Abstract
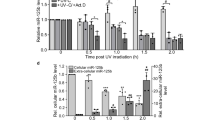
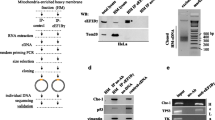
The onco-suppressor p53 protein plays also an important role in the control of various aspects of health and disease. p53 levels are low in normal cells and elevated under stress conditions. While low levels of p53 promote tumor formation, overactive p53 leads to premature aging and cell death. RNA degradation is a critical level of regulation contributing to the control of gene expression. p53, as an RNA-binding protein, exerts 3′ → 5′ exoribonuclease activity, mediating degradation of adenylate/uridylate-rich elements (ARE)–containing ssRNAs. The 3′-UTR of p53-mRNA, which is a target of p53 itself, harbors cis-acting AREs. Our results suggest that p53 controls its own expression through murine double-minute 2 (mdm2)–independent “RNA decay” function in cytoplasm. We demonstrate that p53 expresses an exoribonuclease activity through the binding to ARE sequences of p53-mRNA via translation-independent and translation-dependent polysome-associated pathways. Antagonistic interplay was detected between p53 levels and execution of its exoribonuclease function mirrored in low p53 levels in normal cells, due to the efficient exoribonuclease activity, and in the accumulation of p53 in cells exposed to p53-activating drugs in accordance with the reduced exoribonuclease activity. Apparently, p53, via control of its own mRNA stability and/or translation in cytoplasm, might act as a negative regulator of p53-mRNA levels. The observed connection between exoribonuclease activity and p53 abundance highlights the importance of this function affecting p53 expression, imperative for multiple functions, with implications for the steady-state levels of protein and for the p53 stress response. The modulation in expression of exoribonuclease activity would be translated into the alterations in p53 level.
Key messages
-
p53 controls its own expression through mdm2-independent “RNA decay” function in cytoplasm.
-
p53 expresses an exoribonuclease activity through the binding to ARE sequences of p53-mRNA.
-
Antagonistic interplay exists between stress-induced p53 and execution of its exoribonuclease function.








Similar content being viewed by others
References
Royds JA, Iacopetta B (2006) p53 and disease, when the guardian angel fails. Cell Death Diff. 13:1017–1026
Luo Q, Beaver JM, Liu Y, Zhang Z (2017) Dynamics of p53: a master decider of cell fate. Genes 8:66–82
Vousden KH, Lane DP (2007) p53 in health and disease. Nature Reviews 8:275–283
Aylon Y, Oren M (2016) The paradox of p53: what, how and why? Cold Spring Harb. Perspect. Med. a026328
Zhang G, Xie Y, Zhou Y, Xiang C, Chen L, Zhang C, Hou Z, Chen J, Zong H, Liu G (2017) p53 pathway is involved in cell competition during mouse embryogenesis. Proc Natl Acad Sci U S A 114:498–503
Wu D, Prives C (2018) Relevance of the p53-MDM-2 axis to aging. Cell Death Diff 25:169–179
Comel A, Sorrentino G, Capaci V, Del Sal G (2014) The cytoplasmic side of p53s onco-suppressive activities. FEBS Lett 588:2600–2609
Haupt Y, Maya R, Kazaz A, Oren M (1997) Mdm2 promotes the rapid degradation of p53. Nature 387:296–299
Marine JC, Lozano G (2010) Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Diff. 17:93–102
Pant V, Lozano G (2014) Dissecting the p53-Mdm2 feedback loop in vivo: uncoupling the role in p53 stability and activity. Oncotarget 5:1149–1156
Ringhausen I, O’Shea CC, Finch AJ, Swigart LB, Evan GI (2006) Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell 10:501–514
Montes de Oca Luna R, Wagner DS, Lozano G (1995) Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203–206
Lu X (2010) Tied up in loops: positive and negative autoregulation of p53. Cold Spring Harb. Perspect Biol 2(5)
Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M (2006) A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev 20:2687–2700
Zhou X, Cao B, Lu H (2017) Negative auto-regulators trap p53 in their web. J Mol Cell Biol 9:62–68
Kim H, You S, Foster LK, Farris J, Foster DN (2001) The rapid destabilization of p53 mRNA in immortal chicken embryo fibroblast cells. Oncogene 20:5118–5123
Takagi M, Absalon MJ, McLure KG, Kastan MB (2005) Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123:49–63
Mosner J, Mummenbrauer T, Bauer C, Sczakiel G, Grosse F, Deppert W (1995) Negative feedback regulation of wild-type p53 biosynthesis. EMBO J 14:4442–4449
Garcia-Maurino SM, Rivero-Rodríguez F, Velázquez-Cruz A, Hernández-Vellisc M, Díaz-Quintana A, De la Rosa MA (2017) Díaz-Moreno I. RNA binding protein regulation and cross-talk in the control of AU-rich mRNA fate. Front Mol Biosci 4:71
Pullmann R, Kim HH, Abdelmohsen K, Lal A, Martindale J.L, Yang X, Gorospe M. (2007) Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. MolCell Biol 27: 6265–6278
Balagopal V, Fluch L, Nissan T (2012) Ways and means of eukaryotic mRNA decay. Biochim Biophys Acta 1819:593–603
Schoenberg DR, Maquat LE (2012) Regulation of cytoplasmic mRNA decay. Nat Rev Genet 13:246–259
Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M (2004) Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J 23:3092–3102
