Abstract
Background
Verticillium dahliae is one of the most destructive pathogens in many cotton-growing countries around the globe. There is still the lack of high-resistant upland cotton cultivars against this pathogen, and the usage of fungicides is threatening the environment. Biocontrol is an environment-friendly and a green technique in plant protection. In this regard, the present work was aimed to investigate antagonistic bacterial strains and to introduce them as new bioinoculants against V. dahliae in cotton.
Main body of the abstract
Numerous bacteria were isolated from cotton roots, and their antagonism was examined against V. dahliae. Strain SZAD2 which was identified as Bacillus velezensis on the bases of 16S rRNA gene analysis had significantly high biocontrol potential both in plate assay and under greenhouse conditions. The biocontrol efficacy in seed treatment was 60.31%, while in the soil drench method, it was 79.19%. Transmission electron microscopy (TEM) analysis revealed that the strain systemically colonized the roots and lived within the intercellular space. Hydrogen peroxide staining exposed that the strain increased plant basal defense response by the accumulation of hydrogen peroxide (H2O2) in both roots and leaves. Furthermore, the bacterization of cotton roots triggered induced systemic resistance (ISR) in the roots and caused the activation of the antioxidant enzymes such as phenylalanine ammonia-lyase (PAL), polyphenol oxidase (PPO), peroxidase (POD), and phenol contents. Moreover, the antifungal extract produced by the strain contained the activity of both cellulase and chitinase, which were able to suppress fungal mycelia both in the presence and absence of cotton.
Short conclusions
Strain SZAD2 systemically colonized the plant, showed multiple modes of action against the pathogen which played a vital role in V. dahliae control, and is recommended for commercial exploration as a safe alternative bioinoculant instead of hazardous chemical fungicides.
Similar content being viewed by others
Background
Cotton is a great economically important crop, and it is widely cultivated in many countries because of the economic value of its fiber and seed oil. Verticillium wilt is one of the most destructive diseases in many cotton-producing countries, which is caused by a soil-borne fungus (V. dahliae). Verticillium wilt is notoriously difficult to be controlled as the fungus has a broad host range and produces microsclerotia, which are melanized multicellular survival structures and can remain in the form of dormant in the soil for years (Heale and Isaac 1965).
Conventional and integrate management of Verticillium wilt in cotton include growing resistant cultivars, proper cultural practices, and proper management of water and fertilizer (El-Zik 1985). Even though growing of resistant cultivars are considered as the most effective and economical mean of controlling this disease but there is still the lack of high-resistant upland cotton cultivars against this pathogen (Yang et al. 2008). Crop rotation is effective against the pathogen in a few studies (Xiao et al. 1998), but it is not adopted widely due to the limitation of agricultural areas. Several fungicides have been considered effective and are used against V. dahliae, but no any selective fungicide is currently registered for this pathogen in cotton (Göre et al. 2009). Also, fungicides are not going to be able to affect the pathogen and reduce its severity once it enters the xylem (Fradin and Thomma 2006). Additionally, extensive use of fungicides has raised public concerns on the environment and human health (Dias 2012).
In recent years, research on biocontrol and biochemical products from plants, insects, and microorganisms for pest and pathogen control has been encouraged. Biocontrol is a promising alternative way for soil-borne diseases such as V. dahliae control and management in sustainable and organic agriculture. In this framework, numerous of endophytic bacteria have been isolated from plants and have been used against V. dahliae (Kavino et al. 2007). Furthermore, some endophytic bacterial strains promote plant development by synthesizing phytohormones such as indole-3-acetic acid (IAA) (Santoyo et al. 2016). IAA syntheses inside the plants increase the number of lateral and adventitious roots, which improve root exudation and facilitate access to more nutrients and water (Spaepen and Vanderleyden 2011). The use of antagonistic species of bacillus has been widely researched due to the reality that these bacteria systemically colonize the plants and trigger ISR (Rajendran and Samiyappan 2008a), modulate H2O2, and produce several types of antimicrobial compounds such as cellulase and chitinase. Cellulase and chitinase are responsible for the biological degradation of cellulose and chitin of fungus cell wall, which cause the fungal mycelia physiological abnormal and reduce their infection ability and virulence which is one of the crucial mechanism (Kim et al. 2001). The presence and the specific activity of cellulase and chitinase in the secondary extract produced by the antagonistic bacteria is determined by measuring the amount of reducing sugar liberated dinitrosalicylic acid (DNS), using a substrate of colloidal chitin or carboxy methyl cellulose (CMC) (Annamalai et al. 2011).
The process of triggering physiological changes in plants is termed as “induce systemic resistance” (ISR) (Fatima and Anjum 2017). Among the different physiological changes associated with the ISR, the activity of antioxidant enzymes such as phenyl ammonia-lyase (PAL), polyphenol oxidase (PPO), peroxidase (POD) (Vaikuntapu et al. 2014), and phenol contents production (Rajendran and Samiyappan 2008b) act as good indicators of plant ISR (Han et al. 2016). POD and PPO are reported to be involved in the formation of lignin, which makes the cell wall resistant to fungal degradation and restricts their entrance and their movement inside the infected plant (Saravanakumar et al. 2007). PAL is the main enzyme in the metabolism of phenylpropanoid and in the production of phenolic contents and phytoalexins that prevent pathogens attack and establishment (Daayf et al. 1997). Phenolic compounds mechanically strengthen the cell walls of the plant which inhibits the colonization of the pathogen to the host plant (M’piga et al. 1997).
Oxidative burst or the production of reactive oxygen species (ROS), via consumption of oxygen, is called oxidative burst, a controlled release of O2 and H2O2 at the point of microbe challenge, which is one of the earliest cellular response and is one of the key components of plant defense response. The oxidative burst occurs during compatible interactions (interaction of compatible endophytic bacteria or fungi) result in the production of molecules, which can be employed to create physical barriers to hyphal growth and entrance to the plants and have direct detrimental effects to the cells of invading pathogens (De Gara et al. 2010).
Given the above, the objectives of this study were to isolate high antagonistic bacterial strains against V. dahliae and to characterize them based on morphological, biochemical, and molecular attributes to screen the mode of action and to introduce the most effective bioagent as a new bioinoculant in management and control strategy of Verticillium wilt in cotton.
Materials and methods
Fungal strain and cotton variety
V. dahliae Vd080 (Zhang et al. 2016) which was used in this study was kept on potato dextrose agar (PDA) medium. The mycelia of V. dahliae were obtained from growing the strain in potato dextrose broth (PDB) medium at 28 °C, 160 rpm for 12 days. The susceptible cotton variety (Jimian 11) was used.
