Abstract
Background
Schizophrenia is a psychotic disorder that impacts around 0.5% to 1.2% of the world's population. It has been well established that heredity plays an essential role in the causation of schizophrenia, with genetic heritability of up to 80%. A several new schizophrenia susceptibility genes were identified at the start of the twenty-first century. The aim of this systematic review will be to explore the association between single nucleotide polymorphisms (SNPs) and schizophrenia risk in people all over the world.
Methods
This systematic review collected available data on genetic variants associated with schizophrenia in worldwide populations. A PubMed and Science Direct search was investigated to identify all studies published until December 2020 on genetic susceptibility to schizophrenia in various populations, excluding family studies, transversal studies, cohort studies, experimental studies, and descriptive studies; those that demonstrate an association between repeat polymorphism (CNV, VNTR, etc.). All researches on genetic predispositions of schizophrenia and accepting the predetermined inclusion criteria were included in this systematic review.
Findings
Thirty-six studies focused on the schizophrenia-associated genes were retained in which a total of 44 polymorphisms among 26 susceptibility genes to schizophrenia have been associated in the world populations.
Conclusion
Despite the few number of studies published about genetic of schizophrenia, some genetic variations have been consistently correlated to schizophrenia, particularly in China, as this analysis shows. Further data, especially from genome-wide association studies, might contribute in the development of a reference for schizophrenia genetic susceptibility markers.
Similar content being viewed by others
Introduction
Schizophrenia is conceptualized as a psychotic disorder according to DSM-5. Delusions, hallucinations, and disordered speech are basic "positive symptoms" that may be reliably identified with schizophrenia and may be regarded crucial for a successful diagnosis [1]. With an early onset in late adolescence or early adulthood, the disease affects about 0.5% to 1.2% of the global population. The most common clinical manifestations are positive symptoms, particularly hallucinations and delusions, on the one hand, and negative symptoms, such as reduced mood, speech and interest, as well as language and behavioral disorders, on the other hand [2].
It has been well established that heredity plays a significant role in the causation of schizophrenia, with genetic heritability of up to 80% [3]. Evidence in this context is available from family studies, genetic associations, and genetic linkage studies. Despite the high heritability, the genetic susceptibility factors that contribute to this disease have not been fully elucidated [4].
The scientific community has long sought to understand the pathogenesis of schizophrenia; however, due to high disease's complexity, this objective has proven difficult to achieve. Researchers have browsed the entire human genome for loci and genetic alterations (e.g., single nucleotide polymorphisms [SNPs]) that may be related to schizophrenia and other psychiatric disorders over the last decade [5].
A number of new schizophrenia susceptibility genes were identified at the start of the twenty-first century, and to this day more than 1000 genes have been analyzed for their potential association with schizophrenia. These genes were discovered through association studies based on their chromosomal position or their function. Among the genes that have been suggested to converge functionally on schizophrenia risk, we highlight ACE, GRM3, GSTs, DTNBP1, EFHD2, ERBB4, GABRB2…[5].
A recent systematic review and meta-analysis indicated that AMBRA1, ANK3, ARNTL, CDH13, EFHD1, MHC, PLXNA2, and UGT1A1 were revealed to be linked with SCZ or bipolar disorder (BD) diagnosis in at least two independent samples [6].
Similar to other psychiatric diseases, schizophrenia arises from an interplay between genetic and environmental factors. Studies in Chinese, Spanish, Indian, Iranian, Caucasian, and other populations around the world have shown robust genetic predispositions to this disease. This systematic review aims to collect available information on the genetic determinants of schizophrenia, more precisely to explore the association between single nucleotide polymorphisms (SNPs) and schizophrenia risk in people all over the world.
Methods used for systematic review
This systematic review was reported using the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. This article will discuss the epidemiology and genetics of schizophrenia in the global population. This is a systematic review based on publicly available data. As a result, no ethical approval is needed.
Bibliographic search
We searched Medline through PubMed and Science Direct databases for all existing genome-wide associations studies (GWAS) of schizophrenia that has been published in English from database inception until December 2020. Additionally, the reference lists of pertinent articles and reviews were checked for further articles.
