Abstract
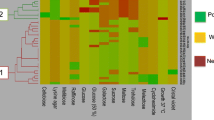
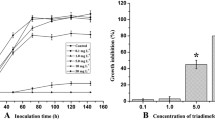
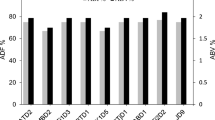
Each brewing yeast has its own unique impact on the formation of aroma compounds, and thus, on the properties of the final beer. The selection of the perfect strain for a specific brewing process results from physiological properties, which can be elucidated in brewing experiments. These properties result from genetic and proteomic features of each yeast strain. In the current study, 23 blind-coded yeasts were analyzed on a genomic level by microsatellite genotyping at 13 loci, on a sub-proteome level by MALDI-TOF MS, and on their phenotypic property by phenolic off flavor (POF) production assessment. These results were compared with the current application profile of each yeast strain. An expanded MALDI-TOF MS database was used to identify the blind-coded samples on species level, which was achieved to 100%. The samples belonged to top-fermenting Saccharomyces (S.) cerevisiae, bottom-fermenting S. pastorianus and S. cerevisiae var. diastaticus. Different groupings below species level were found with microsatellite analyses (classification on strain level) and MALDI sub-proteome (subdivision of yeasts into groups), which provided a prediction of application potential to beer styles for which they are currently used. The test for POF showed a wide variation and appears to be a strain-dependent property. However, this could serve as a starting point for the classification of yeast strains with respect to their usefulness for the production of specific beer styles or non-brewing applications.



Similar content being viewed by others
References
Barnett JA (1992) The taxonomy of the genus Saccharomyces meyen ex reess: A short review for non-taxonomists. Yeast 8(1):1–23. https://doi.org/10.1002/yea.320080102
Verstrepen KJ, Derdelinckx G, Verachtert H, Delvaux FR (2003) Yeast flocculation: what brewers should know. Appl Microbiol Biotechnol 61(3):197–205. https://doi.org/10.1007/s00253-002-1200-8
Donalies UEB, Nguyen HTT, Stahl U, Nevoigt E (2008) Improvement of Saccharomyces yeast strains used in brewing, wine making and baking. In: Stahl U, Donalies UEB, Nevoigt E (eds) Food Biotechnology. Springer, Berlin, pp 67–98 https://doi.org/10.1007/10_2008_099
Yoshida S, Imoto J, Minato T, Oouchi R, Sugihara M, Imai T, Ishiguro T, Mizutani S, Tomita M, Soga T (2008) Development of bottom-fermenting Saccharomyces strains that produce high SO2 levels, using integrated metabolome and transcriptome analysis. Appl Environ Microbiol 74(9):2787–2796
Nakao Y, Kanamori T, Itoh T, Kodama Y, Rainieri S, Nakamura N, Shimonaga T, Hattori M, Ashikari T (2009) Genome sequence of the lager brewing yeast, an interspecies hybrid. DNA Res 16(2):115–129. https://doi.org/10.1093/dnares/dsp003
Hansen J, Bruun SV, Bech LM, Gjermansen C (2002) The level ofMXR1gene expression in brewing yeast during beer fermentation is a major determinant for the concentration of dimethyl sulfide in beer. FEMS yeast research 2(2):137–149. https://doi.org/10.1111/j.1567-1364.2002.tb00078.x
Bokulich NA, Bamforth CW (2013) The microbiology of malting and brewing. Microbiol Mol Biol Rev 77(2):157–172. https://doi.org/10.1128/MMBR.00060-12
Pires EJ, Teixeira JA, Branyik T, Vicente AA (2014) Yeast: the soul of beer’s aroma–a review of flavour-active esters and higher alcohols produced by the brewing yeast. Appl Microbiol Biotechnol 98(5):1937–1949. https://doi.org/10.1007/s00253-013-5470-0
Coghe S, Benoot K, Delvaux F, Vanderhaegen B, Delvaux FR (2004) Ferulic acid release and 4-vinylguaiacol formation during brewing and fermentation: indications for feruloyl esterase activity in Saccharomyces cerevisiae. J Agric Food Chem 52(3):602–608. https://doi.org/10.1021/jf0346556
Mukai N, Masaki K, Fujii T, Iefuji H (2014) Single nucleotide polymorphisms of PAD1 and FDC1 show a positive relationship with ferulic acid decarboxylation ability among industrial yeasts used in alcoholic beverage production. J Biosci Bioeng 118(1):50–55. https://doi.org/10.1016/j.jbiosc.2013.12.017
Schneiderbanger H, Koob J, Poltinger S, Jacob F, Hutzler M (2016) Gene expression in wheat beer yeast strains and the synthesis of acetate esters. J Inst Brew 122(3):403–411
Goncalves M, Pontes A, Almeida P, Barbosa R, Serra M, Libkind D, Hutzler M, Goncalves P, Sampaio JP (2016) Distinct domestication trajectories in top-fermenting beer yeasts and wine yeasts. Curr Biol 26(20):2750–2761. https://doi.org/10.1016/j.cub.2016.08.040
Focke K, Jentsch M (2013) Kleine Zelle—große Wirkung—Hefestämme, Hefelagerung und der Einfluss auf den Biercharakter. Brauindustrie 11:16–18
White C, Zainasheff J (2010) Yeast: the practical guide to beer fermentation. Brewers Publications, Boulder
Dornbusch HD (2010) The Ultimate almanac of world beer recipes: a practical guide for the professional brewer to the world’s classic beer styles from A to Z. Cerevisia Communications, Bamberg, Germany.