Isolation of endophytic bacteria
Cotton plants were collected from Liu he agricultural research farm, Nanjing, China. Endophytic bacteria were isolated, following (Wen et al. 2011) method with some minor modifications. The plants were severed aseptically just above the soil line, washed under tap water and were sterilized with 70% ethanol for 1 min followed by 5% sodium hypochlorite (NaClO) for 5 min then, washed with dH2O for three times. The outer tissues were removed by a knife, and the remaining parts were cut into small fragments. The fragments were sterilized by 70% ethanol and again washed in dH2O for 3 times. The fragments were semi dipped in Luria Bertani (LB) agar medium in the Petri dish and incubated at 28 °C until the colonies appeared around the segments. Distinct colonies were identified based on the following traits: color, form, elevation, margin, diameter, surface, and texture. Every distinct colony was purified by streak culture in Petri dish on LB agar medium in three replicates. A similar procedure without surface sterilization was launched as a negative control. As a positive control, the LB agar medium was inoculated with 0.1 ml of the last rinsed water. The purified colonies were transferred to 2 ml centrifuge tubes had 1 ml LB liquid medium and incubated in a shaker at 37 °C, 160 rpm for 24 h. The isolates were stored at – 20 °C for a short time and in 40% glycerol at – 80°C for long-term storage.
Antagonistic activity and pathogenicity reduction test in plate assay
Antagonistic activity of the isolates was tested on PDA plates in modified agar diffusion assay (Han et al. 2015) with some major modifications. In details, 2 ml of V. dahliae mycelial suspension was mixed with 200 ml of semi-solid (45 °C) 1% PDA medium and was poured into the Petri dish. Two wells were punched into the PDA medium with a sterile Pipette tip. Ten microliters of the overnight culture of isolates were placed into the well, and only LB liquid medium was added as a control. The plates were incubated at 25 °C. Five days later, the zone of inhibition was measured, and the mycelia of fungi from the zone of interaction were removed from the plate carefully, mixed with ddH2O, and the changes in morphology was observed under BX53 Olympus microscope. In the meantime, the mycelia from the interaction zone (treated-mycelia) and the mycelia from isolate-free zone (untreated-mycelia) in Petri dish were transferred to separate tubes, mixed with ddH2O, and their virulence was examined as follows: 2-week-old cotton seedling’s leaves were placed on 0.30% water agar. Ten microliters droplets of bacterial treated or untreated mycelial suspension (106 ml−1) was spotted on the surface of the leaves. Seven days later, the viability of the spotted mycelia was examined under a light microscope, and their germination on the leaves and the necrosis of the spotted area was photographed. Each assay was performed in three replicates.
16S rRNA gene analysis
The isolate was identified by partial analysis of 16S rRNA gene as follows: the total genomic DNA was extracted using TIANamp bacteria DNA Kit (TIANGEN Biotech Co., Ltd., Beijing, China). Universal primers (F27; 5′-AGA GTT TGA TCC TGG CTC AG-3′ and R1492; 5′-GGT TAC CTT GTT ACG ACT T-3′) (Edwards et al. 1989) were used to amplify 16S rRNA gene using PCR (GeneMate series). A total volume of 25 μl reaction mixture was prepared into the PCR tubes as follows: 1 μl DNA template, 0.5 μl forward primer, 0.5 μl reverse primer, 12.5 μl rTaq (Sigma chemical company), and 10.5 μl dH2O. The amplification process was carried out in a thermal cycle with the following reaction conditions: the PCR process began initial denaturation at 95 °C for 1 min, primers annealing 53 °C for 30 s, and primer extension at 72 °C for 1 min. The PCR products were analyzed with 2% gel agarose. The amplified gene was sequenced by (Beijing Liuhe Huada Gene Technology Co. Ltd., China). The sequence was compared with other related sequences in the national center for biotechnology information (NCBI) and was aligned using Clustal X. The phylogenetic tree was constructed using mega X software by the maximum likelihood method with the maximum parsimony options and 1000 bootstrap replicates for the support estimation of each branch. Morphological properties were examined by a general protocol as described by Kamlage (1996). The physiological and biochemical characteristics were examined using the API 20NE system (bioMerieux, Inc., Marcy l’Etoile, France).
Greenhouse experiment
The seeds were washed, sterilized with 5.5% sodium hypochlorite (NaClO) for 1min, then washed with ddH2O for three times and were kept in ddH2O for 24 h for germination initiation. To make the bacterial solution, a fresh culture of bacteria was centrifuged, and the pellets were washed by ddH2O for three times. Then, the pellets were dissolved in 0.5% carboxy methylcellulose (CMC) solution, optical density (OD) = 0.3 for seed treatment. The pellets were dissolved in ddH2O (7 × 109 cell ml−1) for soil drench method. For seed treatment, the seeds were dipped in bacterial solution for 4 h. As a control, the seeds were dipped in the same volume of CMC solution without bacteria and were then grown in 300 ml plastic cups filled with sterile vermiculite medium and watered accordingly. Five days after the treatment, the development of the lateral roots were randomly recorded for 10 plants from each treatment. As a soil drench method, 10 ml of the bacterial solution was added to every cup of 1-week-old seedlings grown in 300 ml plastic cups or the same volume of only dH2O was added (control) following the procedure of Selim et al. (2017).
The seedlings were challenged inoculated at the 6th true leaves stage with V. dahliae spore solution as described by Wang et al. (2015) with some modification. 0.5 ml of fungal spore was inoculated in 200 ml PDB medium in 500 ml flask and incubated in 150 rpm, 37 °C for 7 days. Ten milliliters of spore solution (6.107 ml−1) was applied to the soil around the plant in every cup. Fifteen days after the inoculation, the disease severity was assessed for each plant. A scale from 0 to 4 was used to classify plants according to the percentage of plant leaves effected by chlorosis, necrosis or defoliation (0 = no symptoms, 1 = slight marginal chlorosis, 2 = moderate marginal chlorosis, 3 = moderate wilt and visible necrosis, 4 = severe wilt and defoliation). The leaf wilt index (LWI) was calculated as follows: leaf wilt index = Σ (number of plants with a disease scale × value of the corresponding scale)/(total plants rated × the highest scale value) (Li et al. 2012). Biocontrol efficacy was calculated as follows: Biocontrol efficacy = [(LWI of control plants–LWI of treated plants)/disease incidence of control] × 100 (Huang et al. 2006). The experiment was designed in three replicates with 19 to 25 plants (each cup had 1 or 2 seedlings).
Hydrogen peroxide (H2O2) staining in the leaves
Overnight culture of the isolate was centrifuged at 12000 rpm for 10 min, the supernatants were discarded, the pellets were mixed with ddH2O and centrifuged again in 12000 rpm for 10 min (repeated this step for 3 times). Finally, the pellets were mixed with ddH2O (OD = 0.2). In the 6th true leave stage, the bacterial solution was sprayed on the whole plant (only ddH2O was sprayed on the CK group). Five days later, ROS (H2O2) was assessed as described by Kumar et al. (2009). The experiment was designed in three replicates.
Hydrogen peroxide (H2O2) staining in the roots
For hydrogen peroxide staining and to visualize bacterial internalization and their infections in the roots, the cotton seeds were sterilized with NaClO and were treated with the bacterial solution as described above. The seeds were then grown on a 0.5% water agar medium. One-week-old seedlings roots were stained with a solution of 100 mM potassium phosphate buffer, pH 7.0, 2.5 mM 3, 3′-diaminobenzidine tetrachloride (DAB), and 5 units/mL purpurogallin of horseradish peroxidase (Type VI, Sigma Chemical Company) (Pick and Keisari 1980). The roots were excised, placed on a slide contained aniline blue/lactophenol (0.012 g aniline blue dye, 5 g phenol crystals, 10 ml glycerol, 5 ml lactic acid, 5 ml H2O), and examined under bright field microscopy.