Selection of studies
We included observational studies, particularly cases–control studies that assessed the link between genetic polymorphisms and schizophrenia in various populations.
To focus on genetic as a complex trait of schizophrenia we excluded (1) family studies, transversal studies, cohort studies, experimental studies and descriptive studies; (2) those that demonstrate an association between repeat polymorphism (CNV, VNTR, etc.), mutations and schizophrenia.
For studies reported in several reports, we considered the most detailed reporting on the largest sample size. Three authors (H.B., F.G., and A.H.O.) individually examined articles for inclusion (titles, abstracts, and then full texts).
Data extraction, assessment, and synthesis
Three investigators (H.B., F.G., and A.H.O.) extracted data from qualified articles using an essentialist data extraction form. The gathered information contained the country of the study, study population, sample size, mean age, range and proportion of men for cases and controls, genotyping method, gene and polymorphism studied, and the outcome (effects of studied polymorphism on the disease).
Due to high heterogeneity among included studies, the impact of a genetic variant on schizophrenia (their association, or none association) was identified as linked (YES) or unlinked (NO) based on whether the polymorphism was associated or not with a risk of schizophrenia.
A fourth experienced investigator double-checked all collected evidence for consistency (Hm.B.).
Our decision to conduct a narrative synthesis of the collected data is informed by the small number of qualifying studies and the enormous variation between research pertaining to sample population characteristics and evaluation of the association of genetic polymorphisms and schizophrenia.
Results of the systematic review
Literature search
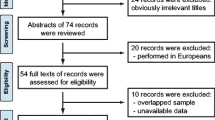
Our initial database search identified 5804 records from PubMed (n = 1632) and Science Direct (n = 4173); after removal of duplicates (n = 352), 5452 records remained. Since checking titles and abstracts, we identified 5220 records to be irrelevant and excluded them. We examined the complete texts of the remaining 232 articles to see if they were appropriate, of which 36 were eventually added [7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42] and 196 were excluded with reasons as indicated in Fig. 1.
Populations, genetic evaluation, and results
Of the 36 studies analyzed, 25 articles enrolled patients with schizophrenia of Asian origin (69,44%) [7,8,9,10, 12, 13, 15,16,17,18,19,20, 22,23,24,25, 28, 29, 33, 36,37,38,39,40, 42]. Patients of European ancestry were included in 6 articles (16,66%) [11, 26, 27, 30, 32, 41], whereas only 2 of African origin (5,55%) [21, 35], and the self-reported ethnic background in the last 3 article is African, African-American, Asian, and European [14, 31, 34]. All studies have been carried out on patients who had schizophrenia. Participants ranged in age from 16 to 76 years, with a men ratio ranging from 46 to 100% based on the population. Across studies, a total of 121 polymorphisms (including single nucleotide polymorphisms (SNPs) and insertion/deletions among 39 different genes were investigated (Table 1).
All SNPs in the selected studies fall in multifunctional scaffold proteins that are expressed in neurons and interact with multiple proteins involved in neuronal migration, glutamatergic, neurotransmission, signal transduction, and hippocampal development, all of which are implicated in the etiology of schizophrenia [15]. Indels were located primarily in the angiotensin converting enzyme (ACE), NQO2, and PAI-1 genes. The aim of all of the researches was to see whether there was a correlation between genetic determinants and schizophrenia in general populations.
Schizophrenia-associated genes
We identified 26 genes and 44 significant genetic polymorphisms implicated in schizophrenia in the research included in this review. Four studies about the 26 genes showing an association with schizophrenia were conducted in the Chinese population DISC1 (rs821616, rs821597), GLRX5 (rs1007814), NRG1 (rs3924999), TCF4 (rs2958182) [18, 20, 40]. Four other studies in Chinese Han population C3 (Complement 3) polymorphism (rs11569514), CACNA2D2 (rs45536634), four polymorphisms of NRXN1 (rs10490168, rs2 024513, rs13382584, rs1558852), and two polymorphisms of OPRM1 (rs1799971. rs2075572) [13, 16, 37, 42]. In the Korean population, a similar associations were detected with GRIA1 (rs1428920, rs2926835), TLR2 (rs3804099, rs3804100), HDAC3 (rs2530223) HDAC4 (rs1063639), DTNBP1 (rs760761, rs2619522) [23,24,25, 33]. Three other studies analyzed previously shown to be associated with schizophrenia in the northern and northeast Chinese han population EFHD2 (rs2473357) [8, 10, 28]. Studies in Iranian population showed that the II genotype of the I/D polymorphism of ACE gene, (rs1816072 (T/C)) polymorphism of GABRB2 and the (Haplotype Val-Lys) of XPC gene have a significant association with the susceptibility of schizophrenia [19, 29, 38].
