Abstract
Background
Periodontitis results from the interaction between a subgingival biofilm and host immune response. Changes in biofilm composition are thought to disrupt homeostasis between the host and subgingival bacteria resulting in periodontal damage. Chronic systemic inflammatory disorders have been shown to affect the subgingival microbiota and clinical periodontal status. However, this relationship has not been examined in subjects with systemic lupus erythematosus (SLE). The objective of our study was to investigate the influence of SLE on the subgingival microbiota and its connection with periodontal disease and SLE activity.
Methods
We evaluated 52 patients with SLE compared to 52 subjects without SLE (control group). Subjects were classified as without periodontitis and with periodontitis. Oral microbiota composition was assessed by amplifying the V4 region of 16S rRNA gene from subgingival dental plaque DNA extracts. These amplicons were examined by Illumina MiSeq sequencing.
Results
SLE patients exhibited higher prevalence of periodontitis which occurred at a younger age compared to subjects of the control group. More severe forms of periodontitis were found in SLE subjects that had higher bacterial loads and decreased microbial diversity. Bacterial species frequently detected in periodontal disease were observed in higher proportions in SLE patients, even in periodontal healthy sites such as Fretibacterium, Prevotella nigrescens, and Selenomonas. Changes in the oral microbiota were linked to increased local inflammation, as demonstrated by higher concentrations of IL-6, IL-17, and IL-33 in SLE patients with periodontitis.
Conclusions
SLE is associated with differences in the composition of the microbiota, independently of periodontal status.
Similar content being viewed by others
Background
Periodontitis is a multifactorial infectious disease that affects tooth supporting tissues [1]. In health, there is equilibrium between the subgingival biofilm and the host immune response. Periodontal bone loss is initiated when there is a disruption that causes the inflammatory response to move closer to the bone inducing osteoclastogenesis and impaired osseous coupling [2]. Changes in bacterial composition or systemic inflammation are thought to disrupt the balance between host and oral microbiota leading to destruction of periodontal tissues [3–5]. Therefore, chronic inflammatory conditions such as diabetes, obesity, rheumatoid arthritis (RA), and systemic lupus erythematosus (SLE) are linked to an increased risk or severity of periodontal disease [6–11]. Furthermore, periodontitis might have an adverse effect by enhancing systemic inflammation and increasing the risk of myocardial infarction [12], preterm birth [7], pneumonia [13], stroke [14], and rheumatoid arthritis [15] as well as worsen SLE severity [16].
SLE is an autoimmune disease characterized by the presence of aberrant antibody responses to nuclear and cytoplasmic antigens. It is a complex multifactorial disease in which genetic and environmental factors contribute to disease susceptibility [17]. An environmental factor that modulates autoimmunity is the bidirectional crosstalk between the human host and microbiota, as reported for rheumatic diseases [18, 19]. However, few studies have addressed the relationship between SLE and the human microbiota [20, 21] or the periodontal condition in SLE patients [10, 16, 22–27]. Moreover, the results have been conflicting, due in part to small sample size.
The influence of SLE on the oral microbiota has not been reported. To address this issue, we analyzed the subgingival microbiota in SLE patients and healthy subjects to draw potential links among systemic immune dysregulation, subgingival microbiota, and periodontal disease. The results indicate that SLE alters the subgingival microbiota. This understanding might be useful in delineating strategies to treat these patients and suggests that these patients should be followed closely for periodontal disease based on their altered microbiota and enhanced susceptibly to develop periodontitis.
Results
Systemic lupus erythematosus results in higher prevalence and severity of periodontal disease
Demographic and clinical characteristics of subjects included in the study are presented in Table 1. Sixty-seven percent of SLE patients had periodontitis, a significantly higher prevalence when compared to healthy controls (53%). The prevalence of chronic periodontitis in healthy Brazilians is consistent with previous reports [28–30]. An interesting finding was the presence of periodontitis at a younger age in SLE patients (40.5 ± 10.1 years) compared to controls (46.3 ± 13.2, p < 0.05). In addition, 29 SLE patients were younger than 35 years, and from these, 52% already had periodontitis (data not shown). SLE subjects had increased probing depth and increased loss of clinical attachment indicating more severe periodontitis (Table 1). Periodontal probing depth was significantly correlated with most of the SLE parameters (duration of SLE rho = 0.32, accumulated dose of prednisone rho = 0.36 and C-reactive protein level rho = 0.30, p < 0.05). Moreover, loss of clinical attachment was correlated with duration of SLE (rho = 0.35, p < 0.05) and the Systemic Lupus International Collaborating Clinics/American College of Rheumatology Damage Index (SLICC/ACR) damage index (rho = 0.32, p < 0.05).
All subjects exhibited similar oral hygiene habits (frequency of tooth brushing and dental flossing per day). No difference in smoking status between groups was found. When evaluating oral hygiene parameters, control subjects with periodontitis had an increased plaque index compared to non-periodontitis subjects (p < 0.05), but this association was not observed in SLE patients (Table 1). It is noteworthy that when 336 SLE patients were evaluated, 62 (24%) were excluded because they did not have at least 8 teeth. Although it is difficult to assess the exact cause of this early tooth loss, many patients reported that they lost their teeth because the teeth became “loose,” consistent with loss from periodontitis.