Transmission electron microscope
To explore systemic colonization and the precise location of the endophytic isolate, the seeds were dipped in bacterial solution for 4h. As a control, the seeds were dipped in the same volume of CMC solution without bacteria as described previously. Then, the seeds were sown in 500 ml glass flask contained sterilized vermiculite medium (all the procedure was performed under laminar flow hood). After 1 week, the seedlings were harvested, the roots were cut into small pieces and kept in 2.5% glutaraldehyde solution in potassium phosphate buffer (PB), pH 7 until transmission electron microscope analysis (Quadt-Hallmann et al. 1997).
Plant systemic resistant enzymes assessment in the roots
The seeds were surface sterilized by NaClO and were treated with a fresh culture of the bacterial solution and were sown in 300 ml plastic cups as described above. One week after cultivation, the seedlings were inoculated with fungal spores and the roots samples were collected at 0, 24, 72, 96, and 168 h time interval, ground in liquid nitrogen, and kept in − 70°C until use.
Determination of phenylalanine ammonia-lyase (PAL) activity
0.5 g nitrogen ground roots were homogenized with 1 ml of 0.1 M phosphate buffer (pH 7.0), centrifuged at 12,000 rpm for 20 min at 4 °C. The supernatants were used as an enzyme source. One hundred microliters of enzyme extract was mixed with 1.2 ml of 0.1 M borate buffer (pH 9) and 1.5 ml 12 M L-phenyl alanine and was incubated at 30 °C for 30 s. 0.1 ml of HCl at a concentration of 5 M was added to terminate the reaction. The absorbance at OD290 was measured by a spectrophotometer. Enzyme activity was expressed as mmol trans-cinnamic acid min−1 gr−1 of tissues (Dickerson et al. 1984). The experiment was set up for three replicates, and each replicate was measured in parallel for three times.
Determination of peroxidase (POD) activity
0.5 g nitrogen ground roots were homogenized with 1 ml of 0.1 M phosphate buffer (pH 7.0). The mixture was shaken and centrifuged at 12,000 rpm for 15 min at 4 °C. The supernatants were used as an enzyme source and were transferred to another centrifuge tube. 1.5 ml of 0.05 M pyrogallol at a concentration of 0.05 M and 0.5 ml of 1% H2O2 were added to 0.5 ml of enzyme extract. The changes in absorbance (OD) at 420 nm were recorded every 60 s for 5 min. The enzyme activity was expressed as changes in OD of the reaction mixture min−1 gr−1 as enzyme activity (Mayer et al. 1966). The experiment was set up three times and every time the optical density was also measured three times.
Determination of polyphenol oxidase (PPO) activity
0.5 g of the stored ground roots were mixed with 1 ml of 0.1 M sodium phosphate buffer (pH 6.5), Shaken and mixed. Afterward, centrifuged at 12,000 rpm for 15 min at 4 °C. The supernatants were transferred to another centrifuge tube and were used as an enzyme source. The reaction mixture contained 200 μl enzyme extract and 1.5 ml of 0.1M sodium phosphate buffer (pH 6.5). To start the reaction, 200 μl of 0.01 M catechol was added, after mixing, the absorbance was measured immediately at 495 nm every 1 min for 5 min. The changes in absorbance at 495 nm min−1 g−1 was used as enzyme activity unit (U) (Hammerschmidt et al. 1982). The experiment was set up for three repetitions and optical density (OD) measurements were also repeated three times.
Total phenolic contents
The total phenolic contents in the roots were assessed as described by Zieslin and Ben-Zaken (1993). In detail, 0.5 g of liquid nitrogen ground roots were homogenized with 5 ml of 80% methanol. One milliliter of methanolic extract was mixed with 5 ml of distilled water and 250 μl of Folin–Cioalteau reagent (1 N), and the solution was kept at 25 °C for 3 min. Then, 1 ml of sodium carbonate saturated solution and 1 ml of sterile distilled water were added and were incubated at 25 °C for 1 h. The OD was measured at 725 nm. The soluble total phenol contents were calculated based on a standard carve from Folin–Cioalteau reagents with the phenol solution and were expressed as catechol equivalent per gram of tissue weight.
Hydrolytic enzymes activity test
To examine cellulase activity of the strain, water agar plates containing 1% CMC were prepared. The strain was spotted in the middle of the plate and incubated at 30 °C for 2 days. The plates were flooded with 0.1% Congo red, and 20 min later, the Congo red was poured off and washed with 1 M NaCl for 10 min. To examine chitinase activity, colloidal chitin was prepared as described by Joe and Sarojini (2017). Agar plates containing 1% colloidal chitin was prepared, and the strain was spotted in the middle of the plate and incubated at 30 °C for 3 days.
Isolation and antifungal activity of the secondary extract
The antifungal compounds produced by the isolate were extracted from the supernatant’s fluid as follows: 500 μl of the bacteria was inoculated in 1 L LB medium and incubated at 37 °C, 160 rpm. The culture was harvested after 48, 72, 96, and 120 h of incubation by centrifugation at 12,000rpm for 15 min. The antifungal ability of the supernatant was tested in Petri dish. Subsequently, the extracts were precipitated by ammonium sulfate 80% saturation and were kept at 4 °C overnight. After centrifugation at 12,000 rpm for 20 min, the precipitates were collected and dissolved in methanol. The methanol was evaporated, and the extracts were dissolved in ddH2O and were freeze-dried. The antifungal activity of the enzyme extracts was examined by agar well diffusion method as described by Balouiri et al. (2016) with some modifications. In details, Petri dishes were filled with PDA medium. Approximately (107 ml−1) V. dahliae spores were spread on the surface of the media with a glass spreader, wells were punched with a sterilized 0.5 cm in diameter Pipette tip. The extracts were filtered (0.2 μm) and were applied to the wells. Only liquid LB medium was applied to the wells as a control. The plates were incubated at 28 °C; 3 days later, the diameter of the inhibited clear zone around the wells was measured and was directed as antifungal activity of the secondary extract. To determine the specific active fractions of the secondary extract, cellulase, and chitinase specific activities were assessed.
Detection of cellulase
The specific activity of the total cellulase was determined by measuring the amount of reducing sugar produced during the enzymatic reaction by dinitrosalicylic acid (DNS) as described by Miller (1959). A reaction mixture contained 0.5 ml enzyme extract, 0.5 ml 0.05 M citrate-phosphate buffer (pH 7.0) and 1.0 ml of 1% (w/v) CMC (Sigma-Aldrich) in 0.05 M citrate-phosphate buffer (pH 7.0) was prepared and incubated in 37 °C for 15 min. 1.5 ml DNS reagent was added to terminate the reaction, boiled for 15 min in a water bath, and the absorbance was measured at 540 nm. The mixture without the reaction enzyme was used as a blank (Saowapar et al. 2014). Each unit of cellulase activity was determined as the amount of the enzyme required for releasing 1 μmole of glucose produce per min per ml of the enzyme solution.