It has already been found that there is a correlation between (GSTT1) polymorphism of GSTs, and Q64R (Gln64Arg) polymorphism of IFNGR2 and the risk of this disease in the Tunisian population [21, 35]. Similar results were observed in Turkish with AUTS2 (rs6943555) [32], Taiwanese with DAOA (rs778292, rs3918342, rs1421292) [12], Pakistanian with DISC (rs1417584) [15], Jordanian with ERBB4 (rs839523) (G/A) [7], and Japanese population with NQO2 (Deletion) [17] gene polymorphisms, respectively (Table 2).
One study conducted in mixed ethnic sample (Caucasian and African-American population) demonstrated an association between schizophrenia [14] and polymorphisms in PDE4B (rs1354064, rs4320761, rs1040716, rs910694, rs1321177, rs2144719, rs783038, rs599381) (Table 2).
Studies with lack of association
Twelve studies from various population did not find a significant association with schizophrenia [9, 11, 12, 22, 26, 27, 30, 31, 34, 36, 39] (Table 2).
Discussion
In this research, we used a systematic review to evaluate the correlation between polymorphisms and schizophrenia in the global population by integrating data from all qualifying studies. These powerful studies serve as informative summaries of the field's progress. Furthermore, a systematic review of smaller research will theoretically help entice and plan pathophysiological hypotheses for further genomic, transcriptomics, proteomics, and drug studies, particularly in unique populations. They can cover the research community's reflection on the diversity and discrepancies in samples, approaches, and conclusions through schizophrenia association, as well as make sense of current and prospective meta- and mega-analysis studies.
Our analysis showed that 75% of the overall pooled polymorphisms of these genes are SNPs (AUTS2, C3, CACNA2D2, DAO, DAOA, DISC, DISC1, DRD2, DTNBP1, EFHD2, ERBB4, GABRB2, GRLX5, GRIA1, GRIK4, HDAC, HSPA1A, HSPA1B, HSPA1L, NRG1, NRXN1, OPRM1, PDE4B, RANBP9, RELN, SAT-1, TCF4, TLR2, VEGFA, VRK2) and 25% of the others types of polymorphisms (ACE, GRM3, GSTs, IFNGR2, MTHFR, NQO2, PAI-1, PPARα, XPC).
After examining several articles across various populations, we selected 25 articles from the Asian population including 21 studies which found a significative association between polymorphism and schizophrenia, and four studies who failed to find an association. There are also 11 other studies from Europe, Africa, and the USA (2 African, 6 European, and 3 mixed African-American-European) among which 5 have found a relation between polymorphism and schizophrenia.
The Iranian study [29] investigated the role of the genotypes of I/D ACE polymorphism by PCR method. The deletion allele (D) was seen as a 191 bp band, whereas the insertion allele (I) was identified as a 478 bp band [29]. When compared to the DD genotype, the II genotype substantially reduced the risk of schizophrenia in females [29]. There was a strong linear relationship between the number of I alleles and the risk of schizophrenia in females. The sexes and the II genotype had a strong interaction [29]. The association between the ACE I/D polymorphism and the risk of schizophrenia has already been established. Two recent studies found contradictory results: the I allele was shown to be a protective factor in a Turkish population [43], although the D allele was found to be protective against schizophrenia and bipolar disorder in a Spanish population [44].
