Abstract
Background
The epidemiology of enteric pathogens has not been well studied in Kenya because of wide disparities in health status across the country. Therefore, the present study describes the prevalence of enteropathogenic bacteria, their seasonal variation, and antibiotic resistance profiles among hospitalized diarrheic children in a suburban region of central Kenya.
Methods
Fecal samples were collected between July 2009 and December 2013 from a total of 1410 children younger than 5 years, hospitalized with acute diarrhea in Kiambu County Hospital, Kenya. Conventional culture, biochemical, and molecular methods were conducted to identify causative bacterial pathogens and their virulence factors. Antimicrobial susceptibility tests were performed using E-test strips and VITEK-2 advanced expert system (AES) to evaluate the drug-resistance pattern of the isolates.
Results
Of the 1410 isolates, bacterial infections were identified in 474 cases. Diarrheagenic Escherichia coli (DEC) was the most frequently isolated pathogen (86.5%). Other pathogens such as Aeromonas (5.5%), Shigella (4%), Salmonella (3.4%), Providencia (3.2%), Vibrio spp. (1.1%), Yersinia enterocolitica (1.1%), and Plesiomonas shigelloides (0.2%) were also identified. Mixed bacterial infection was observed among 11.1% of the cases. The highest infection rate was found during the dry season (59.3%, p = 0.04). Most of the DEC was found to be multidrug resistant to trimethoprim/sulfamethoxazole 97.6%, amoxicillin 97.6%, erythromycin 96.9%, ampicillin 96.6%, and streptomycin 89%.
Conclusions
This study suggests that DEC is the leading diarrhea-causing bacterial pathogen circulating in central Kenya, and seasonality has a significant effect on its transmission. Proper antibiotic prescription and susceptibility testing is important to guide appropriate antimicrobial therapy.
Similar content being viewed by others
Background
Despite significant advances in the understanding of pathogenesis and management, globally acute diarrhea remains the second leading cause of death in children under 5 years of age [1]. It is estimated that globally, 800,000 children under the age of 5 years die annually, mostly in sub-Saharan Africa and South Asia [2]. Currently, the risk of diarrheal diseases has increased fivefold in sub-Saharan Africa as compared to industrialized countries [3]. In sub-Saharan Africa including Kenya, urban migration is on the rise, resulting in rapid expansion of cities. Creation of overcrowding in urban slums, low socioeconomic conditions, poverty, illiteracy, inadequate safe drinking water, poor sanitation, and malnutrition have raised the occurrence of infectious diseases including diarrhea [4].
The major causes of diarrhea differ significantly in developed and developing countries. In the developed countries, diarrhea is seen in the winter with viruses being the primary agents. In the developing world, diarrhea remains a wet season disease with bacteria playing a greater role [5], although rotavirus has been found to be a single dominant enteric pathogen among children in most of the developed and developing countries [6]. Newly virulent enteric pathogens are emerging throughout the world including Kenya. For example, a multidrug-resistant enteroaggregative Escherichia coli (E. coli) O44, which is associated with acute and persistent diarrhea, has been reported in Kenya [7]. Recently, Providencia alcalifaciens was published for the first time as the etiological cause of diarrhea outbreaks in Kiambu [8], and E. coli O157 as a cause of significant outbreaks in Swaziland [9]. However, there is limited information on the surveillance of diarrheagenic pathogens and their antimicrobial resistance pattern. As a result, prevention, active intervention, and response are not well defined at the regional or national level.
The aim of this study was to identify the bacterial pathogens associated with diarrhea, its seasonal burden, and antimicrobial resistance patterns among hospitalized children in a suburban region of central Kenya.