SLE impacts oral microbiota diversity
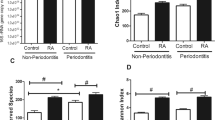
To assess the factors that may lead to increased prevalence and severity of periodontitis in SLE patients, we examined whether the subgingival microbiota was altered. The bacterial load was one log higher in SLE diseased sites compared to control subjects with diseased sites (Fig. 1). In control subjects, the presence of periodontitis resulted in increased microbial diversity (p < 0.05), while in SLE patients, there was decreased microbial diversity at diseased sites (p < 0.05) (Fig. 2a and b).
Subgingival bacterial load of healthy subjects (control) and patients with Systemic Lupus Erythematosus (SLE) with chronic periodontitis (CP) and Non-CP (NCP), determined by real-time PCR using universal primers for 16S rRNA gene. *Statistically different compared to NCP subjects within the same group. #Statistically different compared to control subjects. p < 0.05, Kruskal-Wallis test with Dunn’s post hoc test
Alpha diversity index, a observed species and b Chao1 index of control subjects or Systemic Lupus Erythematosus patients (SLE) with chronic periodontitis (CP) or Non-CP (NCP). *Statistically different compared to NCP subjects within the same group. #Statistically different compared to Control group. p < 0.05, Kruskal-Wallis test with Dunn’s post hoc test. Alpha diversity metrics were calculated after subsampling to obtain equal number of sequences per library
UniFrac analysis was performed to compare the degree of phylogenetic overlap in the microbial communities in SLE and control subjects. Beta diversity, which reflects the relatedness of disease- and health-associated bacterial populations, established that bacterial communities from each group tended to cluster together (PERMANOVA, p = 0.001) (Fig. 3). The bacterial taxa in SLE diseased sites were more similar to each other than bacteria from SLE non-periodontitis and healthy subjects’ sites, indicating lower heterogeneity in subgingival microbial communities in the SLE periodontitis subjects (Fig. 4).
The average weighted UniFrac distance values (the beta diversities) of healthy subjects (control) and patients with Systemic Lupus Erythematosus (SLE) with chronic periodontitis (CP) and Non-CP (NCP).*Statistically different compared to NCP subjects within the same group. #Statistically different compared to control group. p < 0.05, Kruskal-Wallis test with Dunn’s post hoc test
Dysbiosis of oral microbiota in patients with SLE
We investigated the impact of SLE on the subgingival bacterial composition. At healthy periodontal sites in SLE patients, there were increased proportions of a member of Lachnospiraceae family (unclassified Lachnospiraceae A IR009) compared to healthy sites in controls (p < 0.05) (Fig. 5a and b). In contrast, proportions of Prevotella oulorum and Prevotella pleuritidis, Pseudomonas spp, Treponema maltophilum, and Actinomyces IP073, bacteria associated with disease, were elevated at periodontal disease sites in patients with SLE compared to periodontitis sites of non-SLE controls (p < 0.05) (Fig. 5c and d). Sphingomonas were at lower relative levels in healthy periodontal sites of SLE patients (p < 0.05) (Fig. 5a and b) and Rothia aeria, Capnocytophaga gingivalis, Clostridiales, Rasltonia oral taxon 027, Leptotrichia oral taxon A71, Streptococcus sanguinis, and Haemophilus parainfluenzae had decreased relative abundance in periodontitis sites of SLE group (p < 0.05) (Fig. 5c and d)
OTUs and taxa differing between non-periodontitis (upper panel) and periodontitis (lower anel) sites. a, c Heat map with the OTUs showing a p value of less than 0.05 when comparing relative abundances between control and SLE subjects. Subjects are shown in columns, while OTUs appear in rows. Blue asterisks indicate OTUs with decreased proportions in SLE patients, while red asterisks indicated OTUs with increased proportions in SLE in relation to respective controls. b, d The graphic representation of differences in the relative abundances of species represented in the heat maps
Differentially represented OTUs were analyzed via LEfSe, a statistical measure used in metagenomic biomarker discovery [31] (Fig. 6). This analysis revealed that at SLE healthy periodontal sites, species such as Prevotella nigrescens, Prevotella oulorum, Prevotella oris, and Selenomonas noxia exhibited elevated proportions (Fig. 6a). When examining the periodontitis sites, we found again increased relative levels of P. oulorum in SLE patients as well as increased relative levels of Fretibacterium fastidiosum and Fusobacterium oral taxon 360 450, Anaeroglobus geminatus, and TM7 oral taxon 437 (Fig. 6b). Most of the bacterial species found with elevated proportions in SLE patients were anaerobic (Prevotella, Selenomonas, and Treponema), while in healthy non-SLE subjects, there was an increase in the relative abundance of predominantly aerobic species (Rothia, Haemophilus, and Streptococcus).
Microbial intercorrelations
Since periodontitis is a polymicrobial disease, it is important to investigate correlations among subgingival microorganisms. Fig. 7 shows the co-occurrence patterns among bacterial species in subjects with and without SLE. In non-SLE subjects (Fig. 7a), we observed a total of 89 bacterial correlations with positive relationship among pathogenic groups (i.e., Porphyromonas gingivalis and Selenomonas, Treponema vincentii, Fusobacterium nucleatum, and Fretibacterium) and negative correlations between pathogenic bacteria and periodontal healthy-related species (i.e., Streptococcus mitis and Treponema denticola and T. vincentii or between P. oris and Haemophilus parainfluenzae).