The study of C3 polymorphisms is still a major field in schizophrenia research. Association between the SNP rs11569514 in C3 and schizophrenia was investigated in Han Chinese population [37]. Interestingly, in the codominant model (TT vs. AA) the authors found a significant risk effect for rs11569514 on schizophrenia. Recent findings in the Tunisian population, as well as in the Brazilian population, show a significantly higher level of C3 protein in schizophrenia patients [45, 46].
In the research of Zhang et al. [39] among the northern Chinese Han population, there was no significant association between the (rs7116768) and (rs1799732) polymorphisms of the dopamine D2 receptor (DRD2) gene promoter region and schizophrenia [39], these results are consistent with those of previous research in a Spanish population [47], but are in contrast to the results in Brazilian population who found a significant association [48]. However, there are still some uncertainties concerning these associations.
The GRM3 gene was not associated with schizophrenia in the European and American populations [31]. Similar results were observed in the Japanese population [49]. However, other studies have instead reported a link between GRM3 genetic variation and risk of schizophrenia [50, 51].
Among single population studies, 12 variants of NQO2 gene were identified in Japanese population [17] including the insertion/deletion (I/D) polymorphism of the 29 base pair nucleotide sequence in the promoter region. In the schizophrenia group, the frequency of the D allele was significantly higher than in the healthy controls. The current findings show that individuals with the deletion of the 29 bp sequence in the promoter region of the NQO2 gene might be vulnerable to a specific type of schizophrenia [17]. Further research is needed on the implication of this polymorphism on schizophrenia.
One of the most promising potential genes for schizophrenia is neuregulin 1 (NRG1); the rs3924999 G/G genotype was associated with schizophrenia in the early onset subgroup (25 years) in the Chinese population [18]. Similar findings have been reported in Han Chinese and Japanese populations, where the NRG1 gene appears to contribute to schizophrenia susceptibility [52, 53].
In this research [14], 27 single nucleotide polymorphisms (SNPs) across the PDE4B gene were genotyped; several of these SNPs genotyped in the Caucasian population were substantially associated with schizophrenia. Two related intronic SNPs, rs1321177 and rs2144719, were particularly correlated to schizophrenia [14].
Although a number of studies have shown an association between the phosphodiesterase 4B (PDE4B) gene and the risk of schizophrenia, the findings are still inconclusive [54].
Reelin (RELN) is a protein involved in brain development and function and has been related to a variety of neuropsychiatric disorders. Bai et al. reported that the rs362746 of the RELN gene was associated with schizophrenia under the recessive and codominant models, in the Chinese population [10]. In Australia and Turkey, there was also a high correlation between this polymorphism and schizophrenia [55, 56].
The transcription factor 4 (TCF4) gene is highly expressed and plays a critical role in nervous system development, and it has long been implicated in the risk of developing schizophrenia [57].
The rs2958182 on TCF4 gene showed a significant association with schizophrenia in a meta-analysis in Chinese population [20]. This conclusion is in agreement with previous studies, in different ethnicities like Malaysians population, and Han Chinese population [57, 58].
VRK2 is a serine/threonine protein kinase enzyme encoded by the VRK2 gene on human chromosome 2p16.1. The VRK2 rs3732136 polymorphism was shown to be substantially linked with schizophrenia in the study of Zhang et al. [8] in a Northwest Chinese Han population, similar results were reported in other genotype and haplotype association studies. The results suggest that the VRK2 gene may play an important role in the development of schizophrenia in the Han population of northwest China [8]. The polymorphisms of serine/threonine-protein kinase (VRK2) were correlated to schizophrenia in two recent studies performed in Northern European and Asian populations [59, 60].
Several other polymorphisms were genotyped in different populations; for example, AUTS2 (rs6943555) [32], CACNA2D2 (rs45536634) [16], DAOA (rs778292, rs3918342, rs1421292) [12], DISC (rs1417584) [15], DISC1 (rs821616) [18], DNTBP1 (rs3213207, rs1011313, rs760761, and rs2619522) [33], EFHD2 (rs2473357) [28], ERBB4 (rs839523) [7], GABRB2 (rs12187676) [19], GLRX5 (rs1007814) [40], GRIA1 (rs1428920, rs2926835) [23], GSTs (GSTT1) [35], HDAC (rs1063639) [25], IFNGR2 (Q64R) [21], NRXN1 (rs10490168, rs2024513, rs13382584, rs1558852) [42], OPRM1 (rs1799971, rs2075572) [13], TLR2 (rs3804099) [24] and XPC [38] each found a significant association with schizophrenia.