Methods
Study area
This study was conducted between July 2009 and December 2013 at the Kiambu County Hospital (KCH). Kiambu County is located in the central highlands of Kenya in the former central province, close to Kenya’s capital, Nairobi, and has a population of 1,782,083. It has a 40% rural and 60% urban population owing to Nairobi’s consistent growth. KCH is a government referral hospital with a 316-bed capacity that offers a full range of services to people living mostly in the central region of the country. The rainfall pattern in Kenya is seasonally bimodal, with a long rainy season starting from March to May, while a shorter rainy season lasts from October to November.
Study population and specimen collection
A total of 1416 children aged 0–60 months who were hospitalized in the pediatric unit of KCH, with complaints of diarrhea (with or without fever or other accompanying symptoms) and had not taken any antimicrobial agent in the preceding week were recruited into this study. Diarrhea was defined as the passage of three or more watery, loose stools in the previous 24 h, as noted by the mother or the household caretaker. Fecal specimens were obtained from stools/rectal swabs and transported to the laboratory at the Centre for Microbiology Research in sterilized container/Cary-Blair transport medium (Liofilchem, Roseto degli Abruzzi, Italy) and processed within 24 h. Specimens obtained in the evening or at night were kept in a refrigerator at 2–8 °C and transported in a cool box the following day. The hospital staff completed a questionnaire for each patient containing the following information: age, sex, clinical symptoms (fever, vomiting, dehydration, etc.), and medical care sought before hospitalization.
Isolation and identification of pathogenic bacteria
All fecal specimens were cultured on xylose lysine deoxycholate (XLD) agar (Oxoid Ltd., Basingstoke, Hampshire, UK), deoxycholate hydrogen sulfide lactose (DHL) agar, bromothymol blue (BTB) agar, Salmonella-Shigella (SS) agar, thiosulfate-citrate-bile salt-sucrose (TCBS) agar (Eiken Chemical Company Ltd. Tochigi, Japan), and Campylobacter selective Butzler agar (Oxoid Ltd., Basingstoke, Hampshire, UK). In addition to these media, two types of enrichment media were used: alkaline peptone water and selenite broth (HiMedia Laboratories Pvt. Ltd., Mumbai, India). The plates and the broth with the exception of butzler were incubated overnight at 37 °C. Butzler plates were incubated under a micro-aerophilic condition with an Anaeropack® system (Mitsubishi Gas Chemical Company, Inc. Tokyo, Japan) at 42 °C for 48 h. Bacterial isolates were further confirmed by both routine biochemical method [10] and VITEK-2 automated analyzer (bioMérieux, Inc., Durham, NC, USA) system. For each subject, five lactose fermenting and two lactose non-fermenting colonies were subjected to direct PCR for E. coli virulence factor testing.
Serological identification
Serologic identification was performed by the slide agglutination technique with polyvalent and monovalent antisera for serotype identification of Vibrio cholerae, Salmonella, and Shigella spp. (Denka Seiken Co., LTD, Japan). Positive agglutination confirmed the serotype and subtype of the isolates. E. coli strain ATCC 25922 was used as a negative control.
Molecular examination
For genetic analysis, bacterial genomic DNA was extracted by the QIAGEN DNA extraction kit (QIAGEN, Hilden, Germany). Multiplex PCR was performed using published primers specific for genes targeting eae for enteropathogenic Escherichia coli (EPEC); eae and stx for enterohemorrhagic Escherichia coli (EHEC); est and elt for enterotoxigenic Escherichia coli (ETEC); ipaH for enteroinvasive Escherichia coli (EIEC) and Shigella spp.; aggR, CVD432, and aspU for enteroaggregative Escherichia coli (EAEC) [11]; hisJ for Salmonella spp. [12]; and toxR (toxR-F: 5′-TTAACGCTGAATTACATTCA-3′ and toxR-R: 5′-TTAAGATTTACTGAACAGTA-3′) and ctxA (ctxA-F: 5′-TCAATTAGTTTGAGAAGTGC-3′ and ctxA-R: 5′-TCAGATTGATAGCCTGAAAA-3′) for Vibrio spp. The PCR mixture was prepared with puReTaq Ready-To-Go PCR beads kit (GE Healthcare, UK) according to the manufacturer’s instructions.