Detection of chitinase
For chitinase activity measurement, colloidal chitin was used as a substrate and was examined by the release of glucose. One milliliter of enzyme solution was mixed with 1 ml of substrate solution (1% colloidal chitin in phosphate buffer, pH 7.0). The reaction mixture was incubated in 50 °C for 60 min, and then 2 ml of 1% 3, 5-dinitrosalicylic acid (DNS) (sigma) was added to stop the reaction. The mixture was boiled in a water bath for 15 min and was cooled to room temperature. The absorbance was measured at 530 nm.
Antagonistic potential of the secondary extract
In the absence of cotton
Fifty milliliters of liquid LB medium was prepared in 150 ml flasks containing 25 μ/ml and 50 μ/ml concentration of the extract. Only dH2O was added in control. The flasks were inoculated with 50 μl of V. dahliae mycelia and incubated at 28 °C, 80 rpm. Six days later, the number of living mycelia was counted using hemocytometer.
In the presence of cotton plant
The greenhouse experiment was conducted to investigate the antagonistic potential of the extracts against V. dahliae mycelia in the presence of cotton. Sterile soil was mixed with V. dahliae microsclerotia with a concentration of 2% (w/w) and placed in 250 ml plastic cups. Twenty-five micrograms per milliliters and 50 μg/ml concentration of the enzyme extract was dissolved in dH2O and were used as the secondary extract solution. Cotton seeds were surface sterilized as above and were grown in the soil in three replicates then were placed at 27 °C with 14 h photoperiod. After germination, 20 ml of the extract solution was poured into every cup (only dH2O was added for control). Diseases severity and leaf wilt index (LWI) were calculated at 6th true leaves stage as described above. The experiment was designed in 3 replicates with 20 plants (every cup had 1 or 2 plants). Subsequently, 4 cups from each treatment were selected randomly and investigated for the quantification of microsclerotia in the soil and their extension into the plants as follows: 100 mg of the soil was pestled in 1 ml of 0.85% NaCl solution and then was diluted with the same concentration of NaCl solution. The mixture was placed on PDA medium amended with 100 μg ml−1 hygromycin B and the number of microsclerotia was assessed following the procedure of Guo et al. (2011). To recover V. dahliae from the plants, the stem was sectioned in 2-cm-long fragments from the base to the first node and washed under tap water. The fragments were surface sterilized with 75% ethanol, then in 1% NaClO for 1 min, followed by 3 washes with dH2O. The fragments were then cut into 2–3 mm cross-sectional cuttings. Ten cuttings were placed on every PDA plate amended with 100 μg ml−1 hygromycin B and incubated at 28 °C for 5 days. Plant colonization ability of V. dahliae where the soil was amended with different concentrations of the extract was calculated by comparing the number of the stem cuttings from which the fungus germinated with those which the fungus was not germinated (Zhang et al. 2011).
Statistical analysis
Statistical analysis was performed using ANOVA, and the significance of the difference between the treatments was determined by fisher’s protected least significant difference PLSD (statView 5.0.1). P < 0.05 was considered significant.
Results and discussion
Isolation of endophytic bacteria and their antagonistic activity in vitro assay
In the present investigation, overall, 66 endophytic bacteria were extracted from healthy cotton’s roots and their antagonistic potential was evaluated in vitro assay in modified agar diffusion method. Out of them, the isolate, which had the highest suppressive ability on V. dahliae mycelia in vitro was selected for this study. When the isolate suspension was added to the wells containing V. dahliae mycelia, a distinctive zone of inhibition was observed around the bacterial suspension (Fig. 1 a). In contrast, the wells, which were exposed with only liquid LB medium as control, were normal and the mycelia were growing normally. Microscopic observation of fungal mycelia within the isolate interaction zone (treated-mycelia) showed changes in morphology such as enlargement, melanization, and septa were observed in the swollen mycelia (Fig. 1 c). The mycelia from the interaction zone (treated-mycelia) lost their virulence against plant tissue and were non-pathogenic, which could not form germ-tube and seemed to be inactive on the surface of the leaves. In contrast, the mycelia from the isolate-free zone or untreated-mycelia (control) were morphologically normal without any abnormal changes and were highly virulent which germinated on the spotted part of the leaves, germ tube elongated, and formed new hyphae. Because of the high pathogenicity of the untreated-mycelia, the spotted area became black and necrotic around the germinating mycelia (Fig. 1b). The findings were correlated as documented by Kevan et al. (2011) who demonstrated that some endophytic bacteria are antagonistic against plant pathogens, which could be used as effective bioagents in plants pest and diseases control. The obtained results confirm the report of Tjamos (2000) who researched several antagonistic bacteria due to their control capacity of fungal microsclerotia and mycelia which are the primary targets regarding control measures and capacity, and they are essential for the initial infection, host colonization, and fungus survival.
Antifungal activity of the isolate against V. dahliae. a The antagonistic activity of the isolate in plate assay. (a) The black arrowhead indicates the bacterial interaction zone (b) Microscopic image of mycelia from bacteria free zone (CK). Bar = 10 μm. (c) Microscopic image of mycelia in the zone of interaction, the white arrowhead shows the swollen mycelia where septa are appearing. Bar = 10 μm. b Virulence reduction of mycelia against cotton leaves. The black arrow indicates the untreated mycelia germinated on the leaves; the white arrow indicates the necrosis of the spotted part of the leaf. The black arrowhead indicates the treated mycelia (CK) which has no germination ability
Strain identification and characterization
The total length of the partial amplified 16S rRNA gene nucleotide sequence was 980 bp and its identity percentage was 99% with other nucleotides in NCBI. The phylogenetic tree analysis showed the evolutionary relationship between the isolate in our study and other similar strains (Fig. 2). The isolate was identified as B. velezensis based on homology analyses and 16S rRNA gene sequences, which is a well-identified method for bacterial genus and species level identification (Shen et al. 2004) and was adjacent as B. velezensis SZAD2. The physical, morphological, and physiological characterizations of the isolate are shown in Table 1.
Bio-efficacy under greenhouse conditions
As in vitro experiment has certain limitations in biocontrol effectiveness, which could not be used as an alternative way instead of planta conditions (Inam-ul-Haq et al. 2003). Therefore, the biocontrol potential of the strain was assessed under greenhouse conditions. Seeds treatment of cotton with strain SZAD2 caused a remarkable reduction of chlorosis, necrosis, and defoliation. The leaf wilt index (LWI) in control plants was 62.49%, while in seed treated plants, the LWI reduced to 24.8% and the biocontrol efficacy of the isolate was 60.31%. In soil drench method, the LWI was significantly reduced to 13% and the biocontrol efficacy was 79.197%, indicating that strain SZAD2 was more effective in the soil drench method than seed treatment. These results are in accordance with previous studies reporting some B. velezensis strains as potential candidates for biocontrol under greenhouse conditions, such as B. velezensis strain NKG-2 had shown biocontrol potential against Botrytis cinerea in tomatoes (Myo et al. 2019), and our findings had indicated that the correlation between in vitro and in planta was correspondent as documented by Gupta et al. (2010).