In contrast some studies which genotyped other polymorphisms in several genes, for example, DAO [12], GRIK4 [36], HSPA1A [26] HSPA1B [27], HSPA1L [26], MTHFR [34], PAI-1 [41], PPARα [30], RANBP9 [9], SAT-1 [11], VEGFA [22], found no significant association with schizophrenia.
To fully understand the correlation between these polymorphisms and schizophrenia, further research with larger populations and prospective approaches is needed. Therefore, this systematic review still reveals new data of the impact of genes in schizophrenia risk.
A rigorous and systematic literature review, as well as the inclusion of multiple potential genetic risk factors for schizophrenia, is among the review's strengths. As many sample sizes were small, it is possible that the sample sizes were insufficient to detect a true association, especially because schizophrenia is most likely polygenic and influenced by many variants with small effect sizes.
Conclusion
Despite the few number of studies published about genetic of schizophrenia, some genetic variations have been consistently correlated to schizophrenia, particularly in China, as this analysis shows. Further data, especially from genome-wide association studies, might contribute in the development of a reference for schizophrenia genetic susceptibility markers.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- ACE:
-
Polymorphism in angiotensin-converting enzyme
- AMBRA1:
-
Autophagy and beclin 1 regulator 1
- ANK3:
-
Ankyrin 13
- ARNTL:
-
Aryl hydrocarbon receptor nuclear translocator-like
- AUTS2:
-
Autism susceptibility gene 2 protein
- BD:
-
Bipolar disorder
- C3:
-
Complement 3
- CACNA2D2:
-
Calcium voltage-gated auxiliary subunit alpha 2 delta 2
- CDH13:
-
Cadherin-13
- DAO:
-
D-amino acid oxidase.
- DAOA:
-
D-amino acid oxidase activation.
- DISC:
-
Disrupted in schizophrenia
- DISC1:
-
Disrupted in schizophrenia 1
- DNA:
-
Deoxyribonucleic acid
- DRD2:
-
Dopamine receptor D2
- DSM 5:
-
Diagnostic and statistical manual of fifth edition
- DTNBP1:
-
Dystrobrevin-binding protein 1
- EFHD1:
-
EF-hand domain family member D1
- EFHD2:
-
EF-hand domain family member D2
- ERBB4:
-
Erb-b2 receptor tyrosine kinase 4
- GABRB2:
-
Gamma-aminobutyric acid type A receptor subunit beta 2
- GLRX5:
-
Glutaredoxin 5
- GRIA1:
-
Glutamate ionotropic receptor AMPA type subunit 1
- GRIK4:
-
Glutamate ionotropic kainite receptor 4
- GRM3:
-
Glutamate metabotropic receptor 3
- GSTS:
-
The glutathione S-transferase
- GWAS:
-
Genome-wide association study
- HDACHDAC3:
-
Histone deacetylase 3
- HSPA1A:
-
Heat shock protein family A (Hsp70) member 1A
- HSPA1B:
-
Heat shock protein family A (Hsp70) member 1B
- HSPA1L:
-
Heat shock protein family A (Hsp70) member 1-like
- IFNGR2:
-
Interferon gamma receptor 2
- Indel:
-
Insertion/deletion
- MHC:
-
The major histocompatibility complex
- MTHFR:
-
Methylenetetrahydrofolate reductase
- NQO2:
-
N-Ribosyldihydronicotinamide:quinone reductase 2
- NRG1:
-
Neuregulin 1
- NRXN1:
-
Neurexin 1
- OPRM1:
-
µ-Opioid receptor gene
- OR:
-
Odds ratio
- P:
-
P value
- PAI-1:
-
Plasminogen activator inhibitor-1
- PDE4B:
-
Phosphodiesterase 4B
- PLXNA2:
-
Plexin A2
- PPARα:
-
Peroxisome proliferator activated receptor alpha
- PRISMA:
-
Preferred reporting items for the systematic reviews and meta-analysis
- PubMed:
-
Journal database
- RANBP9:
-
Ran-binding protein 9
- RELN:
-
Reelin is a protein coding gene
- SAT-1:
-
Spermidine/spermine N-1 acetyltransférase
- SCZ:
-
Schizophrenia
- SNPs:
-
Single nucleotide polymorphisms
- TCF4:
-