Antimicrobial susceptibility tests
Antimicrobial susceptibility testing of all the 290 randomly selected isolates was performed by E-test (Liofilchem, Roseto degli Abruzzi, Italy) strips and the VITEK-2 advanced expert system (AES) using AST-GN83 cards (bioMérieux, Inc., Durham, NC, USA); minimal inhibitory concentrations (MICs) were determined according to the manufacturer’s instruction manual. The breakpoints used were those recommended by the Clinical and Laboratory Standard Institute [13]. Standard E. coli ATCC 25922 of known susceptibility was used as a control organism. Antibiotics tested included amoxicillin, ampicillin, ampicillin/sulbactam, amoxicillin/clavulanate, cefazolin, cefotaxime, cefoxitin, cefuroxime, cefuroxime/axetil, ceftriaxone, ceftazidime, cefepime, meropenem, aztreonam, erythromycin, doxycycline, tetracycline, ciprofloxacin, ofloxacin, chloramphenicol, amikacin, gentamicin, kanamycin, streptomycin, trimethoprim/sulfamethoxazole, and nitrofurantoin.
Statistical analysis
Frequencies and percentages were calculated for the study variables. Comparisons were drawn using a two-tailed Chi-square test. A p value of less than 0.05 was considered to be statistically significant.
Results
Characteristics of the study population
During the period between July 2009 and December 2013, a total of 1416 fecal samples were obtained from children hospitalized in KCH with acute diarrhea accompanied by vomiting (64.7%), dehydration (40.6%), and fever (28.7%) for rotavirus pre-vaccination surveillance study (Table 1). Six cases were excluded for missing age data. Remaining 1410 (773, 54.8% males and 637, 45.2% females) fecal samples were tested only for rotavirus and enteric bacterial pathogens. At least one enteric bacterial pathogen was detected in 318 (22.6%) diarrheal cases, and mixed pathogens (pathogenic bacteria/rotavirus) were found in 156 (11.1%) cases. Mixed bacterial infections were seen in association with rotaviruses (138 cases) except in 18 cases where two bacterial copathogens were identified (data not shown).
All patients were divided into six age groups (Table 1) with age ranging from 1 month to 5 years, and the highest proportion of bacterial infections (84.8%, 402/474) were observed within 0–18 months old, particularly within the age group of 7–12 months (47.7%). A similar result was also observed among the total number of diarrheal cases (84.1%, 1186/1410). This value was progressively decreased after 18 months.
Prevalence of pathogenic bacteria and seasonal variation
A wide range of bacterial pathogen was detected in 474 cases. Of these, diarrheagenic E. coli (DEC) (86.5%, 410/474) was found to be the most common enteropathogenic bacteria representing EAEC, ETEC, EPEC, EIEC, and EHEC. The distribution of different enteric bacterial pathogen is shown in Table 2. Of the DEC, EAEC (50%, 237/474) was the most prevalent of all bacterial cases of diarrhea, followed by ETEC (24.3%, 115/474), EPEC (11.6%, 55/474), EIEC (0.4%, 2/474), and EHEC (0.2%, 1/474). Among 237 isolated EAEC strains, 105 strains carried both aspU and aggR genes, 45 strains were found to be positive for aspU, aggR, and pCVD432 genes, while those that harbored aspU and aggR gene alone were 76 and 11 strains, respectively. Of the 115 ETEC strains, heat-stable toxin (ST) and heat-labile toxin (LT) genes were detected in 60 and 41 strains, respectively, and both ST and LT genes were found positive in 14 isolates. A total of 55 EPEC strains were positive for eae gene, while ipaH gene was found in all four EIEC strains and EHEC strain was positive for both eae and stx genes.