Meier-Dörnberg T, Michel M, Wager RS, Jacob F, Hutzler M (2017) Genetic and phenotypic characterization of different top-fermenting Saccharomyces cerevisiae Ale Yeast Isolates. Brew Sci 70:9–25
Gonzalez SS, Barrio E, Gafner J, Querol A (2006) Natural hybrids from Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces kudriavzevii in wine fermentations. FEMS Yeast Res 6(8):1221–1234. https://doi.org/10.1111/j.1567-1364.2006.00126.x
Gallone B, Steensels J, Prahl T, Soriaga L, Saels V, Herrera-Malaver B, Merlevede A, Roncoroni M, Voordeckers K, Miraglia L, Teiling C, Steffy B, Taylor M, Schwartz A, Richardson T, White C, Baele G, Maere S, Verstrepen KJ (2016) Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 166(6):1397–1410 e1316. https://doi.org/10.1016/j.cell.2016.08.020
Gibson BR, Storgards E, Krogerus K, Vidgren V (2013) Comparative physiology and fermentation performance of Saaz and Frohberg lager yeast strains and the parental species Saccharomyces eubayanus. Yeast 30(7):255–266. https://doi.org/10.1002/yea.2960
Legras JL, Merdinoglu D, Cornuet JM, Karst F (2007) Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol 16(10):2091–2102. https://doi.org/10.1111/j.1365-294X.2007.03266.x
Legras JL, Erny C, Charpentier C (2014) Population structure and comparative genome hybridization of European flor yeast reveal a unique group of Saccharomyces cerevisiae strains with few gene duplications in their genome. PloS one 9(10):e108089. https://doi.org/10.1371/journal.pone.0108089
Masneuf-Pomarede I, Salin F, Borlin M, Coton E, Coton M, Jeune CL, Legras JL (2016) Microsatellite analysis of Saccharomyces uvarum diversity. FEMS Yeast Res 16(2):fow002. https://doi.org/10.1093/femsyr/fow002
Couto MB, Eijsma B, Hofstra H, van der Vossen J (1996) Evaluation of molecular typing techniques to assign genetic diversity among Saccharomyces cerevisiae strains. Appl Environ Microbio 62(1):41–46
Laidlaw L, Tompkins T, Savard L, Dowhanick T (1996) Identification and differentiation of brewing yeasts using specific and RAPD polymerase chain reaction. J Am Soc Brew Chem 54(2):97–102
Krogerus K, Magalhaes F, Vidgren V, Gibson B (2015) New lager yeast strains generated by interspecific hybridization. J Ind Microbiol Biotechnol 42(5):769–778. https://doi.org/10.1007/s10295-015-1597-6
Sheehan CA, Weiss AS, Newsom IA, Flint V, O’Donnell DC (1991) Brewing yeast identification and chromosome analysis using high resolution CHEF gel electrophoresis. J Inst Brew 97(3):163–167
Timmins EM, Quain DE, Goodacre R (1998) Differentiation of brewing yeast strains by pyrolysis mass spectrometry and Fourier transform infrared spectroscopy. Yeast 14 (10):885–893. (10.1002/(SICI)1097-0061(199807)14:10<885::AID-YEA286>3.0.CO;2-G)
Azumi M, Goto-Yamamoto N (2001) AFLP analysis of type strains and laboratory and industrial strains of Saccharomyces sensu stricto and its application to phenetic clustering. Yeast 18(12):1145–1154. https://doi.org/10.1002/yea.767
de Barros Lopes M, Rainieri S, Henschke PA, Langridge P (1999) AFLP fingerprinting for analysis of yeast genetic variation. Int J Syst Bacteriol 2(2):915–924. https://doi.org/10.1099/00207713-49-2-915
Cappello MS, Bleve G, Grieco F, Dellaglio F, Zacheo G (2004) Characterization of Saccharomyces cerevisiae strains isolated from must of grape grown in experimental vineyard. J Appl Microbiol 97(6):1274–1280. https://doi.org/10.1111/j.1365-2672.2004.02412.x
Gonzalez Flores M, Rodriguez ME, Oteiza JM, Barbagelata RJ, Lopes CA (2017) Physiological characterization of Saccharomyces uvarum and Saccharomyces eubayanus from Patagonia and their potential for cidermaking. Int J Food Microbiol 249:9–17. https://doi.org/10.1016/j.ijfoodmicro.2017.02.018
