Abstract
Inherited retinal dystrophies, such as Leber congenital amaurosis, Stargardt disease, and retinitis pigmentosa, are characterized by photoreceptor dysfunction and death and currently have few treatment options. Recent technological advances in induced pluripotent stem cell (iPSC) technology and differentiation methods mean that human photoreceptors can now be studied in vitro. For example, retinal organoids provide a platform to study the development of the human retina and mechanisms of diseases in the dish, as well as being a potential source for cell transplantation. Here, we describe differentiation protocols for 3D cultures that produce retinal organoids containing photoreceptors with rudimentary outer segments. These protocols can be used as a model to understand retinal disease mechanisms and test potential therapies, including antisense oligonucleotides (AONs) to alter gene expression or RNA processing. This “retina in a dish” model is well suited for use with AONs, as the organoids recapitulate patient mutations in the correct genomic and cellular context, to test potential efficacy and examine off-target effects on the translational path to the clinic.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Keywords
- Retinal organoids
- Induced pluripotent stem cells
- Differentiation
- 3D culture
- Retinal degeneration
- Photoreceptor
- Retina in a dish
1 Introduction
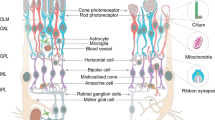
The dysfunction and death of photoreceptor cells are associated with inherited retinal diseases (IRDs), which are a major cause of blindness. The lack of effective treatment to prevent loss of photoreceptors means these diseases are currently irreversible. Recent progress in the differentiation of stem cells to retinal cells has enabled the generation of functional retinal organoids in vitro or a “retina in a dish” [1,2,3,4]. By recapitulating the retina from patient-derived induced pluripotent stem cells (iPSC ), retinal organoids offer a platform for developing therapeutic treatments and modeling patient disease [5, 6].
A dynamic and complex microenvironment is involved in eye development, including direct and indirect cell–cell interaction and specific signaling regulation in different stages of development [7]. Because of this complex microenvironment, retinal organoids have the potential to develop a more mature retina than photoreceptors differentiated in 2D conditions only. Several studies have shown that with defined culture conditions, embryonic stem cells (ESC), and iPSC can be differentiated into retinal organoids in a 3D environment, producing a laminated retina that mimics the in vivo human retina [2, 3, 8]. In addition to recapitulating the structure of native eye development, rudimentary disorganized outer segments can be observed in photoreceptors from retinal organoids.
In this chapter, we describe three different methods to differentiate iPSC to retinal organoids in 3D. Retinal organoids generated from these protocols are well laminated with photoreceptors in their outer layer and develop rudimentary outer segments. Importantly, they also recapitulate photoreceptor mRNA processing and the exquisite pattern of alternative splicing they present [9,10,11]. This makes retinal organoids ideal for studying aberrant splicing events associated with patient variants in several forms of IRDs [9, 12]. Furthermore, they can then be applied to the development of antisense oligonucleotides (AONs) as potential treatments [9, 13].
2 Materials
2.1 General Materials
-
1.
U-bottom ultra-low 96-well plate.
-
2.
25-well plate low attachment.
-
3.
6-well plate.
-
4.
Ultra-low adhesion 6-well plate.
-
5.
Crescent knife.
-
6.
Retinoic acid (RA).
2.2 iPSC Culture
-
1.
Essential 8 Flex medium.
-
2.
Geltrex.
-
3.
Cell dissociation buffer (ThermoFisher).
2.3 EB Suspension Protocol
-
1.
EB2 base medium: GMEM, 20% Knock-Out Serum, 1% Sodium pyruvate, 1% NEAA, and 110 μM 2-Mercaptoethanol (see Note 1).
-
2.
NR media (NRM): DMEM/F12, 1% N2 Supplement, 10% FBS, and 1% NEAA (see Note 1).
-
3.
V-bottom 96-well plate.
-
4.
IWR-1e (Wnt inhibitor).
-
5.
Rock inhibitor (Y-27632).
-
6.
Matrigel (growth factor reduced).
-
7.