Lateral roots stimulation, plant defense response, and internal colonization
Seed treatment caused to remarkably stimulate and developed lateral roots in cotton seedlings (Fig. 3a). Furthermore, the endophytic isolate caused to activate plant defense response by H2O2 accumulation both in the leave and in the roots. Spraying of cotton leaves with bacterial solution caused H2O2 accumulation in the leaves, which was seen as brown coloration under light microscope (Fig. 3b). In addition, roots treatment with bacterial solution caused systemically colonization of the roots and enhanced plant defense response by the accumulation of H2O2 in the roots. ROS staining of cotton roots showed the existence of bacterial infection close to the surface of hair roots and seemed to be located within the cells of the roots. Roots cells showed the internal presence of bacteria stained brown indicative presence of hydrogen peroxide in the tissues (Fig. 3c). In contrast, untreated roots had no evidence of bacterial infection and had no signs of H2O2 production. Transmission electron microscope analysis showed the precise location of the strain, and that B. velezensis SZAD2 cells systemically colonized cotton roots and were mainly located within the intercellular spaces of the roots (Fig. 4). The obtained result was similar to Low and Merida (1996) who reported that plant tissues produce H2O2 during the avirulent interaction of symbiotic endophytic bacteria and fungi with plants, which has a key role in plant defense response. The findings were in accordance with Chin-A-Woeng et al. (1997) who demonstrated that the root colonization is the important factor needed to screen or select strains possessing good biocontrol activity which delivers the antagonistic bacteria at the right place in the plants. The result was parallel to Quadt-Hallmann et al. (1996) who studied bacterial colonization and localization inside the plants using TEM which is a well-known technique for the bacteria internal colonization and localization study and reported that endophytic bacteria live and spread inside the plants. Roy et al. (2013) defined that some beneficial microbes are well-recognized for their ability to modulate root growth and development.
Lateral roots stimulation and plant defense response mediate by the isolate. a Lateral roots stimulation in seedlings. Bar = 100 μm. b H2O2 assessment in the leaves, the white arrowhead shows ROS accumulation along the veins. Bar = 50 μm. c Roots colonization and H2O2 assessment, the black arrow indicates the isolate within the roots. Bar = 100 μm
Increase in plant systemic resistance
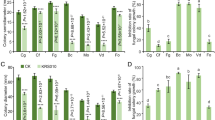
Bacterization of cotton seeds significantly expressed a higher amount of defense enzymes in the root such as PAL, POD, PPO, and phenol contents. Although, the defense enzymes’ activities were also increased in non-bacterized plant inoculated with V. dahliae, but their activities in bacterized plants were much higher than non-bacterized plants (Fig. 5). PAL activity began to increase after 24 h and reached its peak 48 h after inoculation. PAL activity in bacterized plants was transient and was almost two times higher than in non-bacterized plants. POD activity was also higher in bacterized plants then non-bacterized plants, and its activity reached its peak after 96 h of challenging inoculation. PPO activity was significantly higher in bacterized plants then non-bacterized plants, which reached its peak after 72 h. Also, a significant increase in phenol contents was expressed in bacterized plants. The accumulation of phenol contents reached to 332.98 mg/g after 48 h of inoculation. The results were correlated as recorded by Ramamoorthy et al. (2001) who reported that inducing the plants’ resistance by application of a biological inducer is a novel plant protection strategy. Increase in activity of PAL (Raj et al. 2003), PPO (Nandakumar et al. 2002), POD (Chen et al. 2000), and phenol accumulation (Meena et al. 2000) have been well-investigated induced by antagonistic bacteria in plant tissues against various pathogens which are associated with ISR.
Hydrolytic enzyme activity
The isolate was examined for hydrolytic enzyme production. Respectively, it produced both cellulase and chitinase based on the clearance zone on agar plates contained CMC or chitin (Fig. 6). Obtained result was similar as noted by Mohammed (2020) who used CMC and chitin for cellulase and chitinase activity tests.
Antifungal potential of the secondary extracts and their active components in different time of incubation
The antifungal activity of the supernatants began at 48 h and reached its peak in 72 h of incubation but lost their antifungal potential when the isolate was incubated in the culture medium for 96 h, 120 h, or longer (Fig. 7a). The secondary extracts isolated from the supernatants fluid in different time intervals had also different antifungal potential and had different colors. The extract isolated in 48 h of incubation had black yellowish color and had very less antifungal activity but had the highest antifungal potential in 72 h of incubation which had a bright yellowish color. In contrast, the extract isolated in 96 and 120 h of incubation had no antifungal activity, which had gray and dark colors (Fig. 7b). To determine and estimate the active components exist in the secondary extracts obtained in different time intervals, cellulase, and chitinase-specific activities were examined. The specific activity of cellulase enzyme 48 h after incubation was 0.058 U/ml and reached to its peak by day 3 (72 h after incubation) (3.1 U/ml), then continuously reduced its activity by day 4 and by day 5. Chitinase activity also began 48 h after the culture (0.57 U/ml) and reached to its peak by day 3 (72 h) (1.69 U/ml). Then, reduced its activity by day 4 and by day 5 (Fig. 8). As the secondary extract obtained from the supernatant fluid of the isolate in 72 h of incubation had the highest activity of hydrolytic enzymes such as cellulase and chitinase and also had the highest antagonistic potential against V. dahliae in Petri dish, so we speculated them as active secondary extract which is complex hydrolytic enzymes. In this study, the specific activities of cellulase and chitinase indicated that the active components of the secondary extract are complex hydrolytic enzymes such as cellulase and chitinase as recorded by Ashwini and Srividya (2014).
Antifungal potential of the supernatants and their crude extracts in different time interval of incubation. a Antifungal ability of the supernatants after incubating the strain in LB medium for different times. The clear area indicates the inhibited area. b The color difference of the crude extracts in different time interval
Suppressive potential of the hydrolytic enzyme extract against V. dahliae
Number of living spores was much lower in V. dahliae liquid culture contained hydrolytic enzyme extract than control (CK). After 5 days, the number of living spores/ml in control where no active extract was added reached to 15. 103 spores/ml, but when 25 μg ml−1 and 50 μg ml−1 active enzyme extracts were added the number of living spores was 4 (103) and 1.5 (103)/ml, which indicated that the active extract from B. velezensis SZAD2 can directly suppress V. dahliae spores and reduce its germination and viability (Fig. 9a).