McMurrough I, Madigan D, Donnelly D, Hurley J, Doyle AM, Hennigan G, McNulty N, Smyth MR (1996) Control of ferulic acid and 4-vinyl guaiacol in brewing. J Inst Brew 102(5):327–332
Vanbeneden N, Gils F, Delvaux F, Delvaux FR (2008) Formation of 4-vinyl and 4-ethyl derivatives from hydroxycinnamic acids: Occurrence of volatile phenolic flavour compounds in beer and distribution of Pad1-activity among brewing yeasts. Food Chem 107(1):221–230. https://doi.org/10.1016/j.foodchem.2007.08.008
Mertens S, Steensels J, Gallone B, Souffriau B, Malcorps P, Verstrepen K (2017) Rapid screening method for phenolic off-flavor (POF) production in yeast. https://doi.org/10.1094/ASBCJ-2017-4142-01
Joubert R, Brignon P, Lehmann C, Monribot C, Gendre F, Boucherie H (2000) Two-dimensional gel analysis of the proteome of lager brewing yeasts. Yeast 16 (6):511–522. (10.1002/(SICI)1097-0061(200004)16:6<511::AID-YEA544>3.0.CO;2-I)
Trabalzini L, Paffetti A, Scaloni A, Talamo F, Ferro E, Coratza G, Bovalini L, Lusini P, Martelli P, Santucci A (2003) Proteomic response to physiological fermentation stresses in a wild-type wine strain of Saccharomyces cerevisiae. Biochem J 370(Pt 1):35–46. https://doi.org/10.1042/BJ20020140
Kobi D, Zugmeyer S, Potier S, Jaquet-Gutfreund L (2004) Two-dimensional protein map of an “ale"-brewing yeast strain: proteome dynamics during fermentation. FEMS Yeast Res 5(3):213–230. https://doi.org/10.1016/j.femsyr.2004.07.004
Zuzuarregui A, Monteoliva L, Gil C, del Olmo M (2006) Transcriptomic and proteomic approach for understanding the molecular basis of adaptation of Saccharomyces cerevisiae to wine fermentation. Appl Environ Microbiol 72(1):836–847. https://doi.org/10.1128/AEM.72.1.836-847.2006
Hansen R, Pearson SY, Brosnan JM, Meaden PG, Jamieson DJ (2006) Proteomic analysis of a distilling strain of Saccharomyces cerevisiae during industrial grain fermentation. Appl Microbiol Biotechnol 72(1):116–125. https://doi.org/10.1007/s00253-006-0508-1
Kern CC, Vogel RF, Behr J (2014) Differentiation of lactobacillus brevis strains using matrix-assisted-laser-desorption-ionization-time-of-flight mass spectrometry with respect to their beer spoilage potential. Food Microbiol 40:18–24
Wieme AD, Spitaels F, Aerts M, De Bruyne K, Van Landschoot A, Vandamme P (2014) Identification of beer-spoilage bacteria using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Int J Food Microbiol 185:41–50. https://doi.org/10.1016/j.ijfoodmicro.2014.05.003
Guo L, Ye L, Zhao Q, Ma Y, Yang J, Luo Y (2014) Comparative study of MALDI-TOF MS and VITEK 2 in bacteria identification. J Thorac Dis 6(5):534–538. https://doi.org/10.3978/j.issn.2072-1439.2014.02.18
Demirev P, Sandrin TR (2016) Applications of Mass Spectrometry in Microbiology. Springer, Heidelberg
Croxatto A, Prod’hom G, Greub G (2012) Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol Rev 36(2):380–407. https://doi.org/10.1111/j.1574-6976.2011.00298.x
Kern CC, Vogel RF, Behr J (2014) Identification and differentiation of brewery isolates of Pectinatus sp. by matrix-assisted-laser desorption–ionization time-of-flight mass spectrometry (MALDI-TOF MS). Eur Food Res Technol 238(5):875–880. https://doi.org/10.1007/s00217-014-2173-4
Nacef M, Chevalier M, Chollet S, Drider D, Flahaut C (2017) MALDI-TOF mass spectrometry for the identification of lactic acid bacteria isolated from a French cheese: The Maroilles. Int J Food Microbiol 247:2–8. https://doi.org/10.1016/j.ijfoodmicro.2016.07.005