Correlations among bacteria species in control (a) and SLE subjects (b) with chronic periodontitis (CP) (green) and Non-CP (NCP) (yellow). In white area are the bacteria that are common to subjects with CP and NCP. Spearman Rank test with a cutoff rho value of 0.4 and p < 0.05. Blue lines indicate positive correlations and red lines negative correlations
In SLE patients, we observed a wider net of correlations than in the healthy subjects, with a total of 111 correlations (Fig. 7b). Positive correlations were found among periodontal pathogens (T. denticola, P. gingivalis, Fretibacterium fastidiosum, and Tannerella forsythia) [32]. In contrast, negative correlations were observed among pathogenic bacteria (F. fastidiosum, P. gingivalis, T. forsythia, T. denticola, and Prevotella intermedia) and health-related species (H. parainfluenzae and S. mitis) in SLE group.
Microbiota correlations with SLE index, salivary, and serum concentration of cytokines
We investigated the relationship between bacteria and local inflammation by measuring cytokines in saliva. There was a significant increase in inflammatory cytokines IL-6, IL-17, and IL-33 in saliva of SLE patients with periodontitis compared to control group with periodontitis. No significant differences in cytokine levels were found in non-SLE subjects with periodontitis compared to non-SLE subjects without periodontitis (Fig. 8). The production of these cytokines in saliva of SLE patients was correlated with relative abundance of Selenomonas (IL-6 rho = 0.42, IL-33 rho = 0.44, p < 0.05) Prevotella denticola (IL-33 rho = 0.40, p < 0.05), Veilonella atypica (IL-33 rho = 0.50, p < 0.05), and Leptotrichia (IL-17 rho = 0.4, p < 0.05). Leptotrichia was also correlated with IL-17 in healthy periodontal sites from SLE patients (rho = 0.7, p < 0.05). In the serum, there was a significant increase in the levels of IL-6 in the SLE patients with periodontitis, while no difference was found in other analyzed cytokines TNF-α, IL-33, and IFN-γ (Additional file 1: Table S1). No correlations were observed between IL-6 and relative abundance of bacteria.
Concentration of IL-6 (a), IL-17 (b), and IL-33 (c) in saliva of healthy subjects (control) and patients with Systemic Lupus Erythematosus (SLE) with chronic periodontitis (CP) and Non-CP (NCP). Cytokine measurement was determined by CBA and ELISA, and values were normalized by stimulated salivary flux.*Statistically different compared to NCP subjects within the same group. #Statistically different compared to healthy subjects with CP. p < 0.05, Kruskal-Wallis test with Dunn’s post hoc test
Spearman Rank correlations were calculated to establish relationships between the relative abundance of bacterial taxa found in subgingival sites of SLE patients and parameters of SLE severity. Table 2 shows that six species of Prevotella correlated with SLE disease index and inflammatory markers. Fretibacterium oral taxon BB62 and F. fastidiosum correlated with SLE duration, while S. noxia correlated with CRP levels. P. gingivalis was positively correlated with SLE duration and accumulated dose of prednisone.
Discussion
The major findings of this study are that SLE patients had a dysbiotic subgingival microbiota with higher subgingival bacterial load, reduced microbial diversity, and changes in bacterial composition with a shift toward greater proportions of pathogenic bacteria. The results suggest that SLE has a significant impact on periodontal health as SLE patients have more severe periodontitis. Reinforcing this finding, we also noticed that 24% of the initial SLE patients evaluated were excluded because they did not have at least eight teeth. In comparison, none of the healthy subjects were excluded for this reason. This finding is likely to be important since periodontal disease is an important cause of tooth loss in adults [33]. Moreover, periodontal probing depth, a measure of periodontal destruction, was correlated with the duration of SLE, accumulated dose of prednisone and the level of systemic inflammation measured by serum C-reactive protein.
We found a high prevalence of periodontitis in SLE patients, which was observed in almost 70% of these patients, in agreement with previous studies [16, 26, 34]. In our study, the experimental and control groups were similar regarding age and gender, ruling out these potential confounding factors [30]. Other factors may also contribute to the premature development of periodontal disease in SLE subjects, such as medications used to treat SLE. However, most are anti-inflammatory drugs consisting of steroids and immunosuppressive medications [35] that typically reduce gingival inflammation [36, 37]. We found that the dose of prednisone was correlated with periodontal destruction and also with the presence of pathogenic bacteria species such as Fretibacterium and P. gingivalis. Whether this is due to more severe SLE in subjects with higher prednisone or a direct effect of the drug is open to question. Reports that corticosteroids have little direct impact on the periodontium would argue for the former [38].
Our study is the first to analyze the bacterial composition of the subgingival microbiota in SLE patients using 16S rRNA gene sequencing. This approach allowed a comprehensive view of the subgingival microbial communities associated with SLE. We found that the bacterial load was one log higher in SLE patients with periodontitis compared to control subjects with periodontitis. Alterations in the inflammatory status of the periodontium induced by SLE may impact the subgingival bacterial composition, which in turn may further enhance inflammatory changes and mediate tissue destruction in SLE patients [39]. Consistently higher levels of IL-6, IL-17, and IL-33 were detected in SLE patients with periodontitis. These cytokines are increased in the serum of SLE patients [39–43] and non-SLE individuals with inflamed gingival tissues [44–47]. IL-33 exacerbates periodontal disease [47], increases IL-6 expression [48], and enhances a Th17 response [42]. IL-6 promotes activation and differentiation of T cells, B cells, macrophages, neutrophils, and osteoclasts and promotes formation of Th17 cells [49]. The levels of IL-6, IL-17, and IL-33 were positively correlated with the relative abundance of pathogenic bacteria such as Selenomonas, P. denticola, Veilonella, and Leptotrichia. Accordingly, increased inflammation may provide a source of nutrients in the form of tissue breakdown products and alter the redox environment favoring the growth of anaerobic bacteria [3]. In turn, changes in the microbiota might be important to amplify local inflammation and periodontal tissue damage.