Suppressive effects of the active extracts in the presence and absence of cotton a Antagonistic potential of the active extract in vitro. CK has white color which shows the high number of mycelia. In contrast, the treated group has a small number of mycelia. b Antagonistic potential of the active extract in the presence of cotton. The number of stem cuttings from which fungus grew was used to determine the extent of fungal colonization (a) CK (b), and (c) 25 and 50 μg ml−1 active extract was poured into the soil. The white arrows indicate leaf defoliation; the black arrows indicate the stems from which the fungi grew
The greenhouse experiment showed that the active extract had highly suppressive potential against V. dahliae microsclerotia in the presence of cotton. The number of microsclerotia in the soil amended with 25 and 50 μg ml−1 active extract was significantly lower than the control soil. In the control group, the amount of fungal microsclerotia reached to 202 × 102 CFU/g of soil, while the number of microsclerotia was 141 CFU/g × 102 and 7 × 1002 CFU/g, when the soil was amended with 25 and 50 μg ml−1 active enzyme extract. Further analysis showed that soil treatment with the active extract significantly reduced the extent of V. dahliae colonization in cotton plants, which caused the symptoms and disease severity reduction in plants, and only the plants, which were grown in control soil (without the active enzyme extract) showed severe wilt and defoliation. The number of stem fragments from which the fungus grew was much lower in treated soil plants than in the CK group. In control plants, 9 out of 10 stem fragments showed colonies appearance. While in treated plants where the soil was amended with 25 μg ml−1 and 50 μg ml−1 active enzyme extract, only 4 and 1 stem segments showed fungal colonies appearance (Fig. 9b). In addition, treatment of soil with 25 μg ml−1 and 50 μg ml−1 of active extract reduced significantly leaf wilt symptoms, which demonstrated 61.90 and 87.96% biocontrol efficacies against V. dahliae in cotton. These results in the present study indicated that the active extract from the strain SZAD2, which is a complex hydrolytic enzyme, caused to suppress the microsclerotia and reduce their number both in vitro and soil or in the presence/absence of cotton plants, which has been proved to be an effective mechanism in controlling the fungal pathogens (Chet et al. 1990). Ahmadi et al. (2008) reported that a wide range of organisms including bacteria are important for their hydrolytic enzymes production such as cellulase and chitinase, which are produced by bacteria and have the most important roles in biological control of pathogenic fungi and play a major role in degrading fungal cell wall. These findings were in a close correlation with Rais et al. (2018) who reported that the most important factor for a better bioagent against soil-borne diseases is to produce antibiosis and lysis, which kill the pathogen and inhibit its proliferation in host tissue.
Conclusion
The study revealed that the strain SZAD2, which was isolated from cotton roots and was identified by 16S rRNA gene sequencing analysis had a significantly high biocontrol potential against V. dahliae. The strain systemically colonized the roots and caused the production of H2O2 both in the roots and leaves, and also activated antioxidant enzyme such as PAL, PPO, POD, and the accumulation of phenol contents in the roots, which indicated that it can stimulate the basal defense response and induced systemic resistance. In addition, the secondary extract produced by the strain had the highest activity of cellulase and chitinase which reduced the number of mycelia both in the presence and absence of cotton. This study is the first report regarding B. velezensis SZAD2 biocontrol potential against Verticillium wilt in cotton, which demonstrated multiple modes of action, and it could be a potential candidate as a new bioagent in management of this critical soil-borne fungus and could be used for commercial exploration.
Availability of data and materials
The datasets analyzed during the current study are available from the corresponding author on a reasonable request.
Abbreviations
- TEM:
-
Transmission electron microscope
- ISR:
-
Induced systemic resistance
- PAL:
-
Phenylalanine ammonia-lyase
- PPO:
-
Polyphenol oxidase
- POD:
-
Peroxidase
- PDA:
-
Potato dextrose agar
- PDB:
-
Potato dextrose agar broth
- LB:
-
Luria Bertani
- dH2O:
-
Distilled water
- NCBI:
-
National Center for Biotechnology Information
- CMC:
-
Carboxy methylcellulose
- LWI:
-
Leaf wilt index
- ROS:
-
Reactive oxygen species
- H2O2 :
-
Hydrogen peroxide
- DAB:
-
Diaminobenzidine tetrachloride
- OD:
-
Optical density
- DNS:
-
Dinitrosalicylic acid
- IAA:
-
Indole-3-acetic acid
- rTaq:
-
ReadyMixTM Taq
- ANOVA:
-
One-way analysis of variance
- PLSD:
-
Protected least significant difference
References
Ahmadi K, Yazdi MT, Najafi MF, Shahverdi A, Faramarzi M, Zanini G, Behrava J (2008) Isolation and characterization of a chitionolytic enzyme producing microorganism, Paenibacillus chitinolyticus JK2 from Iran. Res J Microbiol 3(6):395–404
Annamalai N, Veeramuthu Rajeswari M, Vijayalakshmi S, Balasubramanian T (2011) Purification and characterization of chitinase from Alcaligenes faecalis AU02 by utilizing marine wastes and its antioxidant activity. Ann Microbiol 61(4):801–807. https://doi.org/10.1007/s13213-011-0198-5
Ashwini N, Srividya S (2014) Potentiality of Bacillus subtilis as biocontrol agent for management of anthracnose disease of chilli caused by Colletotrichum gloeosporioides OGC1. 3. Biotech 4(2):127–136. https://doi.org/10.1007/s13205-013-0134-4
Balouiri M, Sadiki M, Ibnsouda SK (2016) Methods for in vitro evaluating antimicrobial activity: A review. J Pharm Anal 6(2):71–79. https://doi.org/10.1016/j.jpha.2015.11.005
Chen C, Belanger RRV, Benhamou N, Paulitz T (2000) Defense enzymes induced in cucumber roots by treatment with plant growth promoting rhizobacteria (PGPR) and Pythium aphanidermatum. Physiological and Molecular Plant Pathology 56:13–23
Chet I, Ordentlich A, Shapira R, Oppenheim A (1990) Mechanisms of biocontrol of soil-borne plant pathogens by Rhizobacteria. Plant and Soil 129(1):85–92. https://doi.org/10.1007/BF00011694
Chin-A-Woeng TFC, de Priester W, van der Bij AJ, Lugtenberg BJJ (1997) Description of the colonization of a gnotobiotic tomato rhizosphere by Pseudomonas fluorescens Biocontrol Strain WCS365, Using Scanning Electron Microscopy. Molecular Plant-Microbe Interactions® 10(1):79–86. https://doi.org/10.1094/MPMI.1997.10.1.79
Daayf F, Bel-Rhlid R, Bélanger RR (1997) Methyl ester ofp-coumaric acid: a phytoalexin-like compound from long English cucumber leaves. J Chem Ecol 23(6):1517–1526. https://doi.org/10.1023/B:JOEC.0000006419.22408.65
De Gara L, Locato V, Dipierro S, de Pinto MC (2010) Redox homeostasis in plants. The challenge of living with endogenous oxygen production. Respiratory Physiology & Neurobiology 173: S13-S19. doi:https://doi.org/10.1016/j.resp.2010.02.007
Dias MC (2012) Phytotoxicity: an overview of the physiological responses of plants exposed to fungicides. Journal of Botany 1-4. doi:https://doi.org/10.1155/2012/135479
Dickerson DP, Pascholati SF, Hagerman AE, Butler LG, Nicholson RL (1984) Phenylalanine ammonia-lyase and hydroxycinnamate: CoA ligase in maize mesocotyls inoculated with Helminthosporium maydis or Helminthosporium carbonum. Physiological Plant Pathology 25(2):111–123. https://doi.org/10.1016/0048-4059(84)90050-X
Edwards U, Rogall T, Blocker H, Emde M, Bottger EC (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17(19):7843–7853. https://doi.org/10.1093/nar/17.19.7843
El-Zik KM (1985) Integrated Control of Verticillium Wilt of Cotton. Plant Disease 69 (12):1025–1032.