Haronikova L, Olivares-Ilana V, Wang L, Karakostis K, Chen S, Fahraeus R (2019) The p53 mRNA: and integral part of the cellular stress response. Nucl Acids Res 47:3257–3271
Mazan-Mamczarz K, Galbán S, López de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M (2003) RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci U S A 100:8354–8359
Galban S, Martindale JL, Mazan-Mamczarz K, López de Silanes I, Fan J, Wang W, Decker J, Gorospe M (2003) Influence of the RNA-binding protein HuR in Pvhl-regulated p53 expression in renal carcinoma. Mol Cell Biol 23:7083–7095
Ibrahim H, Wilusz J, Wilusz CJ (2008) RNA recognition by 3′-to-5′ exonucleases: the substrate perspective. Biochim Biophys Acta 1779:256–265
Bakhanashvili M, Gedelovich R, Grinberg S, Rahav G (2008) Exonucleolytic degradation of RNA by the tumor suppression protein p53 in cytoplasm. J Mol Med 86:75–88
Derech-Haim S, Teiblum S, Kadosh R, Rahav G, Bonda E, Sredni B, Bakhanashvili M (2012) Ribonuclease activity of p53 in cytoplasm in response to various stress signals. Cell Cycle 11:1400–1413
Janus F, Albrechtsen N, Knippschild U, Wiesmuller L, Grosse F, Deppert W (1999) Different regulation of the p53 core domain activities 3′-to5′ exonuclease and sequence-specific DNA binding. Mol Cell Biol 19:2155–2168
Riley KJL, Maher LJ 3rd (2007) p53-RNA interactions: new clues in an old mystery. RNA 13:1825–1833
Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA (2004) In vivo activation of the p53 pathway by small molecule antagonists of mdm2. Science 303:844–848
Cox ML, Meek DW (2010) Phosphorylation of serine 392 in p53 is a common and integral event during p53 induction by diverse stimuli. Cell Signal 22:564–571
Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Yoshida T, Itoh H, Kohno K (2004) Binding of RNA to p53 regulates its oligomerization and DNA binding activity. Oncogene 23:4371–4379
Bai J, Cederbaum AI (2006) Cycloheximide protects HepG2 cells from serum withdrawal- induced apoptosis by decreasing p53 and phosphorylated p53 levels. J Pharmacol Exp Ther 319:1435–1443
Lopez I, Tournillon AS, Nylander K, Fahraeus R (2015) p53 mediated control gene expression via mRNA translation during endoplasmic reticulum stress. Cell Cycle 14:3373–3378
Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J (2009) Co-translational mRNA decay in saccharomyces cerevisiae. Nature 46:225–229
Giaccia AJ, Kastan MB (1998) The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev 12:2973–2983
Vilborg A, Glahder JA, Wilhelm MT, Bersani C, Corcoran M, Mahmoudi S, Rosenstierne M, Grandér D, Farnebo M, Norrild B, Wiman KG (2009) The p53 target Wig-1 regulates p53 mRNA stability through an AU-rich element. Proc Natl Acad Sci U S A 106:15756–15761
Vilborg A, Wilhelm MT, Wiman KG (2010) Regulation of tumor suppressor p53 at the RNA level. J Mol Med 88:645–652
Zhang T, Kruys V, Huez G, Gueydan C (2002) AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem Soc Trans 30:952–958
Wang H, Ding N, Guo J, Xia J, Ruan Y (2016) Dysregulation of TTP and HuR plays an important role in cancers. Tumour Biol 37:14451–14461
Haimovich G, Choder M, Singer H, Trcek T (2013) The fate of the messenger is pre-determined: a new model for regulation of gene expression. Bioch Bioph Acta 1829:643–653
Ahuja D, Goyal A, Ray PS (2016) Interplay between RNA-binding protein HuR and microRNA-125b regulates p53 mRNA translation in response to genotoxic stress. RNA Biol 13:1152–1165
Yin X, Fontoura BM, Morimoto T, Carroll RB (2003) Cytoplasmic complex of p53 and eEF2. J Cell Physiol 196:474–482
Fontoura BM, Atienza CA, Sorokina EA, Morimoto T, Carroll RB (1997) Cytoplasmic p53 polypeptide is associated with ribosomes. Mol Cell Biol 17:3146–3154
Fontoura BM, Sorokina EA, David E, Carroll RB (1992) p53 is covalently linked to 5.8S rRNA. Mol Cell Biol 12:5145–5151
Marcel V, Catez F, Diaz JJ (2015) p53, a translational regulator: contribution to its tumor- suppressor activity. Oncogene 34:5513–5523
Marcel V, Van Long FN, Diaz JJ (2018) 40 years of research put p53 in translation. Cancers 10:152
Abdelmohsen K, Panda AC, Kang MJ, Guo R, Kim J, Grammatikakis I, Yoon JH, Dudekula DB, Noh JH, Yang X et al (2014) 7SL represses p53 translation by competing with HuR. Nucleic Acids Res 42:10099–10111
Acknowledgments
MDA-MB-468 cells were provided by Prof. R. Yerushalmi from Rabin Medical Center, Davidoff Cancer Center, Petach Tiqva, Israel. HCT116 isogenic cells were provided by Prof. M. Oren from Weizmann Institute, Rechovot, Israel.
Author information
Authors and Affiliations
Contributions
S.D. and Y.F. performed the experiments; A.H. reviewed the results and thoroughly edited the manuscript; M.B. designed the study and wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Derech-Haim, S., Friedman, Y., Hizi, A. et al. p53 regulates its own expression by an intrinsic exoribonuclease activity through AU-rich elements. J Mol Med 98, 437–449 (2020). https://doi.org/10.1007/s00109-020-01884-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-020-01884-0