeed T, Almenhali HA, Azzam F, Hamzah AIAH, Khan TA (2024) Melatonin improved efficiency of 24-epibrassinolide to counter the collective stress of drought and salt through osmoprotectant and antioxidant system in pea plants. Sci Hortic 323:112453
Zahedi SM, Hosseini MS, Abadía J, Marjani M (2020) Melatonin foliar sprays elicit salinity stress tolerance and enhance fruit yield and quality in strawberry (Fragaria× ananassa Duch.). Plant Physiol Biochem 149:313–323
Zandalinas SI, Balfagón D, Gómez-Cadenas A, Mittler R (2022) Plant responses to climate change: metabolic changes under combined abiotic stresses. J Exp Bot 73(11):3339–3354
Zhang Y (2005) miRU: an automated plant miRNA target prediction server. Nucl Acids Res 33:W701–W704
Zhang J, Sun A (2009) Genome-wide comparative analysis of the metalloprotease ftsH gene families between Arabidopsis thaliana and rice. Sheng Wu Gong Cheng Xue Bao Chinese J Biotechnol 25(9):1402–1408
Zhang F, Liu S, Li L, Zuo K, Zhao L, Zhang L (2016) Genome-wide inference of protein-protein interaction networks identifies crosstalk in abscisic acid signaling. Plant Physiol 171(2):1511–1522
Zhang K, Li Y, Huang T, Li Z (2022) Potential application of TurboID-based proximity labeling in studying the protein interaction network in plant response to abiotic stress. Front Plant Sci 13:974598
Zhao H, Zhang K, Zhou X, Xi L, Wang Y, Xu H, Pan T, Zou Z (2017) Melatonin alleviates chilling stress in cucumber seedlings by upregulation of CsZat12 and modulation of polyamine and abscisic acid metabolism. Sci Rep 7(1):4998
Zhao Y, Kuang Z, Wang Y, Li L, Yang X (2021) MicroRNA annotation in plants: current status and challenges. Brief Bioinform 22(5):bbab075
Zhao X, Qu D, Wang L, Gao Y, An N, Wang A, Li Y, Yang J, Wu F, Su H (2022) Genome-wide identification of cysteine-rich receptor-like kinases in sweet cherry reveals that PaCRK1 enhances sweet cherry resistance to salt stress. Plant Cell Rep 41(10):2037–2088
Zhou J, Liu M, Jiang J, Qiao G, Lin S, Li H, Xie L, Zhuo R (2012) Expression profile of miRNAs in Populus cathayana L. and Salix matsudana Koidz under salt stress. Mol Biol Rep 39:8645–8654
Zhou G, Ren Y, Ma L, Cheng C, Wang B, Yao Z, Lı S, Tao M, Zhao Y, Lı Z, Zhang H (2023) Identification and transcriptome analysis of the R2R3-MYB gene family in Haloxylon ammodendron. Not Sci Biol 15(4):11649–11649
Zhu X, Yu A, Zhang Y, Yu Q, Long R, Kang J, Yang Q, Guo C, Li M (2023) Genome-wide identification and characterization of filamentation temperature-sensitive H (FtsH) genes and expression analysis in response to multiple stresses in Medicago truncatula. Mol Biol Rep 50(12):10097–10109
Zhou G, Ren Y, Ma L, Cheng C, Wang B, Yao Z, Lı S, Tao M, Zhao Y, Lı Z, Zhang H (2023) Identification and transcriptome analysis of the R2R3-MYB gene family in Haloxylon ammodendron. Not Sci Biol 15(4):11649–11649
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK). The Scientific and Technological Research Council of Türkiye (TÜBİTAK) is the source of open-access financing.
Author information
Authors and Affiliations
Contributions
All authors conceptualized the manuscript. Behcet Inal, Selman Muslu, Esma Yigider, Ayse Gul Kasapoglu, and Emre Ilhan contributed to preparing the first draft of the writing. Behcet Inal, Selman Muslu, Ayse Gul Kasapoglu, and Emre Ilhan were responsible for the laboratory experiments and bioinformatics analysis. All authors contributed to the development and revision of the manuscript. The last version of the manuscript has been checked and approved by entire authors.
Corresponding author
Ethics declarations
Conflict of interests
None of the writers have any conflicts of interest to assert.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Inal, B., Muslu, S., Yigider, E. et al. In silico analysis of Phaseolus vulgaris L. metalloprotease FtsH gene: characterization and expression in drought and salt stress. Genet Resour Crop Evol (2024). https://doi.org/10.1007/s10722-024-02031-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-024-02031-1