SLE influenced microbial diversity, as patients with SLE had lower diversity in sites with periodontal breakdown compared to equivalent sites in matched control subjects. It is possible that the lower diversity facilitates microbial changes that increase susceptibility to periodontitis based on findings of decreased bacterial diversity in inflammatory diseases such as psoriatic arthritis [50] and Crohn’s disease [51]. We also found that bacteria commonly found in periodontal healthy, such as Capnocytophaga [52–56], Rothia [54, 56–58], H. parainflunzae [56, 59], and Streptococcus [60] were reduced in proportional abundance in SLE patients with periodontitis, and bacterial species frequently detected in periodontal disease were observed in higher proportions in SLE patients, even in periodontal healthy sites (i.e., Prevotella oulorum, P. nigrescens, P. oris, S. noxia, and Leptotrichia). In addition, bacterial samples from diseased sites in subjects with SLE were more similar to each other than bacterial samples from healthy sites. Shi et al. [61] also found that bacteria from periodontal disease sites were more similar to each other than bacteria from healthy sites. This was observed in different sites within an individual and in comparison to sites between individuals. These findings suggest that the ecological environment is more consistent and similar in disease than in healthy conditions.
The subgingival microbial community in the SLE patients differs not only in the taxonomic composition but also in the co-occurrence patterns of periodontal microorganisms. Co-occurrence analysis indicates that in the SLE patients, the presence of pathogenic bacteria was positively correlated with other pathogenic bacteria (T. denticola, P. gingivalis, T. forsythia, and F. fastidiosum) [32]. Shi et al. similarly reported that periodontitis-associated microorganisms were highly correlated in the diseased state but poorly correlated after treatment, suggesting that there are coordinated interactions among the pathogenic microorganisms. It is possible that SLE favors bacterial interactions that lead to increased periodontal disease.
Periodontal disease in SLE patients shows distinct features compared to systemic healthy subjects. SLE may disrupt the balance between host and microbiota in favor of a dysbiotic condition, resulting in increased periodontal damage or an enhanced inflammatory response leading to an increased risk of periodontitis. It is also possible that periodontitis aggravates SLE disease severity, as a worsened periodontal condition was correlated with increased systemic inflammation in subjects with SLE. This result is consistent with reports that periodontal treatment improves clinical outcomes in rheumatic diseases such as rheumatoid arthritis and SLE [16, 26, 62, 63]. Moreover, increases in the proportion of some subgingival bacteria such as P. nigrescens has been related to preterm birth [64], rheumatoid arthritis [9, 65, 66], and atherosclerosis [67]. In our study P. nigrescens was correlated to higher values of SLE index (SLEDAI and SLICC/ACR), P. denticola with increased neutrophil levels, and Prevotella melaninogenica with increased serum CRP levels in SLE patients. Other studies in rheumatoid arthritis have found that Prevotella species [52, 68] and A. geminatus [52] were found in higher levels in RA patients than in healthy controls. These results suggest a possible two-way cycle in which systemic inflammation enhanced by SLE induces dysbiosis of the subgingival microbiota and progression to periodontitis. Periodontitis may then affect the systemic immune response, leading to increased activity of SLE [69].
The present study represents the first comprehensive evaluation of the subgingival microbiota associated with periodontal status in SLE patients and provides a strong basis for further study on the relationship between the two diseases. As a cross-sectional study, the present work has limitations as it is not able to define cause and effect relationship. A longitudinal study in which the same individual was followed just after SLE diagnosis could provide evidence for a cause and effect relationship between the two diseases. This would be particularly useful in identifying bacterial changes associated with the onset of periodontal disease in SLE patients. An intervention trial in which patients are treated for periodontitis would further enhance our understanding as to whether periodontitis has an impact of SLE disease severity.
Conclusions
SLE patients exhibit a higher prevalence of periodontal disease and increased periodontal disease severity. SLE patients had increased local inflammation and an altered subgingival microbiota, which may account, in part for the periodontal changes. Periodontal inflammation was also associated with more severe SLE scores. The results point to the need to closely monitor the periodontal health of subjects with SLE and for periodontal treatment at early stages, including the removal of subgingival dental plaque.
Methods
Subjects
Three hundred thirty-six patients diagnosed with SLE (according to ACR 1982/1997 revised classification criteria) from Rheumatology Outpatient Clinic of Clinics Hospital of Federal University of Minas Gerais (UFMG), Belo Horizonte, Brazil, were initially evaluated. Based on a power analysis at alpha=1 and 5%, 50 subjects will reach a power of 99% when comparing bacterial taxa frequencies between subgingival samples as reported by La Rosa et al. [70]. Fifty-two patients were included in the study after matched the inclusion criteria: Subjects included were at least 18 years of age, no other rheumatic diseases (except for secondary Sjögren syndrome), no treatment for periodontal disease within the last 6 months, no use of orthodontic appliances, no use of antibiotics within the last 3 months, no need for antibiotics for infective endocarditis prophylaxis during dental procedures, no chronic renal insufficiency requiring dialysis or after kidney transplantation, pregnancy or lactation, no acute or chronic infectious conditions at the time of the study visit, no diagnosis of neoplastic disease within the last 5 years, and the presence of at least eight teeth. Additionally, we have excluded from the study patients that did not agree to participate, did not understand the study, or decided to leave the study after the first clinic visit. SLE group was matched for age and gender with 52 subjects without SLE or other known rheumatic diseases (control group).