Transcription factor 4
- TLR2:
-
Toll-like receptor 2
- UGT1A1:
-
UDP glucuronosyltransferase family 1 member A1
- VEGFA:
-
Vascular endothelial growth factor A
- VNTR:
-
Variable number tandem repeat
- VRK2:
-
Vaccinia-related kinase 2
- XPC:
-
Xeroderma pigmentosum, complementation group C
References
Tandon R et al (2013) Definition and description of schizophrenia in the DSM-5. Schizophr Res 150:3–10
Jin L et al (2019) The association between rs12807809 polymorphism in neurogranin gene and risk of schizophrenia: a meta-analysis. Med (United States) 98:1–9
Sullivan PF, Kendler KS, Neale MC (2003) Schizophrenia as a complex trait. Arch Gen Psychiatry 60:1187
Kaur G et al (2019) An association study of dopaminergic (DRD2) and serotoninergic (5-HT2) gene polymorphism and schizophrenia in a North Indian population. Asian J Psychiatr 39:178–184
Zhuo C et al (2019) The genomics of schizophrenia: Shortcomings and solutions. Prog Neuro-Psychopharmacology Biol Psychiatry 93:71–76
Prata DP, Costa-Neves B, Cosme G, Vassos E (2019) Unravelling the genetic basis of schizophrenia and bipolar disorder with GWAS: A systematic review. J Psychiatr Res 114:178–207
Al-Eitan L, Al-Habahbeh S, Alkhatib R (2017) Genetic association analysis of ERBB4 polymorphisms with the risk of schizophrenia susceptibility in a Jordanian population of Arab descent. Turk J Med Sci 47:542–553
Zhang B et al (2015) Association of the VRK2 gene rs3732136 polymorphism with schizophrenia in a Northwest Chinese Han population. Genet Mol Res 14:9404–9411
Bae JS et al (2015) Investigating the potential genetic association between RANBP9 polymorphisms and the risk of schizophrenia. Mol Med Rep 11:2975–2980
Bai W et al (2019) Association between RELN polymorphisms and schizophrenia in a Han population from Northeast China. Psychiatr Genet 29:232–236
Bermudo-Soriano CR et al (2009) SAT-1 -1415T/C polymorphism and susceptibility to schizophrenia. Prog Neuro-Psychopharmacology Biol Psychiatry 33:345–348
Chu CS et al (2017) The DAOA gene is associated with schizophrenia in the Taiwanese population. Psychiatry Res 252:201–207
Ding S et al (2013) Association study of OPRM1 polymorphisms with Schizophrenia in Han Chinese population. BMC Psychiatry 13:107
Fatemi SH et al (2008) PDE4B polymorphisms and decreased PDE4B expression are associated with schizophrenia. Schizophr Res 101:36–49
Fatima W et al (2020) Chromosomal region 1q24.1 is associated with increased risk of schizophrenia in Pakistani population. Gene 734:144390
Fu Y et al (2020) Association of the CACNA2D2 gene with schizophrenia in Chinese Han population. PeerJ 2020:1–11
Harada S, Tachikawa H, Kawanishi Y (2003) A possible association between an insertion/deletion polymorphism of the NQO2 gene and schizophrenia. Psychiatr Genet 13:205–209
He BS et al (2016) Association of the DISC1 and NRG1 genetic polymorphisms with schizophrenia in a Chinese population. Gene 590:293–297
Heidari Nia M, Sargazi S, Saravani R, Mirinejad S, Jahantigh D, Shakiba M (2020) Relationship between GABRB2 gene polymorphisms and schizophrenia susceptibility: a case-control study and in silico analyses. Int J Neurosci. https://doi.org/10.1080/00207454.2020.1830087
Gao JY, Ma P, Li Y, Yan CX, Zhang Q, Yang XX, Shi Q, Zhang B, Wen XP (2020) Association between a TCF4 Polymorphism and Susceptibility to Schizophrenia. Biomed Res Int. 2020:1216303. https://doi.org/10.1155/2020/1216303
Jemli A et al (2017) IFNGR2 genetic polymorphism associated with sex-specific paranoid schizophrenia risk. Nord J Psychiatry 71:42–47