Hedgehog smoothened agonist (SAG).
-
8.
TrypLE.
2.4 EB Adherent Protocol
-
1.
Neural induction medium (NIM): DMEM/F-12 (1:1), 1% N2 supplement, 1% NEAA, and Heparin 2 μg/ml (see Note 1).
-
2.
Retinal Differentiation Medium (RDM): DMEM/F12 (3:1), 2% B27 (without Vitamin A), 1% NEAA, and 1% Pen/Step (see Note 1).
-
3.
Neural Retina Maturation Medium 1 (RMM1): DMEM/F12 (3:1), 2% B27 (without vitamin A), 1% NEAA, 1% Pen/Strep, 10% FBS, 100 μM Taurine, and 1% Glutamax (see Note 1).
-
4.
Neural Retina Maturation Medium 2 (RMM2): DMEM/F12 (3:1), 1% N2, 1% NEAA, 1% Pen/Strep, 10% FBS, 100 μM Taurine, and 1% Glutamax (see Note 1).
-
5.
Blebbistatin.
2.5 Non-EB Adherent Protocol
-
1.
Essential 6 medium (ThermoFisher).
-
2.
Neural induction Medium (NIM): Advanced DMEM/F12, 1% N2 supplement, 1% NEAA, 1% Glutamax, and 1% Pen/Strep (see Note 1).
-
3.
Retinal Differentiation Media (RDM): DMEM/F12 (3:1), 1% Pen/Strep, 1% NEAA, and 2% B27 (see Note 1).
-
4.
Neural Retina Maturation Medium 1 (RMM1): DMEM/F12 (3:1), 1% Pen/Strep, 2% B27, 10% FBS, 100 μM Taurine, 1% NEAA, and 1% Glutamax (see Note 1).
-
5.
Neural Retina Maturation Medium 2 (RMM2): DMEM/F12 (3:1), 1% Pen/Strep, 2% B27 (without vitamin A), 1% N2, 10% FBS, 100 μM Taurine, 1% NEAA, and 1% Glutamax (see Note 1).
2.6 RNA Extraction
-
1.
RNA mini kit.
-
2.
PBS.
-
3.
Micropestle.
3 Methods
iPSC are maintained with Essential 8 Flex (E8F) in Geltrex coated 6-well plates (see Note 2). Once they reach 70% confluence, iPSC are treated with 500 μl cell dissociation buffer for 2 min at 37 °C in the incubator. After the incubation, remove the cell dissociation buffer and add 1 ml of E8F into a well. Use 1 ml tip scraping the well to collect iPSC in small clumps. Cell clumps are collected and transferred into a new Geltrex-coated plate with 1 ml tip. Medium is changed every other day, and iPSC can be double-fed with 4 ml E8F to cover the weekend (see Note 3).
3.1 EB Suspension Protocol
This protocol is adapted from the method initially described by Sasai and colleagues [3].
-
1.
Maintain iPSC in a 6-well plate as described earlier. Use 1 ml TrypLE to disperse cells into single cells. Collect the cell pellet after centrifugation at 300 × g for 5 min and resuspend in 2 ml E8F with 10 μM Rock inhibitor (ROCKi).
-
2.
Place 10,000 cells per well with 100 μl E8F with ROCKi in a V-bottom low attachment 96-well plate.
-
3.
The next day, add 100 μl E8F with ROCKi (Day 1).
-
4.
Change half medium, 100 μl, with EB2 with 10 μM ROCKi, 3 μM IWRe-1, and 2% Matrigel twice a week until Day 12.
-
5.
Change half medium, 100 μl, with EB2 with 3 μM IWRe-1, 2% Matrigel, 10% FBS, and 100 nM SAG twice a week until Day 18.
-
6.
Transfer cells into U-bottom ultra-low attachment 96-well plates, and the medium is switched to NRM supplemented with 0.5 μM RA from Day 20 until Day 100. Change medium three times a week (Fig. 1) (see Note 4).
-
7.