Other pathogens included Aeromonas (5.5%, 26/474), Shigella (4%, 19/474), Salmonella (3.4%, 16/474), Providencia spp. (3.2%, 15/474), Vibrio spp. (1.1%, 5/474), Yersinia enterocolitica (1.1%, 5/474), and Plesiomonas shigelloides (0.2%, 1/474). Among Shigella, S. flexneri (57.9%, 11/19), S. sonnei (26.3%, 5/19), S. boydii (10.5%, 2/19), and S. dysenteriae (5.3%, 1/19) were identified. Three of the five cases of Vibrio species were V. cholerae O1, and two were V. fluvialis confirmed by biochemical test and PCR amplification. There were 16 serotypes of Salmonella enterica strains including enteritidis (n = 14) and typhimurium (n = 2) serotypes. Campylobacter spp. was not found during the study period.
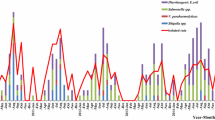
Seasonal variation in the occurrence of diarrheal infection was reported. It was observed that a significantly higher percentage (56.6%, 798/1410) of diarrheal cases were detected during the dry season (Dec–Feb and June–Sep) (p = 0.001). Also, the prevalence of bacterial infection was found to be higher in the dry season, 59.3% (281/474) than in the rainy season, 40.7% (193/474) (p = 0.04). A higher proportion of EAEC (60.3%; p = 0.002) and ETEC infection (65.2%; p = 0.124) was observed more frequently during the dry season. Relatively high prevalence of EPEC (56.4%; p = 0.297) infection occurred during the rainy season (Oct–Nov and Mar–May), but this difference was not statistically significant. Moreover, two EIEC and one EHEC strains were detected during the rainy season. Salmonella strains were isolated at almost similar frequencies in both seasons, but a higher rate of Shigella spp. (63.2%; p = 0.043) were detected during the dry season (Table 2).
Antimicrobial resistance
Table 3 summarizes the antimicrobial resistance pattern of different bacterial pathogens. Lower resistance rate was observed in carbapenems (3.4%), followed by quinolone (4.8%) and cephalosporin antibiotic groups (6.2–23.5%). Higher resistance was seen in penicillin groups such as amoxicillin (97.2%) and ampicillin (96.6%), followed by macrolide groups such as erythromycin (96.9%), and sulfonamides group such as trimethoprim/sulfamethoxazole (97.6%). Among these, most of the bacterial pathogens showed multidrug-resistant patterns.
Discussion
Diarrheal diseases are a major health concern for children in Kenya as well as other countries in sub-Saharan Africa. Knowledge of the circulating enteropathogen is essential for the implementation of appropriate public health measures to control diarrheal diseases. Currently, Global Enteric Multicenter Study (GEMS) is conducting a comprehensive study of childhood diarrheal diseases at seven sites in Africa and Asia, including western Kenya [14]. As a part of the rotavirus pre-vaccination surveillance study conducted in the suburban region of central Kenya, this is the first report investigating the prevalence, seasonal variation, and the antimicrobial resistance patterns of enteric bacterial pathogens, isolated from the hospitalized diarrheal children younger than 5 years old.
In our study, diarrheal infection was higher among the children in age groups within 0–18 months. This could be due to their underdeveloped immune system that is incapable of mounting an effective immunological response [15]. Also, risk of placing contaminated fingers and fomites in the mouth is greatly increased due to physiological phenomenon like teething and crawling which begins at this age [16]. On the other hand, less diarrheal infection was observed with increasing age groups which is consistent with previous studies [17]. This meant that poor immunity among children invariably due to under nutrition increases susceptibility to diarrheal diseases.