Gao K et al (2015) Association study of VEGFA polymorphisms with schizophrenia in Han Chinese population. Neurosci Lett 590:121–125
Kang WS et al (2012) Genetic variants of GRIA1 are associated with susceptibility to schizophrenia in Korean population. Mol Biol Rep 39:10697–10703
Kang WS et al (2013) Association between genetic polymorphisms of Toll-like receptor 2 (TLR2) and schizophrenia in the Korean population. Gene 526:182–186
Kim T, Park JK, Kim HJ, Chung JH, Kim JW (2010) Association of histone deacetylase genes with schizophrenia in Korean population. Psychiatry Res 178:266–269
Kowalczyk M et al (2018) Association studies of HSPA1A and HSPA1L gene polymorphisms with schizophrenia. Arch Med Res 49:342–349
Kowalczyk M et al (2020) Association of HSPA1B polymorphisms with paranoid schizophrenia in a polish population. NeuroMol Med 22:159–169
Gao M et al (2020) Association between EFHD2 gene polymorphisms and schizophrenia among the Han population in northern China. J Int Med Res 48:10
Mazaheri H, Saadat M (2015) Association between insertion/deletion polymorphism in angio-tension converting enzyme and susceptibility to schizophrenia. Iran J Public Health 44:369–373
Nadalin S, Giacometti J, Buretić-Tomljanović A (2014) PPARα-L162V polymorphism is not associated with schizophrenia risk in a Croatian population. Prostaglandins Leukot Essent Fat Acids 91:221–225
Norton N et al (2005) No evidence for association between polymorphism in GRM3 and schizophrenia. BMC Psychiatry 5:1–6
Ozsoy F, Balcı N, Yigit S, Kulu M (2020) Effect of AUTS2 gene rs6943555 variant in male patients with schizophrenia in a Turkish population. Gene 756:144913
Pae CU et al (2009) Dysbindin gene (DTNBP1) and schizophrenia in Korean population. Eur Arch Psychiatry Clin Neurosci 259:137–142
Philibert R et al (2006) No association of the C677T methylenetetrahydrofolate reductase polymorphism with schizophrenia. Psychiatr Genet 16:221–223
Raffa M et al (2013) Relationship between GSTM1 and GSTT1 polymorphisms and schizophrenia: a case-control study in a Tunisian population. Gene 512:282–285
Ren D et al (2017) No association of GRIK4 polymorphisms with schizophrenia in the Chinese Han population. Psychiatr Genet 27:159–160
Zhang S et al (2018) Association between polymorphisms of the complement 3 gene and schizophrenia in a Han Chinese Population. Cell Physiol Biochem 46:2480–2486
Taghipour N, Saadat I, Saadat M (2019) Association between polymorphisms of Xeroderma pigmentosum complementation group C gene (XPC) and susceptibility to schizophrenia. Gene 695:99–100
Zhang X cen et al (2019) No association between polymorphisms in the promoter region of dopamine receptor D2 gene and schizophrenia in the northern Chinese Han population: a case–control study. Brain Behav 9:1–8
Yang B et al (2017) Association study of the GLRX5 rs1007814 polymorphism with schizophrenia in the Han Chinese population. Psychiatr Genet 27:76–77
Yenilmez C et al (2017) A study of the possible association of plasminogen activator inhibitor type 1 4G/5G insertion/deletion polymorphism with susceptibility to schizophrenia and in its subtypes. J Clin Pharm Ther 42:103–107
Yue W et al (2011) A case-control association study of NRXN1 polymorphisms with schizophrenia in Chinese Han population. Behav Brain Funct 7:3–8
Kucukali CI et al (2010) Angiotensin-converting enzyme polymorphism in schizophrenia, bipolar disorders, and their first-degree relatives. Psychiatr Genet 20:14–19