From Day 100, select and maintain laminated organoids in 25-well plates in NRM with no RA till collection day (Fig. 1) (see Note 5).
EB suspension protocol. Top row: Schematic diagram of EB suspension protocol steps and media. Lower row: Representative organoids at different stages of differentiation are shown. Visible lamination can be observed at approximately Day 35 and good organoids can maintain the lamination and mature during differentiation to form an outer nuclear layer of photoreceptors with inner segment and outer segment (which can be seen by the “brush border,” arrowhead). Scale bar is 250 μm. The cartoon images are made with BioRender
3.2 EB Adherent Protocol
This protocol is adapted from the method initially described by Canto-Soler and colleagues [2].
-
1.
Collect the iPSC clusters, as described earlier, from three confluent wells (or one T25 flask) with E8F + 10 μM Blebbistatin (see Note 6) and transfer the cell clumps into three wells (2 ml per well) of ultra-low adhesion 6-well plate to form the embryoid bodies (EB).
-
2.
After 24 h (Day 1), use a 10-ml pipette to collect the EB into 15-ml falcon and centrifuge at 110 × g for 2 min. Remove the supernatant and collect the EB with 6 ml medium of 75% E8F + 10 μM Blebbistatin and 25% neural induction medium (NIM). Transfer the EB back to the wells, 2 ml in each well (see Note 7).
-
3.
With the same technique described in step 2, change the medium to 50% E8F + 10 μM Blebbistatin and 50% NIM on Day 2 and 100% NIM on Day 3 and Day 5 (see Note 8).
-
4.
On Day 7, transfer EB from three wells to six wells of Geltrex-coated 6-well plate in NIM, 4 ml in each well. Gently mix the medium in the wells to let the EB equally distributed in the wells (Fig. 2) (see Note 2).
-
5.
Change 4 ml NIM medium twice a week until Day 15.
-
6.
Feed the cells daily with retinal differentiation medium (RDM) from Day 16 until neural retina (NR) domains [9] are formed (Fig. 2).
-
7.
Pick individual NR mechanically with a crescent knife using an inverted brightfield microscope (EvosXL Core) in the safety cabinet in between Day 28 and Day 35 and culture in suspension in U-bottom ultra-low 96-well plates with RDM, one NR per well. Change medium three times a week (see Notes 4 and 11).
-
8.
Switch medium to RMM1 from Day 42. Of note, 1 μM RA is introduced from Day 63.
-
9.
Switch medium to RMM2 with 0.5 μM RA from Day 90 and RMM2 only from Day 100.
-
10.
On Day 100, laminated retinal organoids can be observed with a microscope and are selected and transferred to 25-well plates with 1 ml medium (see Note 5).
-
11.
Maintain retinal organoids in RMM2 until collection day.
EB adherent protocol. Top row: Schematic diagram of EB adherent protocol steps and media. Lower row: Representative images at different stages of differentiation are shown. EB formation in suspension is followed by attachment to Geltrex-coated wells and formation of NR. Picked NR successfully mature through the differentiation form an outer nuclear layer of photoreceptors with inner segment and outer segment (which can be seen by the “brush border,” arrowhead). Scale bar is 250 μm. The cartoon images are made with BioRender
3.3 Non-EB Adherent Protocol
This protocol is adapted from the method initially described by Ali and colleagues [8].
-
1.
iPSC are maintained in 6-well plates as described earlier. E8F is switched to Essential 6 medium for 2 days when the cells reach 90–100% confluence (see Note 9).
-
2.
Introduce 4 ml NIM from Day 3 and change the medium three times a week until the NR are formed (Fig. 3) (see Note 10).
-
3.
Pick NR, as described in EB Adherent Protocol (see Subheading 3.2, step 7), and maintain in U-bottom ultra-low attachment 96-well plates with RDM up to 1 week (see Notes 4 and 11).
-
4.
Switch medium from RDM to RMM1 for 4 weeks.
-
5.
After 4 weeks with RMM1 only, introduce 1 μM RA in RMM1 for 2 weeks.
-
6.
Switch medium to RMM2 with 0.5 μM RA until Day 100.