In this study, DEC was the predominant pathogen (86.5%, 410/474) identified in the screened fecal samples. We amplified eight targeted genes by PCR to identify five types of DEC. Among the strains of DEC isolates, EAEC (50%) was the most frequently identified potential pathogen followed by ETEC (24.3%), EPEC (11.6%), EIEC (0.4%), and EHEC (0.2%). The relative frequency of these categories of DEC was similar to that observed in Nigeria [18], Tanzania [19], and Mozambique [20]. Contrary to our findings, a study previously conducted in Kenya [21] and Ghana [22] showed that EPEC was the most frequently identified bacterial pathogen. This could have been due to the divergence of DEC pathotype, since it varies globally from region to region, and even between and within countries in an identical geographic location [23].
The aggR gene is a transcriptional activator gene required for the expression of the anti-aggregation protein (dispersin) gene aap (also called aspU) and an anti-aggregation protein transporter gene aatA (also known as CVD432 or AA prove) which have been commonly used for the detection of EAEC [11]. Furthermore, pathogenic studies have suggested that aggR gene controls the virulence functions in EAEC [24]. Therefore, strains expressing the aggR regulon are denoted as typical EAEC [25]. In most epidemiological studies on EAEC, typical EAEC strain has been mainly targeted because many investigators have found a strong association between these and diarrhea [19, 26, 27], which is consistent with our study. ETEC was reported as the second most common DEC isolated in this study. It is well known that ETEC strains cause diarrhea through the action of the enterotoxin LT and ST. Our findings showed a higher prevalence of ST- than LT-producing ETEC, which was also observed in previous studies [28]. Additionally, eae and the ipaH genes were used to detect EPEC and EIEC pathotypes, respectively, while both eae and stx genes were for EHEC [11].
Shigella, Salmonella, and Campylobacter spp. are the well-known pathogens that cause bacterial gastroenteritis in the world. Surprisingly, in the present study, an infrequent number of Shigella (4%) and Salmonella spp. (3.4%) were isolated than in a previously reported study carried out between May 1997 and April 1998 in western Kenya, which showed higher percentages of Shigella (44%), Campylobacter (30%), and Salmonella (14%) infection [29]. In a more recent study carried out in four provinces of Kenya, the isolation rate of Shigella (2.3%) and Salmonella (3.5%) were found similar to our study [21]. This highlights the declining proportion of diarrhea due to a probable improvement of water supply and sanitation in most parts of the country. Among bacterial pathogens, we also detected Aeromonas and Providencia spp., and Plesiomonas shigelloides which had not been reported previously in this study area.
Mixed infections were observed among 11.1% patients, and most of the cases, bacterial infections were associated with rotaviruses (data not shown). In developing countries, it is not uncommon to isolate more than one enteric pathogen from the same patient. The prevalence of co-infection in our patients is less than in Madagascar (22.8%) [30], but higher than in Egypt (5.9%) [31]. Therefore, the interpretation of data on mixed infection is complicated because not all the potential enteropathogens detected necessarily contribute to the etiology of the patient’s diarrheal disease. Due to the limitation in the study design, we were not able to analyze indicators of severe diarrhea in patients with co-infections that would have prompted a more extensive investigation.
Our results also brought out the effect of seasonal variation on the occurrence of diarrheal pathogens in the environment. A higher infection rate (56.6%) was observed during the dry seasons as compared to the rainy seasons (43.4%), which is in agreement with previous studies [28, 32]. During the dry seasons, shortage of water, high temperature, and dusty conditions are favorable to the proliferation of infectious agents [32]. The highest infection rate of diarrhea caused by ETEC (75/115, 65.2%), EAEC (143/237, 60.3%), and Shigella (12/19, 63.2%) was observed more frequently during the dry season while infection caused by EPEC (31/55, 56.4%) was observed during the rainy season. These findings are in agreement with the study conducted in Tanzania except that EPEC was isolated at similar frequencies in both seasons [28]. In contrast, a study in Bangladesh found a higher prevalence of ETEC and EPEC during the dry season [33]. However, it is documented that distinct seasonal patterns of diarrhea occur in many geographical areas. In a temperate climate, bacterial diarrhea occurred more frequently during the warm season, whereas viral diarrhea, particularly diarrhea caused by rotavirus, peaked during the winter. In tropical areas, rotavirus diarrhea occurred throughout the year, increasing in frequency during the drier, cold months, whereas bacterial diarrheas peaked during the warmer, rainy seasons [34].