Fatima S, Anjum T (2017) Identification of a potential ISR determinant from pseudomonas aeruginosa PM12 against fusarium wilt in tomato. Front Plant Sci 8:848. https://doi.org/10.3389/fpls.2017.00848
Fradin EF, Thomma BP (2006) Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Molecular plant pathology 7(2):71–86
Göre ME, Caner ÖK, Altın N, Aydın MH, Erdoğan O, Filizer F, Büyükdöğerlioğlu A (2009) Evaluation of cotton cultivars for resistance to pathotypes of Verticillium dahliae. Crop Protection 28(3):215–219. https://doi.org/10.1016/j.cropro.2008.10.004
Guo M, Chen Y, Du Y, Dong Y, Guo W, Zhai S, Zheng X (2011) The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog 7(2):e1001302. https://doi.org/10.1371/journal.ppat.1001302
Gupta A, Kishore K, Bhardwaj S, Aman T, Sapna D, Jarial R, Laj R (2010) Biological control of crown gall on peach and cherry rootstock colt by native Agobacterium radiobacter isolates. Open Horticult J 3:1–10
Hammerschmidt R, Nuckles EM, Kuć J (1982) Association of enhanced peroxidase activity with induced systemic resistance of cucumber to Colletotrichum lagenarium. Physiological Plant Pathology 20(1):73–82. https://doi.org/10.1016/0048-4059(82)90025-X
Han Q, Wu F, Wang X et al (2015) The bacterial lipopeptide iturins induce Verticillium dahliae cell death by affecting fungal signalling pathways and mediate plant defense responses involved in pathogen-associated molecular pattern-triggered immunity. Environ Microbiol 17(4):1166–1188. https://doi.org/10.1111/1462-2920.12538
Han Y, Li P, Gong S, Yang L, Wen L, Hou M (2016) Defense Responses in Rice Induced by Silicon Amendment against Infestation by the Leaf Folder Cnaphalocrocis medinalis. PLoS One 11(4):e0153918. https://doi.org/10.1371/journal.pone.0153918
Heale JB, Isaac I (1965) Environmental factors in the production of dark resting structures in Verticillium alboatrum, V. dahliae and V. tricorpus. Transactions of the British Mycological Society 48(1): 39-IN37. doi:https://doi.org/10.1016/S0007-1536(65)80005-5
Huang J, Li H, Yuan H (2006) Effect of organic amendments on Verticillium wilt of cotton. Crop Protection 25(11):1167–1173. https://doi.org/10.1016/j.cropro.2006.02.014
Inam-ul-Haq M, Javed M, Ahmad R, Rehman A (2003). Evaluation of different strains of Pseudomonas fluorescens for the biocontrol of fusarium wilt of chick pea. Pakistan J. Plant Pathol 2: 65-74. Retrieved from DOI: https://doi.org/10.3923/ppj.2003.65.74
Joe S, Sarojini S Mapana (2017) An Efficient Method of Production of Colloidal Chitin for Enumeration of Chitinase Producing Bacteria. Journal of Sciences 4 (16) 37-45. Doi: org/10.12723/mjs.43.4
Kamlage B (1996) Methods for general and molecular bacteriology. Edited by P. Gerhardt, RGE Murray, WA Wood and NR Krieg 791 pages, numerous figures and tables. American Society for Microbiology Washington, D.C, 1994. Price: 55.00 £. Food / Nahrung, 40(2), 103-103. doi:https://doi.org/10.1002/food.19960400226
Kavino M, Harish S, Kumar N, Saravanakumar D, Samiyappan R (2007) Induction of systemic resistance in banana (Musa spp.) against Banana bunchy top virus (BBTV) by combining chitin with root-colonizing Pseudomonas fluorescens strain CHA0. European Journal of Plant Pathology 120(4):353–362. https://doi.org/10.1007/s10658-007-9223-8
Kevan P-G, Shipp L (2011), Biological control as biotechnological amelioration and ecosystem intensification in managed ecosystems. Comprehensive Biotechnology 4: 757-761. doi.org/https://doi.org/10.1016/B978-0-12-809633-8.09246-3
Kim S-K, Kim Y-T, Byun H-G, Park P-J, Ito H (2001) Purification and characterization of antioxidative peptides from bovine skin. BMB Reports 34(3):219–224
Kumar V, Parkhi V, Kenerley CM, Rathore KS (2009) Defense-related gene expression and enzyme activities in transgenic cotton plants expressing an endochitinase gene from Trichoderma virens in response to interaction with Rhizoctonia solani. Planta 230(2):277–291. https://doi.org/10.1007/s00425-009-0937-z
Li CH, Shi L, Han Q, Hu HL, Zhao MW, Tang CM, Li SP (2012) Biocontrol of verticillium wilt and colonization of cotton plants by an endophytic bacterial isolate. J Appl Microbiol 113(3):641–651. https://doi.org/10.1111/j.1365-2672.2012.05371.x
Low PS, Merida JR (1996) The oxidative burst in plant defense: Function and signal transduction. Physiologia Plantarum 96(3):533–542. https://doi.org/10.1111/j.1399-3054.1996.tb00469.x
Mayer AM, Harel E, Ben-Shaul R (1966) Assay of catechol oxidase—a critical comparison of methods. Phytochemistry 5(4):783–789. https://doi.org/10.1016/S0031-9422(00)83660-2
Meena B, Radhajeyalakshmi R, Marimuthu T, Vidhyasekaran P, Doraiswamy S, & Velazhahan R (2000) Induction of pathogenesis-related proteins, phenolics and phenylalanine ammonia-lyase in groundnut by Pseudomonas fluorescens / Induktion von Pathogenese-assoziierten Proteinen, Phenolen und Phenylalaninammonium-Lyase in Erdnüssen durch Pseudomonas fluorescens. Zeitschrift für Pflanzenkrankheiten und Pflanzenschutz. Journal of Plant Diseases and Protection 107(5): 514-527. Retrieved from www.jstor.org/stable/43215355
Miller GL (1959) Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Analytical Chemistry 31(3):426–428. https://doi.org/10.1021/ac60147a030
Mohammed AF (2020) Optimization of cellulase and chitinase enzymes production by plant growth promoting rhizobacteria. Novel Research in Microbiology Journal 4(1): 641-652. DOI: 10.21608/nrmj.2020.73437
M'piga P, Belanger R, Paulitz T, Benhamou N (1997) Increased resistance to Fusarium oxysporumf. sp. radicis-lycopersiciin tomato plants treated with the endophytic bacterium Pseudomonas fluorescens strain 63-28. Physiol Molecular Plant Pathol 50(5):301–320
Myo EM, Liu B, Ma J, Shi L, Jiang M, Zhang K, Ge B (2019) Evaluation of Bacillus velezensis NKG-2 for bio-control activities against fungal diseases and potential plant growth promotion. Elsevier BV 134:23–313. https://doi.org/10.1016/j.biocontrol.2019.03.017.