-
7.
Visually confirmed laminated organoids are transferred to 25-well plates and maintained in RMM2 without B27 until collection day (Fig. 3) (see Note 12).
Non-EB adherent protocol. Top row: Schematic diagram of non-EB adherent protocol steps and media. Lower row: Representative images at different stages of differentiation are shown. NR are formed in NIM medium, and picked NR cultured in suspension going through differentiation mature to form organoids with an outer nuclear layer of photoreceptors with inner segment and outer segment projecting outwards (which can be seen by the “brush border,” arrowhead). Scale bar is 250 μm. The cartoon images are made with BioRender
3.4 AON Treatment of Organoids
-
1.
Mature organoids are generated from protocols described in the above sections.
-
2.
Dilute AONs into working concentration (e.g., 0.1–10 μM) with culture medium, depending on the methods (see Note 13).
-
3.
Remove the medium and treat organoids with media containing AONs (see Note 14).
-
4.
Treat organoids with AONs two times a week with a full change of the medium containing the AON (see Note 15).
-
5.
On the collection day, transfer the organoids into 1.5-ml microcentrifuge tubes with 1 ml PBS individually.
-
6.
Remove PBS and keep the microcentrifuge tube on dry ice for 10 min.
-
7.
RNA can be extracted immediately or the samples can be stored at −80 °C (see Note 16).
3.5 RNA Extraction from Organoids
-
1.
Samples are prepared as described in the previous step.
-
2.
Homogenize organoids individually with micropestle in the microcentrifuge tube.
-
3.
Add lysis buffer from RNA mini kit, in this case from Qiagen, and homogenize organoids again (see Note 17).
-
4.
Follow the instruction of RNA extraction kit to finish RNA extraction and cDNA can then be synthesized.
3.6 Read-out
These methods can be used to produce laminated retinal organoids for the study of RNA processing and morphological changes associated with genomic variants and their potential correction with AONs. The assays used for downstream analyses are dependent on the specific questions being asked. Routine analyses would usually involve RT-PCR and qPCR, but the organoids are also amenable to RNAseq, single-cell sorting, next-generation sequencing, or long-range sequencing. This can provide a unique insight into human photoreceptor splicing and its manipulation for discovery science or therapeutic benefit.
4 Notes
-
1.
General. Once supplemented, the complete medium is stable for up to 2 weeks when stored in the fridge at 4 °C. Freshly made medium can be aliquoted and stored in the freezer at −20 °C for longer storage.
-
2.
Geltrex from stock solution is diluted 50 times in DMEM/F12 medium and 1 ml diluted Geltrex is used to coat a well in a 6-well plate. Plate is coated at 37 °C in the incubator for an hour. EB will attach in the wells from this step.
-
3.
General. Different iPSC clones might have different efficiency of differentiation using these methods. It is recommended to start with at least two of the protocols to test which method is more efficient for that specific clone. The retinal identity and correct lamination of organoids produced by any of the three described methods can be predicted by careful visual inspection under a microscope, but it must be verified by expression of mature retinal markers (e.g., recoverin, cone arrestin, rhodopsin, LM opsin) by immunofluorescence staining and/or gene expression assays.
-
4.
Medium is changed three times a week for 96-well plates and two times a week for 25-well plates.
-
5.
Cut the end of a 1-ml pipette tip off to transfer organoids from 96-well plates to 25-well plates. One retinal organoid per well of 25-well plates. More than one organoid in a well might cause them to merge together.
-
6.
Three wells from a 6-well plate or one T25 flask are optimized conditions we use, but this vary depending on the size of the clumps and confluence of the iPSC . So this step may need to be optimized in each lab.
-
7.
Be gentle while collecting and transferring EB from and into wells. Avoid breaking the EB into single cells.
-
8.
Gamm and colleagues reported that a single dose of BMP4 at day 6 of differentiation, followed by one-half media changes every 3 days until day 16, improved NR production [14].