Pavlovic M, Mewes A, Maggipinto M, Schmidt W, Messelhausser U, Balsliemke J, Hormansdorfer S, Busch U, Huber I (2014) MALDI-TOF MS based identification of food-borne yeast isolates. J Microbiol Methods 106:123–128. https://doi.org/10.1016/j.mimet.2014.08.021
Moothoo-Padayachie A, Kandappa HR, Krishna SBN, Maier T, Govender P (2013) Biotyping Saccharomyces cerevisiae strains using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). Eur Food Res Technol 236(2):351–364. https://doi.org/10.1007/s00217-012-1898-1
Blattel V, Petri A, Rabenstein A, Kuever J, Konig H (2013) Differentiation of species of the genus Saccharomyces using biomolecular fingerprinting methods. Appl Microbiol Biotechnol 97(10):4597–4606. https://doi.org/10.1007/s00253-013-4823-z
Gutierrez C, Gomez-Flechoso MA, Belda I, Ruiz J, Kayali N, Polo L, Santos A (2017) Wine yeasts identification by MALDI-TOF MS: Optimization of the preanalytical steps and development of an extensible open-source platform for processing and analysis of an in-house MS database. Int J Food Microbiol 254:1–10. https://doi.org/10.1016/j.ijfoodmicro.2017.05.003
Usbeck JC, Wilde C, Bertrand D, Behr J, Vogel RF (2014) Wine yeast typing by MALDI-TOF MS. Appl Microbiol Biotechnol 98(8):3737–3752. https://doi.org/10.1007/s00253-014-5586-x
Lauterbach A, Usbeck JC, Behr J, Vogel RF (2017) MALDI-TOF MS typing enables the classification of brewing yeasts of the genus Saccharomyces to major beer styles. PloS one 12(8):e0181694. https://doi.org/10.1371/journal.pone.0181694
Legras JL, Ruh O, Merdinoglu D, Karst F (2005) Selection of hypervariable microsatellite loci for the characterization of Saccharomyces cerevisiae strains. Int J Food Microbiol 102(1):73–83. https://doi.org/10.1016/j.ijfoodmicro.2004.12.007
Perez M, Gallego F, Martinez I, Hidalgo P (2001) Detection, distribution and selection of microsatellites (SSRs) in the genome of the yeast Saccharomyces cerevisiae as molecular markers. Lett Appl Microbiol 33(6):461–466
Cavalli-Sforza LL, Edwards AW (1967) Phylogenetic analysis: models and estimation procedures. Evolution 21(3):550–570
Yamauchi H, Yamamoto H, Shibano Y, Amaya N, Saeki T (1998) Rapid methods for detecting Saccharomyces diastaticus, a beer spoilage yeast, using the polymerase chain reaction. J Am Soc Brew Chem 56(2):58–63
Bayly JC, Douglas LM, Pretorius IS, Bauer FF, Dranginis AM (2005) Characteristics of Flo11-dependent flocculation in Saccharomyces cerevisiae. FEMS Yeast Res 5(12):1151–1156. https://doi.org/10.1016/j.femsyr.2005.05.004
Michel M, Kopecká J, Meier-Dörnberg T, Zarnkow M, Jacob F, Hutzler M (2016) Screening for new brewing yeasts in the non-Saccharomyces sector with Torulaspora delbrueckii as model. Yeast 33:129–144
Usbeck JC, Kern CC, Vogel RF, Behr J (2013) Optimization of experimental and modelling parameters for the differentiation of beverage spoiling yeasts by matrix-assisted-laser-desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS) in response to varying growth conditions. Food Microbiol 36(2):379–387. https://doi.org/10.1016/j.fm.2013.07.004
Jombart T, Collins C (2015) A tutorial for discriminant analysis of principal components (DAPC) using adegenet 2.0.0. Imperial College London, MRC Centre for Outbreak Analysis and Modelling, London
Jespersen L, Jakobsen M (1996) Specific spoilage organisms in breweries and laboratory media for their detection. Int J Food Microbiol 33(1):139–155. https://doi.org/10.1016/0168-1605(96)01154-3
Heresztyn T (1986) Metabolism of volatile phenolic compounds from hydroxycinnamic acids by Brettanomyces yeast. Arch Microbiol 146(1):96–98