Nandakumar R, Babu S, Radjacommare R, Raguchander T, Samiyappan R (2002) Pseudomonas fluorescens mediated antifungal activity against Rhizoctonia solani causing sheath blight in rice. Phytopathologia Mediterranea 41(2):109–119
Pick E, Keisari Y (1980) A simple colorimetric method for the measurement of hydrogen peroxide produced by cells in culture. Journal of immunological methods 38(1-2):161–170
Quadt-Hallmann A, Hallmann J, Kloepper JW (1996) Bacterial endophytes in cotton: location and interaction with other plant-associated bacteria. Can J Microbiol 43(3):254–259
Quadt-Hallmann A, Kloepper JW, Benhamou N (1997) Bacterial endophytes in cotton: mechanisms of entering the plant. Canadian Journal of Microbiology 43(6):577–582. https://doi.org/10.1139/m97-081
Rais A, Shakeel M, Malik K, Hafeez FY, Yasmin H, Mumtaz S, Hassan MN (2018) Antagonistic Bacillus spp. reduce blast incidence on rice and increase grain yield under field conditions. Microbiological Research 208:54–62. https://doi.org/10.1016/j.micres.2018.01.009
Raj SN, Deepak SA, Basavaraju P, Shetty HS, Reddy MS, Kloepper JW (2003) Comparative performance of formulations of plant growth promoting rhizobacteria in growth promotion and suppression of downy mildew in pearl millet. Crop protection 22(4):579–588. https://doi.org/10.1016/s0261-2194(02)00222-3
Rajendran L, Samiyappan R (2008a) Endophytic Bacillus Species Confer Increased Resistance in Cotton Against Damping off Disease Caused by Rhizoctonia solani. Plant Pathology Journal 7(1):1–12
Rajendran L, Samiyappan R (2008b) Endophytic Bacillus species confer increased resistance in cotton against damping off disease caused by Rhizoctonia solani. Plant Pathology Journal 7(1):1–12. https://doi.org/10.3923/ppj.2008.1.12
Ramamoorthy V, Viswanathan R, Raguchander T, Prakasam V, Samiyappan R (2001) Induction of systemic resistance by plant growth promoting rhizobacteria in crop plants against pests and diseases. Crop protection 20(1):1–11
Roy D, Panchal S, Rosa BA, Melotto M (2013) Escherichia coli O157:H7 induces stronger plant immunity than Salmonella enterica Typhimurium SL1344. Phytopathology 103(4):326–332. https://doi.org/10.1094/PHYTO-09-12-0230-FI
Santoyo G, Moreno-Hagelsieb G, Orozco-Mosqueda Mdel C, Glick BR (2016) Plant growth-promoting bacterial endophytes. Microbiol Res 183:92–99. https://doi.org/10.1016/j.micres.2015.11.008
Saowapar K, P-o Y, Taweesak T, Somboon T (2014) Screening and identification of cellulase producing bacteria isolated from oil palm meal. J App Pharm Sci 4(04):090–096. https://doi.org/10.7324/JAPS.2014.40416
Saravanakumar D, Vijayakumar C, Kumar N, Samiyappan R (2007) PGPR-induced defense responses in the tea plant against blister blight disease. Crop Protection 26(4):556–565. https://doi.org/10.1016/j.cropro.2006.05.007
Selim HMM, Gomaa NM, Essa AMM (2017) Application of endophytic bacteria for the biocontrol of Rhizoctonia solani (Cantharellales: ceratobasidiaceae) damping-off disease in cotton seedlings. Biocontrol Science and Technology 27(1):81–95. https://doi.org/10.1080/09583157.2016.1258452
Shen DX, Du J Fau - Feng Z.-c, Feng ZC (2004). Rapid diagnosis of common pathogenic bacteria infection in newborn infants by 16SrDNA oligonucleotide array. Zhonghua Er Ke Za Zhi 42(9): (0578-1310 (Print)) 668-672
Spaepen S, Vanderleyden J (2011) Auxin and plant-microbe interactions. Cold Spring Harb Perspect Biol 3(4). https://doi.org/10.1101/cshperspect.a001438
Tjamos E (2000) Strategies in developing methods and applying techniques for the biological control of Verticillium dahliae. Advences in Verticillium Research and Disease Management:227–231
Vaikuntapu PR, Dutta S, Samudrala RB, Rao VR, Kalam S, Podile AR (2014) Preferential promotion of Lycopersicon esculentum (Tomato) growth by plant growth promoting bacteria associated with tomato. Indian J Microbiol 54(4):403–412. https://doi.org/10.1007/s12088-014-0470-z
Wang T, Liang Y, Wu M, Chen Z, Lin J, Yang L (2015) Natural products from Bacillus subtilis with antimicrobial properties. Chinese Journal of Chemical Engineering 23(4):744–754. https://doi.org/10.1016/j.cjche.2014.05.020
Wen C-Y, Yin Z-G, Wang K-X, chen J-G, Shen S-S (2011) Purification and structural analysis of surfactin produced by endophytic Bacillus subtilis EBS05 and its antagonistic activity against Rhizoctonia cerealis. Plant Pathol. J 27(4): 342-348.
Xiao CL, Subbarao KV, Schulbach KF, Koike ST (1998) Effects of Crop Rotation and irrigation on Verticillium dahliae microsclerotia in soil and wilt in cauliflower. Phytopathology™ 88(10):1046–1055. https://doi.org/10.1094/PHYTO.1998.88.10.1046
Yang C, Guo W, Li G, Gao F, Lin S, Zhang T (2008) QTLs mapping for Verticillium wilt resistance at seedling and maturity stages in Gossypium barbadense L. Plant Science 174(3):290–298. https://doi.org/10.1016/j.plantsci.2007.11.016
Zhang Y, Wang X, Yang S, Chi J, Zhang G, Ma Z (2011) Cloning and characterization of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana. Plant Cell Reports 30(11):2085. https://doi.org/10.1007/s00299-011-1115-x
Zhang Y-L, Li Z-F, Feng Z-L, Feng H-J, Shi Y-Q, Zhao L-H et al (2016) Functional analysis of the pathogenicity-related gene VdPR1 in the vascular wilt fungus Verticillium dahliae. PLoS ONE 11(11):e0166000. https://doi.org/10.1371/journal.pone.0166000
Zieslin N, Ben-Zaken R (1993) Peroxidase activity and presence of phenolic substances in peduncles of rose flowers. Plant physiology and biochemistry (Paris) 31(3):333–339
Acknowledgements
N/A
Funding
The project was funded by Nanjing Agricultural University and was fully supported by the Chinese Scholarship Council (CSC) (2016GXYE71) for providing the master scholarship for this study.
Author information
Authors and Affiliations
Contributions
SZ performed most of the experimental work and analyzed the data. CT critically revised the article. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethic approval and consent to participate
N/A
Consent for publication
N/A
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sherzad, Z., Canming, T. A new strain of Bacillus velezensis as a bioagent against Verticillium dahliae in cotton: isolation and molecular identification. Egypt J Biol Pest Control 30, 118 (2020). https://doi.org/10.1186/s41938-020-00308-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41938-020-00308-y