-
9.
iPSCs need to reach almost 100% confluence, this is crucial for non-EB adherent protocol. Lower density might cause cell death and failure of the protocol. Essential 6 medium is changed daily.
-
10.
For weekend feeding, 6 ml of NIM is used on Friday instead of 4 ml. NR are usually formed between week 4 and week 6.
-
11.
At the NR picking step, it is recommended to pick as many as possible (or needed) to increase the number of mature organoids. Between 50% and 90% of the NR picked will not make it to mature laminated retinal organoids (dependent on cell line). Some fail to form organized neuroepithelium in suspension and some collapse in a later stage forming a ball of neuro-retinal rosettes that will not develop the full outer and inner retinal layers. It is necessary to account for this when designing experiments.
-
12.
We find media without B27 from this stage may improve the organization of the inner retinal cell layers in organoids.
-
13.
0.1–10 μM is the concentration range that we have tested for AONs (with phosphorothioate backbone and either 2′-O-methyl or 2′-O-methoxyethyl modifications). The working concentration might vary with different AONs, as this is empirical.
-
14.
Gymnotic treatment with phosphorothioate backbone AONs is effective for retinal organoids. Addition of 6 μM EndoPorter will assist morpholino uptake. To treat the organoids, 200 μl of total volume is used in 96-well plates and 1 ml in 25-well plates.
-
15.
Treatment time is empirical and will depend on the specific target or assay being used. We have used treatment times between 72 h and 4 weeks.
-
16.
Nonsense mediated decay can be inhibited with emetine prior to sample collection, if it is suspected this is affecting the detection of aberrant transcripts.
-
17.
A 30-gauge needle can be used to help homogenize organoids. 200–500 ng of RNA can be extracted per organoid .
References
Gonzalez-Cordero A, West EL, Pearson RA, Duran Y, Carvalho LS, Chu CJ, Naeem A, Blackford SJI, Georgiadis A, Lakowski J, Hubank M, Smith AJ, Bainbridge JWB, Sowden JC, Ali RR (2013) Photoreceptor precursors derived from three-dimensional embryonic stem cell cultures integrate and mature within adult degenerate retina. Nat Biotechnol 31(8):741–747. https://doi.org/10.1038/nbt.2643
Zhong X, Gutierrez C, Xue T, Hampton C, Vergara MN, Cao LH, Peters A, Park TS, Zambidis ET, Meyer JS, Gamm DM, Yau KW, Canto-Soler MV (2014) Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun 5:4047. https://doi.org/10.1038/ncomms5047
Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, Sasai Y (2012) Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10(6):771–785. https://doi.org/10.1016/j.stem.2012.05.009
Meyer JS, Shearer RL, Capowski EE, Wright LS, Wallace KA, McMillan EL, Zhang SC, Gamm DM (2009) Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci U S A 106(39):16698–16703. https://doi.org/10.1073/pnas.0905245106
Gamm DM, Phillips MJ, Singh R (2013) Modeling retinal degenerative diseases with human iPS-derived cells: current status and future implications. Expert Rev Ophthalmol 8(3):213–216. https://doi.org/10.1586/eop.13.14
Sasai Y (2013) Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell Stem Cell 12(5):520–530. https://doi.org/10.1016/j.stem.2013.04.009
Bassett EA, Wallace VA (2012) Cell fate determination in the vertebrate retina. Trends Neurosci 35(9):565–573. https://doi.org/10.1016/j.tins.2012.05.004