In this study, 26 antibiotics were tested against randomly selected 290 DEC pathogens (Table 3). Most tested isolates showed more than 90% sensitivity to ciprofloxacin, ofloxacin, amikacin, kanamycin, and cefoxitin probably due to their seldom use [29]. On the contrary, a high proportion of amoxicillin (97.2%), ampicillin (96.6%), erythromycin (96.9%), trimethoprim/sulfamethoxazole (97.6%), streptomycin (89%), ampicillin/sulbactam (67%), and tetracycline (61.7%) resistance was observed. The high rate of resistance towards these antibiotics may be due to their indiscriminate use because they are readily available over the counter [29]. Furthermore, the majority of the E. coli isolates in our study displayed resistance to one or more antibiotics including amoxicillin, ampicillin, trimethoprim/sulfamethoxazole, erythromycin, and streptomycin, which was also observed in previous studies in Kenya [7].
Conclusions
The present study describes the burden of enteric bacterial pathogens in suburban settings of central Kenya. It highlights the involvement of EAEC diarrheal cases in Kiambu regions in a significant proportion and that EAEC may be an emerging pathogen. A high seasonal variation of bacterial infections is also reported. This study further provides updated information on the prevalence of other enteropathogens that have been circulating in this region, which can be useful in setting suitable health intervention strategies or informing future epidemiological studies on diarrheal pathogens in Kenya. The emergence of multidrug-resistant bacteria raises a broader health issue. Therefore, antibiotic treatment should be strictly controlled by the health authority. Continuous monitoring of the resistance pattern is mandatory for the appropriate selection of antimicrobial drugs.
Abbreviations
- BTB:
-
Bromothymol blue
- CLSI:
-
Clinical and Laboratory Standard Institute
- DEC:
-
Diarrheagenic E. coli
- DHL:
-
Deoxycholate Hydrogen Sulfide Lactose
- EAEC:
-
Enteroaggregative Escherichia coli
- EHEC:
-
Enterohemorrhagic Escherichia coli
- EIEC:
-
Enteroinvasive Escherichia coli
- ELISA:
-
Enzyme-linked immunosorbent assay
- EPEC:
-
Enteropathogenic Escherichia coli
- ETEC:
-
Enterotoxigenic Escherichia coli
- GEMS:
-
Global Enteric Multicenter Study
- KCH:
-
Kiambu County Hospital
- PCR:
-
Polymerase chain reaction
- SS:
-
Salmonella-Shigella
- TCBS:
-
Thiosulfate-citrate-bile salt sucrose
- XLD:
-
Xylose lysine deoxycholate
References
Walker CL, Aryee MJ, Boschi-Pinto C, Black RE. Estimating diarrhea mortality among young children in low and middle-income countries. Plos one. 2012;7:e29151.
Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–22.
Schiller LR. Management of diarrhea in clinical practice: strategies for primary care physicians. Rev gastrointest disord. 2007;7:27–38.
Ronak BP, Thomas FB. Urbanization—an emerging humanitarian disaster. N engl j med. 2009;361:741–3.
Mackenjee MKR, Coovadia YM, Coovadia HM, Hewitt J, Robins-Browne RM. Aetiology of diarrhea in adequately nourished African children in Durban, South Africa. Ann trop paediatr. 1984;4:183–7.
Nyaga MM, Jere KC, Esona MD, Seheri ML, Stucker KM, Halpin RA, et al. Whole genome detection of rotavirus mixed infections in human, porcine and bovine samples co-infected with various rotavirus strains collected from sub-Saharan Africa. Infect genet evol. 2015;31:321–34.