Medical records were collected for SLE parameters, Systemic Lupus Erythematosus Disease Activity Index 2000 (SLEDAI 2 K) measured SLE activity [71] and Systemic Lupus International Collaborating Clinics/American College of Rheumatology Damage Index (SLICC/ACR) evaluated SLE-associated damage [72]. Periodontal status was assessed by two calibrated examiners. The following parameters were recorded: plaque index, probing depth (PD), clinical attachment level (CAL), and bleeding on probing (BOP). Periodontitis was defined as the presence of two or more interproximal sites with probing depth ≥4 mm or one site with probing depth ≥5 mm [73].
Sample collection
Subgingival dental plaque samples were collected as described elsewhere [14]. Sterile endodontic paper points (ISO40) (Tanariman, Manacaparu, AM, Brazil) were inserted in the five sites with deepest periodontal pockets and kept there for 1 min. All paper points with subgingival plaque samples were pooled together. After removal, the material was stored in a sterile tube containing 500 μL of sterile distilled water and centrifuged at ×3000g for 5 min. The paper points were discharged, and the pellet was kept at −80 °C until DNA extraction.
For saliva collection, patients could not eat or drink an hour before collection procedure. They washed the mouth with pure water and swallowed whole saliva before collection. Saliva was collected by continuous drool into a sterile 50 mL tube for 5 min. The non-stimulated salivary flow was measured in milliliters per minute (ml/min). The saliva samples were subsequently diluted (1:1) in a phosphate-buffered saline (PBS) solution containing protease inhibitors and subsequently frozen at −80 °C until analysis.
Blood samples were collected using a vacutainer with EDTA and immediately placed on ice, clarified by centrifugation at ×3000g for 5 min at 4 °C, and kept frozen at −80 °C until assayed.
DNA extraction and sequencing
Genomic DNA was extract from the samples using the Quick-gDNA MicroPrep kit (Zymo Research, Irvine, CA, USA) and 50 μL (10 mg/ml) of lysozyme per sample for maximal bacterial cell lysis. All procedures were completed in a laminar flow hood with RNase free materials. The quantity and quality of DNA was measured using a spectrophotometer method (Tecan, Männedorf, Switzerland).
The primers 515 F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT −3′) which target the hypervariable V4 region of the 16S rRNA gene were used for amplification [74] followed by gel purification and ethanol precipitation to remove any remaining contaminants and PCR artifacts and subjected to Illumina MiSeq Plataform at the Next-Generation Sequencing Core of University of Pennsylvania.
Microbiological analysis
Taxonomy was assigned using “Quantitative Insights Into Microbial Ecology” (QIIME) software package [75] with UCLUST against CORE [76]. Alpha rarefaction was performed using the Observed Species and Chao1 metrics. Beta diversity was calculated using UniFrac. Quantification of total bacterial load was determined by real-time PCR using universal primers for 16S rRNA gene (F:AGAGTTTGATCCTGGCTCAG; R: ACGGCTACCTTGTTACGACTT) (IDT, Coralville, Iowa, USA).
Cytokines measurement
Analyses of IL-6 and IL-17 cytokines were determined using a BD CBA Human Th1/Th2/Th17 Cytokine Kit (Becton, Dickinson and Company, BD Biosciences, San Diego, CA) and analyzed on a BD FACSCalibur flow cytometer (Becton, Dickinson and Company). The concentration of the cytokine IL-33 were measured by enzyme-linked immunosorbent assay (ELISA) using commercially available kits (R&D Systems, Minneapolis, MN, USA). The assays were performed according to the manufacturer’s instructions. The results were expressed as picograms of cytokines and adjusted accordingly salivary flux for salivary samples.
Statistics
Clinical, demographic, alpha diversity, and bacterial load data were compared using Kruskal-Wallis test with Dunn’s post hoc test. PERMANOVA was performed to compare beta diversity. Correlations between relative abundance of taxa and clinical parameters of periodontal disease and SLE were calculated using Spearman correlation coefficients. For co-occurrence analysis Spearman rank correlation was calculated with cutoff value of rho ≥0.4 and p < 0.05 as previously described [77]. Relative abundances of Operation Taxonomy Units (OTU) were compared among SLE and control subjects and tested for statistical significance using DESeq2 [78] and LEfSe [31]. Briefly, LEfSe is a metagenomic biomarker-discovery approach based on an algorithm that first performs a nonparametric Kruskal-Wallis test in order to identify bacterial taxa whose relative abundance is significantly different in a group of interest compared to controls. Subsequently, LEfSe applies linear discriminant analysis (LDA) to those bacterial taxa identified as significantly different (p < 0.05) and further assesses the effect size of each differentially abundant taxon. Only those taxa that obtain a log LDA score 2 are ultimately considered. As a result, LEfSe indicates those taxa and OTUs that better discriminate groups. P values <0.05 were considered to indicate statistical significance.