Mukai N, Masaki K, Fujii T, Kawamukai M, Iefuji H (2010) PAD1 and FDC1 are essential for the decarboxylation of phenylacrylic acids in Saccharomyces cerevisiae. J Biosci Bioeng 109(6):564–569. https://doi.org/10.1016/j.jbiosc.2009.11.011
Chen P, Dong J, Yin H, Bao X, Chen L, He Y, Wan X, Chen R, Zhao Y, Hou X (2015) Single nucleotide polymorphisms and transcription analysis of genes involved in ferulic acid decarboxylation among different beer yeasts. J Inst Brew 121(4):481–489
Li YC, Korol AB, Fahima T, Beiles A, Nevo E (2002) Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol 11(12):2453–2465. https://doi.org/10.1046/j.1365-294X.2002.01643.x
Marklein G, Josten M, Klanke U, Muller E, Horre R, Maier T, Wenzel T, Kostrzewa M, Bierbaum G, Hoerauf A, Sahl HG (2009) Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J Clin Microbiol 47(9):2912–2917. https://doi.org/10.1128/JCM.00389-09
Usbeck JC, Behr J (2012) Vogel RF Differentiation of top-and bottom-fermenting brewing yeasts and insight into their metabolic status by MALDI-TOF MS, In: Poster World Brewing Congress, Portland
Schurr BC, Behr J, Vogel RF (2015) Detection of acid and hop shock induced responses in beer spoiling Lactobacillus brevis by MALDI-TOF MS. Food Microbiol 46:501–506. https://doi.org/10.1016/j.fm.2014.09.018
Christner M, Trusch M, Rohde H, Kwiatkowski M, Schluter H, Wolters M, Aepfelbacher M, Hentschke M (2014) Rapid MALDI-TOF mass spectrometry strain typing during a large outbreak of Shiga-Toxigenic Escherichia coli. PloS one 9(7):e101924. https://doi.org/10.1371/journal.pone.0101924
Pham T, Wimalasena T, Box W, Koivuranta K, Storgårds E, Smart K, Gibson B (2011) Evaluation of ITS PCR and RFLP for differentiation and identification of brewing yeast and brewery ‘wild’ yeast contaminants. J Inst Brew 117(4):556–568
Walther A, Hesselbart A, Wendland J (2014) Genome sequence of Saccharomyces carlsbergensis, the world’s first pure culture lager yeast. Genes Genomes Genet 4(5):783–793 https://doi.org/10.1534/g3.113.010090
Spencer JF, Spencer DM (1983) Genetic improvement of industrial yeasts. Annu Rev Microbiol 37(1):121–142. https://doi.org/10.1146/annurev.mi.37.100183.001005
Meier-Dörnberg T, Jacob F, Michel M, Hutzler M (2017) Incidence of Saccharomyces cerevisiae var. diastaticus in the beverage industry: cases of contamination, 2008–2017. Master Brew Assoc Am 54(4):140–148. https://doi.org/10.1094/TQ-54-4-1130-01
Acknowledgements
Part of this work was supported by the German Ministry of Economics and Technology and the Wifö (Wissenschaftsförderung der Deutschen Brauwirtschaft e.V., Berlin, Germany) in project AiF 17698 N. We would like to thank our technical assistant Sabine Forster, and Dr Tobias Fischborn (Lallemand Inc.) for their support with the experiment, as well as Dr Damien Biot-Pelletier (Lallemand Inc.) and Viktor Eckel (Technische Mikrobiologie Weihenstephan) for critical reading of the manuscript. We are also grateful for the information about the origin of the new yeast strains for the MALDI-TOF MS library expansion from Dr. Mathias Hutzler and Tim Meier-Dörnberg.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Compliance with Ethics requirements
This communication does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lauterbach, A., Wilde, C., Bertrand, D. et al. Rating of the industrial application potential of yeast strains by molecular characterization. Eur Food Res Technol 244, 1759–1772 (2018). https://doi.org/10.1007/s00217-018-3088-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-018-3088-2