Crescenti A et al (2009) Insertion/deletion polymorphism of the angiotensin-converting enzyme gene is associated with schizophrenia in a Spanish population. Psychiatry Res 165:175–180
Nsaiba MJ et al (2015) C3 Polymorphism influences circulating levels of C3, ASP and lipids in schizophrenic patients. Neurochem Res 40:906–914
Sória L dos S, Gubert C de M, Ceresér KM, Gama CS, Kapczinski F (2012) O aumento dos níveis séricos de C3 e C4 em pacientes com esquizofrenia em comparação com pacientes com transtorno bipolar eutímico e saudáveis. Rev Bras Psiquiatr 34, 119–120
Parsons MJ et al (2007) A dopamine D2 receptor gene-related polymorphism is associated with schizophrenia in a Spanish population isolate. Psychiatr Genet 17:159–163
Cordeiro Q, Siqueira-Roberto J, Zung S, Vallada H (2009) Association between the DRD2-141C insertion/deletion polymorphism and schizophrenia. Arq Neuropsiquiatr 67:191–194
Tochigi M et al (2006) No association between the metabotropic glutamate receptor type 3 gene (GRM3) and schizophrenia in a Japanese population. Schizophr Res 88:260–264
Fujii Y et al (2003) Positive associations of polymorphisms in the metabotropic glutamate receptor type 3 gene (GRM3) with schizophrenia. Psychiatr Genet 13:71–76
Saini SM et al (2017) Meta-analysis supports GWAS-implicated link between GRM3 and schizophrenia risk. Transl Psychiatry 7:e1196
Wen Z et al (2016) Genetic association between NRG1 and schizophrenia, major depressive disorder, bipolar disorder in Han Chinese population. Am J Med Genet Part B Neuropsychiatr Genet 171:468–478
Shiota S et al (2008) Association and interaction analyses of NRG1 and ERBB4 genes with schizophrenia in a Japanese population. J Hum Genet 53:929–935
Feng Y et al (2016) Association of PDE4B polymorphisms with susceptibility to schizophrenia: A meta-analysis of case-control studies. PLoS ONE 11:1–14
Ovadia G, Shifman S (2011) The genetic variation of RELN expression in schizophrenia and bipolar disorder. PLoS ONE 6:5
Sozuguzel MD, Sazci A, Yildiz M (2019) Female gender specific association of the Reelin (RELN) gene rs7341475 variant with schizophrenia. Mol Biol Rep 46:3411–3416
Mohamed ZI et al (2019) Functional characterization of two variants in the 3’-untranslated region (UTR) of transcription factor 4 gene and their association with schizophrenia in sib-pairs from multiplex families. Asian J Psychiatr 40:76–81
Hui L et al (2015) TCF4 gene polymorphism is associated with cognition in patients with schizophrenia and healthy controls. J Psychiatr Res 69:95–101
Tesli M et al (2016) VRK2 gene expression in schizophrenia, bipolar disorder and healthy controls. Br J Psychiatry 209:114–120
Chang H et al (2016) Further evidence of VRK2 rs2312147 associated with schizophrenia. World J Biol Psychiatry 17:457–466
Acknowledgements
Not applicable.
Funding
No funding was received for the present systematic review.
Author information
Authors and Affiliations
Contributions
HB and AR designed research; FG, HB and AHO conducted the bibliographic research and selected the studies involved in this systematic review under the supervision of HB and AR; FG, HB and AHO performed the data extraction, assessment, and synthesis; FG, HB and AHO wrote the paper under the supervision of HB and AR; HB and AR had primary responsibility for final content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Boulenouar, H., Benhatchi, H., Guermoudi, F. et al. An actualized screening of schizophrenia-associated genes. Egypt J Med Hum Genet 23, 81 (2022). https://doi.org/10.1186/s43042-022-00269-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-022-00269-x