Abbreviations
- BOP:
-
Bleeding upon probing
- CP:
-
Chronic periodontitis
- CRP:
-
C-reactive protein
- IL:
-
Interleukin
- LDA:
-
Linear discriminant analysis
- NCP:
-
Non-periodontitis
- RA:
-
Rheumatoid arthritis
- SLE:
-
Systemic lupus erythematosus
- SLEDAI:
-
Systemic Lupus Erythematosus Disease Activity Index
- SLICC/ACR:
-
Systemic Lupus International Collaborating Clinics/American College of Rheumatology Damage Index
References
Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4:1–6.
Xiao W, Li S, Pacios S, Wang Y, Graves DT. Bone remodeling under pathological conditions. Front Oral Biol. 2015;18:17–27.
Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15:30–44.
Andriankaja OM, Galicia J, Dong G, Xiao W, Alawi F, Graves DT. Gene expression dynamics during diabetic periodontitis. J Dent Res. 2012;91:1160–5.
Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44:3665–73.
Wu Y-Y, Xiao E, Graves DT. Diabetes mellitus related bone metabolism and periodontal disease. Int J Oral Sci. 2015;7:63–72.
Polyzos NP, Polyzos IP, Zavos A, Valachis A, Mauri D, Papanikolaou EG, et al. Obstetric outcomes after treatment of periodontal disease during pregnancy: systematic review and meta-analysis. BMJ. 2010;341:c7017-c7017.
Linden GJ, Herzberg MC. Periodontitis and systemic diseases: a record of discussions of working group 4 of the Joint EFP/AAP Workshop on Periodontitis and Systemic Diseases. J Periodontol. 2013;40:S20–23.
Scher JU, Bretz WA, Abramson SB. Periodontal disease and subgingival microbiota as contributors for rheumatoid arthritis pathogenesis: modifiable risk factors? Curr Opin Rheumatol. 2014;26:424–9.
Kobayashi T, Ito S, Yasuda K, Kuroda T, Yamamoto K, Sugita N, et al. The combined genotypes of stimulatory and inhibitory Fc gamma receptors associated with systemic lupus erythematosus and periodontitis in Japanese adults. J Periodontol. 2007;78:467–74.
Corrêa JD, Saraiva AM, Queiroz-Junior CM, Madeira MFM, Duarte PM, Teixeira MM, et al. Arthritis-induced alveolar bone loss is associated with changes in the composition of oral microbiota. Anaerobe. 2016;39:91–6.
Otomo-Corgel J, JJ P, MP R, MA R. State of the science: chronic periodontitis and systemic health. J Evid Based Dent Pract. 2012;12:20–8.
Linden GJ, Lyons A, Scannapieco FA. Periodontal systemic associations: review of the evidence. J Clin Periodontol. 2013;40 Suppl 1:S8–19.
Ghizoni JS, Antônio L, Taveira DA, Garlet GP, Flávio M, Pereira JR, et al. Increased levels of Porphyromonas gingivalis are associated with ischemic and hemorrhagic cerebrovascular disease in humans: an in vivo study. J Appl Oral Sci. 2012;20:104–12.
Ribeiro J, Leão A, Novaes AB. Periodontal infection as a possible severity factor for rheumatoid arthritis. J Clin Periodontol. 2005;32:412–6.
Fabbri C, Fuller R, Bonfá E, Guedes LKN, D’Alleva PSR, Borba EF. Periodontitis treatment improves systemic lupus erythematosus response to immunosuppressive therapy. Clin Rheumatol. 2014;33:505–9.
Rekvig OP, Van Der Vlag J. The pathogenesis and diagnosis of systemic lupus erythematosus: still not resolved. Semin Immunopathol. 2014;36:301–11.
Phillips R. Rheumatoid arthritis: microbiome reflects status of RA and response to therapy. Nat Rev Rheumatol. 2015;11:502-502. Nature Publishing Group.
Proal AD, Albert PJ, Marshall TG. The human microbiome and autoimmunity. Curr Opin Rheumatol. 2013;25:234–40.
Zhang H, Liao X, Sparks JB, Luo XM. Dynamics of gut microbiota in autoimmune lupus. Appl Environ Microbiol. 2014;80:7551–60.
Hevia A, Milani C, López P, Cuervo A, Arboleya S, Duranti S, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. MBio. 2014;5:1–10.
Mutlu S, Richards A, Maddison P, Scully C. Gingival and periodontal health in systemic lupus erythematosus. Community Dent Oral Epidemiol. 1993;21:158–61.
de Araújo Navas EAF, Sato EI, Pereira DFA, Back-Brito GN, Ishikawa JA, Jorge AOC, et al. Oral microbial colonization in patients with systemic lupus erythematous: correlation with treatment and disease activity. Lupus. 2012;21:969–77.
Al-Mutairi K, Al-Zahrani M, Bahlas S, Kayal R, Zawawi K. Periodontal findings in systemic lupus erythematosus patients and healthy controls. Saudi Med J. 2015;36:463–8.
Calderaro DC, Ferreira GA, de Mendonça SMS, Corrêa JD, Santos FX, Sanção JGC, et al. Há associação entre o lúpus eritematoso sistêmico e a doença periodontal? Rev Bras Reumatol. 2016;56:280–4.
Novo E, Garcia-macgregor E, Viera N, Chaparro N, Crozzoli Y. Periodontitis and anti-neutrophil cytoplasmic antibodies in systemic lupus erythematosus and rheumatoid arthritis: a comparative study. J Periodontol. 1999;70:185–8.