Sang WK, Oundo JO, Mwituria JK, Waiyaki PG, Yoh M, Iida T, et al. Multidrug-resistant enteroaggregative Escherichia coli associated with persistent diarrhea in Kenyan children. Emerg infect dis. 1997;3:373–4.
Shah MM, Odoyo E, Larson PS, Apondi E, Kathiiko C, Miringu G, et al. First report of a foodborne Providencia alcalifaciens outbreak in Kenya. Am j trop med hyg. 2015;93:497–500.
Effler E, Isaacson M, Arntzen L, Heenan R, Canter P, Barrett T, et al. Factors contributing to the emergence of Escherichia coli O157 in Africa. Emerg infect dis. 2001;7:812–9.
Kelly MT, Brenner DJ, Farmer JJ III. Enterobacteriaceae. In: Manual of clinical microbiology. 4th ed. Washington DC: ASM Press; 1985
Toma C, Lu Y, Higa N, Nakasone N, Chinen I, Baschkier A, et al. Multiplex PCR assay for identification of human diarrheagenic Escherichia coli. J clin microbiol. 2003;41:2669–71.
Cohen ND, Neibergs HL, Mcgruder ED, Whitford HW, Behle RW, Ray PM, et al. Genus-specific detection of salmonellae using the polymerase chain reaction (PCR). J vet diagn invest. 1993;5:368–71.
Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. Seventeenth Informational Supplement. CLSI Document M100-S17. Wayne: Clinical and Laboratory Standards Institute; 2007.
Kotloff KL, Blackwelder WC, Nasrin D, Nataro JP, Farag TH, van Eijk A, et al. The Global Enteric Multicenter Study (GEMS) of diarrheal disease in infants and young children in developing countries: epidemiologic and clinical methods of the case/control study. Clin infect dis. 2012;55:S232–45.
MAL-ED Network Investigators. The MAL-ED study: a multinational and multidisciplinary approach to understand the relationship between enteric pathogens, malnutrition, gut physiology, physical growth, cognitive development, and immune responses in infants and children up to 2 years of age in resource-poor environments. Clin infect dis. 2014;59:S193–206.
Shah MS, Ahmad A, Khalique N, Afzal S, Ansari MA, Khan Z. Home-based management of acute diarrhoeal disease in an urban slum of Aligarh, India. J infect dev ctries. 2012;6:137–42.
Jones TF, Mcmillian MB, Scallan E, Frenzen PD, Cronquist AB, Thomas S, et al. A population-based estimate of the substantial burden of diarrhoeal disease in the United States; FoodNet, 1996–2003. Epidemiol infect. 2007;135:293–301.
Onanuga A, Igbeneghu O, Lamikanra A. A study of the prevalence of diarrhoeagenic Escherichia coli in children from Gwagwalada, Federal Capital Territory, Nigeria. Pan afr med j. 2014;17:146.
Moyo SJ, Maselle SY, Matee MI, Langeland N, Mylvaganam H. Identification of diarrheagenic Escherichia coli isolated from infants and children in Dar es Salaam, Tanzania. BMC infect dis. 2007;7:92.
Rappelli P, Folgosa E, Solinas ML, Dacosta JL, Pisanu C, Sidat M, et al. Pathogenic enteric Escherichia coli in children with and without diarrhea in Maputo, Mozambique. FEMS immunol med microbiol. 2005;43:67–72.
Sang WK, Oundo V, Schnabel D. Prevalence and antibiotic resistance of bacterial pathogens isolated from childhood diarrhoea in four provinces of Kenya. J infect dev ctries. 2012;6:572–8.
Addy PA, Antepim G, Frimpong EH. Prevalence of pathogenic Escherichia coli and parasites in infants with diarrhoea in Kumasi, Ghana. East afr med j. 2004;81:353–7.
Shetty VA, Kumar SH, Shetty AK, Karunasagar I, Karunasagar I. Prevalence and characterization of diarrheagenic Escherichia coli isolated from adults and children in Mangalore, India. J lab physicians. 2012;4:24–9.