Gonzalez-Cordero A, Kruczek K, Naeem A, Fernando M, Kloc M, Ribeiro J, Goh D, Duran Y, Blackford SJI, Abelleira-Hervas L, Sampson RD, Shum IO, Branch MJ, Gardner PJ, Sowden JC, Bainbridge JWB, Smith AJ, West EL, Pearson RA, Ali RR (2017) Recapitulation of human retinal development from human pluripotent stem cells generates transplantable populations of cone photoreceptors. Stem Cell Rep 9(3):820–837. https://doi.org/10.1016/j.stemcr.2017.07.022
Parfitt DA, Lane A, Ramsden CM, Carr AJ, Munro PM, Jovanovic K, Schwarz N, Kanuga N, Muthiah MN, Hull S, Gallo JM, da Cruz L, Moore AT, Hardcastle AJ, Coffey PJ, Cheetham ME (2016) Identification and correction of mechanisms underlying inherited blindness in human iPSC-derived optic cups. Cell Stem Cell 18(6):769–781. https://doi.org/10.1016/j.stem.2016.03.021
Ling JP, Wilks C, Charles R, Leavey PJ, Ghosh D, Jiang L, Santiago CP, Pang B, Venkataraman A, Clark BS, Nellore A, Langmead B, Blackshaw S (2020) ASCOT identifies key regulators of neuronal subtype-specific splicing. Nat Commun 11(1):137. https://doi.org/10.1038/s41467-019-14020-5
Kim S, Lowe A, Dharmat R, Lee S, Owen LA, Wang J, Shakoor A, Li Y, Morgan DJ, Hejazi AA, Cvekl A, DeAngelis MM, Zhou ZJ, Chen R, Liu W (2019) Generation, transcriptome profiling, and functional validation of cone-rich human retinal organoids. Proc Natl Acad Sci U S A 116(22):10824–10833. https://doi.org/10.1073/pnas.1901572116
Buskin A, Zhu L, Chichagova V, Basu B, Mozaffari-Jovin S, Dolan D, Droop A, Collin J, Bronstein R, Mehrotra S, Farkas M, Hilgen G, White K, Pan KT, Treumann A, Hallam D, Bialas K, Chung G, Mellough C, Ding Y, Krasnogor N, Przyborski S, Zwolinski S, Al-Aama J, Alharthi S, Xu Y, Wheway G, Szymanska K, McKibbin M, Inglehearn CF, Elliott DJ, Lindsay S, Ali RR, Steel DH, Armstrong L, Sernagor E, Urlaub H, Pierce E, Luhrmann R, Grellscheid SN, Johnson CA, Lako M (2018) Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa. Nat Commun 9(1):4234. https://doi.org/10.1038/s41467-018-06448-y
Dulla K, Aguila M, Lane A, Jovanovic K, Parfitt DA, Schulkens I, Chan HL, Schmidt I, Beumer W, Vorthoren L, Collin RWJ, Garanto A, Duijkers L, Brugulat-Panes A, Semo M, Vugler AA, Biasutto P, Adamson P, Cheetham ME (2018) Splice-modulating oligonucleotide QR-110 restores CEP290 mRNA and function in human c.2991+1655A>G LCA10 models. Mol Ther Nucleic Acids 12:730–740. https://doi.org/10.1016/j.omtn.2018.07.010
Capowski EE, Samimi K, Mayerl SJ, Phillips MJ, Pinilla I, Howden SE, Saha J, Jansen AD, Edwards KL, Jager LD, Barlow K, Valiauga R, Erlichman Z, Hagstrom A, Sinha D, Sluch VM, Chamling X, Zack DJ, Skala MC, Gamm DM (2019) Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development 146(1). https://doi.org/10.1242/dev.171686
Acknowledgments
This work is supported by Wellcome Trust, Fight for Sight, Foundation Fighting Blindness, Retina UK, Moorfields Eye Charity and NC3Rs. We would like to thank the other members of the Cheetham, Hardcastle, and van der Spuy groups past and present for their support, encouragement and help in iPSC and organoid maintenance. We would also like to thank Anai Gonzalez-Cordero for advice on the non-EB adherent protocol.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2022 The Author(s)
About this protocol
Cite this protocol
Hau, KL., Lane, A., Guarascio, R., Cheetham, M.E. (2022). Eye on a Dish Models to Evaluate Splicing Modulation. In: Arechavala-Gomeza, V., Garanto, A. (eds) Antisense RNA Design, Delivery, and Analysis. Methods in Molecular Biology, vol 2434. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2010-6_16
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2010-6_16
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2009-0
Online ISBN: 978-1-0716-2010-6
eBook Packages: Springer Protocols