Wang C-Y, Chyuan I-T, Wang Y-L, Kuo MY-P, Chang C-W, Wu K-J, et al. β2-glycoprotein I-dependent anti-cardiolipin antibodies associated with periodontitis in patients with systemic lupus erythematosus. J Periodontol. 2015;86:995–1004.
Susin C, Valle P, Oppermann RV, Haugejorden O, Albandar JM. Occurrence and risk indicators of increased probing depth in an adult Brazilian population. J Clin Periodontol. 2005;32:123–9.
Susin C, Dalla Vecchia CF, Oppermann RV, Haugejorden O, Albandar JM. Periodontal attachment loss in an urban population of Brazilian adults: effect of demographic, behavioral, and environmental risk indicators. J Periodontol. 2004;75:1033–41.
Silva-Boghossian CM, Luiz RR, Colombo APV. Periodontal status, sociodemographic, and behavioral indicators in subjects attending a public dental school in Brazil: analysis of clinical attachment loss. J Periodontol. 2009;80:1945–54.
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60.
Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent Jr RL. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44.
Montandon A, Zuza E, Toledo BE. Prevalence and reasons for tooth loss in a sample from a dental clinic in Brazil. Int J Dent. 2012;2012:8–10.
Kobayashi T, Ito S, Yamamoto K, Hasegawa H, Sugita N. Risk of periodontitis in systemic lupus erythematosus is associated with Fcy receptor polymorphisms. J Periodontol. 2003;74:378–84.
Mok CC, Lau CS. Pathogenesis of systemic lupus erythematosus. J Clin Pathol. 2003;56:481–90.
Tollefsen T, Saltvedt E, Koppang HS. The effect of immunosuppressive agents on periodontal disease in man. J Periodontal Res. 1978;13:240–50.
Been V, Engel D. The effects of immunosuppressive drugs on periodontal inflammation in human renal allograft patients. J Periodontol. 1982;53:245–8.
Seymour RA. Effects of medications on the periodontal tissues in health and disease. Periodontol. 2006;40:120–9.
Jacob N, Stohl W. Cytokine disturbances in systemic lupus erythematosus. Arthritis Res Ther. 2011;13:228.
Tackey E, Lipsky PE, Illei GG. Rationale for interleukin-6 blockade in systemic lupus erythematosus. Lupus. 2004;13:339–43.
Nalbandian A, Crispín JC, Tsokos GC. Interleukin-17 and systemic lupus erythematosus: current concepts. Clin Exp Immunol. 2009;157:209–15.
Li P, Lin W, Zheng X. IL-33 neutralization suppresses lupus disease in lupus-prone mice. Inflammation. 2014;37:824–32.
Yang Z, Liang Y, Xi W, Li C, Zhong R. Association of increased serum IL-33 levels with clinical and laboratory characteristics of systemic lupus erythematosus in Chinese population. Clin Exp Med. 2011;11:75–80.
Stefani FA, Viana MB, Dupim AC, Brito JAR, Gomez RS, da Costa JE, et al. Expression, polymorphism and methylation pattern of interleukin-6 in periodontal tissues. Immunobiology. 2013;218:1012–7.
Corrêa JD, Madeira MFM, Resende RG, Correia-Silva JDF, Gomez RS, de Souza DDG, et al. Association between polymorphisms in interleukin-17A and -17 F genes and chronic periodontal disease. Mediators Inflamm. 2012;2012:846052.
Saraiva AM, Alves e Silva MRM, Correia Silva JDF, da Costa JE, Gollob KJ, Dutra WO, et al. Evaluation of IL17A expression and of IL17A, IL17F and IL23R gene polymorphisms in Brazilian individuals with periodontitis. Hum. Immunol. 2013;74:207–14.
Malcolm J, Awang RA, Oliver-Bell J, Butcher JP, Campbell L, Adrados Planell A, et al. IL-33 exacerbates periodontal disease through induction of RANKL. J Dent Res. 2015;94:968–75.
Balato A, Lembo S, Mattii M, Schiattarella M, Marino R, De Paulis A, et al. IL-33 is secreted by psoriatic keratinocytes and induces pro-inflammatory cytokines via keratinocyte and mast cell activation. Exp Dermatol. 2012;21:892–4.
Naka T, Nishimoto N, Kishimoto T. The paradigm of IL-6: from basic science to medicine. Arthritis Res. 2002;4 Suppl 3:S233–42.
Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67:128–39.
Walker AW, Sanderson JD, Churcher C, Parkes GC, Hudspith BN, Rayment N, et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. BioMed Central Ltd.
Scher JU, Ubeda C, Equinda M, Khanin R, Buischi Y, Viale A, et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis Rheum. 2012;64:3083–94.
Ge X, Rodriguez R, Trinh M, Gunsolley J, Xu P. Oral microbiome of deep and shallow dental pockets in chronic periodontitis. PLoS One. 2013;8:2–11.
Galimanas V, Hall M, Singh N, Lynch MD, Goldberg M, Tenenbaum H, et al. Bacterial community composition of chronic periodontitis and novel oral sampling sites for detecting disease indicators. Microbiome. 2014;2:32.
Wang J, Qi J, Zhao H, He S, Zhang Y, Wei S, et al. Metagenomic sequencing reveals microbiota and its functional potential associated with periodontal disease. Sci Rep. 2013;3:1843.