Harrington SM, Dudley EG, Nataro JP. Pathogenesis of enteroaggregative Escherichia coli infection. FEMS microbiol lett. 2006;254:12–8.
Nataro JP. Enteroaggregative Escherichia coli, Emerging infections 6. Scheld WM, Murray BE, Hughes JM, ed. Washington DC: ASM Press; 2004.
Sarantuya J, Nishi J, Wakimoto N, Erdene S, Nataro JP, Sheikh J, et al. Typical enteroaggregative Escherichia coli is the most prevalent pathotype among E. coli strains causing diarrhea in Mongolian children. J clin microbiol. 2004;42:133–9.
Hebbelstrup Jensen B, Olsen KE, Struve C, Krogfelt KA, Petersen AM. Epidemiology and clinical manifestations of enteroaggregative Escherichia coli. Clin microbiol rev. 2014;27:614–30.
Vargas M, Gascon J, Casals C, Schellenberg D, Urassa H, Kahigwa E, et al. Etiology of diarrhea in children less than five years of age in Ifakara, Tanzania. Am j trop med hyg. 2004;70:536–9.
Shapiro RL, Kumar L, Phillips-Howard P, Wells JG, Adcock P, Brooks J, et al. Antimicrobial-resistant bacterial diarrhea in rural western Kenya. J infect dis. 2001;183:1701–4.
Randremanana R, Randrianirina F, Gousseff M, Dubois N, Razafindratsimandresy R, Hariniana ER, et al. Case-control study of the etiology of infant diarrheal disease in 14 districts in Madagascar. Plos one. 2012;7:e44533.
El-Shabrawi M, Salem M, Abou-Zekri M, El-Naghi S, Hassanin F, El-Adly T, et al. The burden of different pathogens in acute diarrhoeal episodes among a cohort of Egyptian children less than five years old. Prz gastroenterol. 2015;10:173–80.
Podewils LJ, Mintz ED, Nataro JP, Parashar UD. Acute, infectious diarrhea among children in developing countries. Semin pediatr infect dis. 2004;15:155–68.
Albert MJ, Faruque SM, Faruque AS, Neogi PK, Ansaruzzaman M, Bhuiyan NA, et al. Controlled study of Escherichia coli diarrheal infections in Bangladeshi children. J clin microbiol. 1995;33:973–7.
WHO: Readings on diarrhoea, Student manual. 1992. http://apps.who.int/iris/bitstream/10665/40343/1/9241544449.pdf.
Acknowledgements
We extend our gratitude to the Director of KEMRI (Kenya Medical Research Institute) for his support during the entire study period. We are grateful to the children and their families for participation in the study, and we thank the staff members of the Kiambu County Hospital (KCH) and the NUITM-KEMRI project. We also thank Amina Galata, Ernest Wandera, and Betty Muriithi for technical assistance.
Funding
This study was supported by Grant-in-Aid for Scientific Research, Japan (Research B: 15H05286).
Availability of data and materials
Please contact author for data requests.
Authors’ contributions
MS developed the research plan with the help of YI, which was supported by Grant-in-Aid for Scientific Research, Japan. CK, EO, MB, GM, and SG collected the stool samples and performed the research work. MS and AW worked on data analysis. MS prepared the manuscript. MK and YI edited the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Ethical clearance was obtained from the Kenya Medical Research Institute (KEMRI) Scientific Steering Committee (KEMRI/SSC/1323). Written informed consent was obtained from parents/guardians of the children before enrollment.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Shah, M., Kathiiko, C., Wada, A. et al. Prevalence, seasonal variation, and antibiotic resistance pattern of enteric bacterial pathogens among hospitalized diarrheic children in suburban regions of central Kenya. Trop Med Health 44, 39 (2016). https://doi.org/10.1186/s41182-016-0038-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41182-016-0038-1