Colombo APV, Bennet S, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J Periodontol. 2012;83:1279–87.
Moutsopoulos NM, Chalmers NI, Barb JJ, Abusleme L, Greenwell-Wild T, Dutzan N, et al. Subgingival microbial communities in leukocyte adhesion deficiency and their relationship with local immunopathology. PLoS Pathog. 2015;11:e1004698.
Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson J a, Strausbaugh LD, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–25.
Lourenço TGB, Heller D, Silva-Boghossian CM, Cotton SL, Paster BJ, Colombo APV. Microbial signature profiles of periodontally healthy and diseased patients. J Clin Periodontol. 2014;41:1027–36.
Wade WG. Characterisation of the human oral microbiome. J Oral Biosci. 2013;55:143–8. Elsevier.
Shi B, Chang M, Martin J, Mitreva M, Lux R, Klokkevold P, et al. Dynamic changes in the subgingival microbiome and their potential for diagnosis and prognosis of periodontitis. MBio. 2015;6:e01926–14.
Monsarrat P, JN V, Cantagrel A, Algans N, Cousty S, Kémoun P, et al. Effect of periodontal treatment on the clinical parameters of patients with rheumatoid arthritis: study protocol of the randomized, controlled ESPERA trial. Trials. 2013;14:253.
de Pablo P, Chapple ILC, Buckley CD, Dietrich T, IL C, et al. Periodontitis in systemic rheumatic diseases. Nat Rev Rheumatol. 2009;5:218–24.
Kumar PS. Oral microbiota and systemic disease. Anaerobe. 2013;24:90–3. Elsevier Ltd.
Yeoh N, Burton JP, Suppiah P, Reid G, Stebbings S. The role of the microbiome in rheumatic diseases. Curr Rheumatol Rep. 2013;15:314.
de Aquino SG, Abdollahi-Roodsaz S, Koenders MI, van de Loo F a J, Pruijn GJM, Marijnissen RJ, et al. Periodontal pathogens directly promote autoimmune experimental arthritis by inducing a TLR2- and IL-1-driven Th17 response. J. Immunol. 2014;192:4103–11.
Yakob M, Söder B, Meurman JH, Jogestrand T, Nowak J, Söder P-Ö. Prevotella nigrescens and Porphyromonas gingivalis are associated with signs of carotid atherosclerosis in subjects with and without periodontitis. J Periodontal Res. 2011;46:749–55.
Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2013:1–20.
Golub L, Payne J, Reinhardt R, Nieman G. Can systemic diseases co-induce (not just exacerbate) periodontitis? A hypothetical “two-hit” model. J Dent Res. 2006;85:102–5.
Rosa PS La, Brooks JP, Deych E, Boone EL, Edwards DJ, Wang Q, et al. Hypothesis Testing and Power Calculations for Taxonomic-Based Human Microbiome Data. Plos One. 2012;7:1–13.
Ibañez D, Gladman DD, Urowitz MB. Adjusted mean Systemic Lupus Erythematosus Disease Activity Index-2 K is a predictor of outcome in SLE. J Rheumatol. 2005;32:824–7.
Gladman D, Ginzler E, Goldsmith C, Fortin P, Liang M, Urowitz M, et al. The development and initial validation of the systemic lupus international collaborating clinics American College of Rheumatology Damage Index for Systemic Lupus Erythematosus. Arthritis Rheum. 1996;39:363–9.
Eke PI, Page RC, Wei L, Thornton-Evans G, Genco RJ. Update of the case definitions for population-based surveillance of periodontitis. J Periodontol. 2012;83:1449–54.
Caporaso JG, Lauber CL, Walters W a, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. Nature Publishing Group; 2012;6:1621–4.
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6.
Griffen AL, Beall CJ, Firestone ND, Gross EL, DiFranco JM, Hardman JH, et al. CORE: a phylogenetically-curated 16S rDNA database of the core oral microbiome. PLoS One. 2011;6:1–10.
Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med Nature. 2015;21:895–905. Publishing Group.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Acknowledgements
We thank Daniele Souza, Janine Silva, Mayra L. Albiero, and Marcelo Mattos for their contributions to this work.
Funding
This study was supported by CAPES, CNPq, and FAPEMIG, Brazil, as well as the NIDCR, R01DE017732.
Availability of data and materials
All Illumina sequence data from this study were submitted to the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA325500. The dataset (OTU table) supporting the conclusions of this article is included within the article as additional file (Additional file 2: Table S2).
Authors’ contributions
The project was conceived by TAS, DTG, GAF, and ALT. The clinical material was collected by JDC, DCC, and SMM. All laboratory work was conducted by JDC, TAS with the help of SMM, and EX. Bioinformatics analysis was conducted by EJL, GRF, EX, and JDC. JDC, DTG, and TAS interpreted the data and wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The subjects gave informed consent, and the study protocol was approved by the Institution Ethics Committee (CAAE; 03128012.0.0000.5149/2012).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Table S1.
Levels of inflammatory cytokines in serum of control subjects and SLE patients. (XLSX 9 kb)
Additional file 2: Table S2.
OTU table. (TXT 278 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Corrêa, J.D., Calderaro, D.C., Ferreira, G.A. et al. Subgingival microbiota dysbiosis in systemic lupus erythematosus: association with periodontal status. Microbiome 5, 34 (2017). https://doi.org/10.1186/s40168-017-0252-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40168-017-0252-z