Abstract
RghBNG, a gene of unknown function, was cloned from Rehmannia glutinosa by reverse transcription PCR and rapid amplification of cDNA ends. The full-length cDNA of RghBNG was 548 bp with a282-bp open reading frame. It encoded a polypeptide of 93 amino acids with a predicted molecular weight of 10.5 kDa and a theoretical isoelectric point of 9.25. Bioinformatics analysis indicated that RghBNG had no homology to any known plant genes, whereas the RghBNG polypeptide was highly similar to other plant proteins and possessed one conserved B12D protein family functional domain. Phylogenetic analysis revealed that RghBNG encoded for a dicot protein. RghBNG spatial and temporal expression patterns and responses to abiotic stresses and plant growth regulators were investigated by qRT-PCR. RghBNG transcripts were detected in roots, stems, leaves, petals, receptacles, stamens and pistils with the highest and lowest levels respectively observed in petals and leaves of mature plants. Additionally, RghBNG transcripts were detected at three developmental stages of roots, stems and leaves; the highest levels were observed in roots at seedling stage; Transcript levels changed to varying degrees in different tissues and stages; We also studied the effects of abiotic stress and plant growth regulators in roots and leaves. RghBNG expression was significantly increased (p < 0.01) by chromium, gibberellic acid and NaCl, with the highest levels induced by chromium stress; In contrast, 6-benzyladenine reduced expression. These results strongly suggest that RghBNG is involved in R. glutinosa growth, development and response to plant growth regulators and abiotic stresses.
Similar content being viewed by others
Introduction
Abiotic stresses reduce average yields of most crops by more than 50% (Wang et al. 2003; Bhatnagar-Mathur et al. 2008). Drought, salinity and extreme temperature are among the major abiotic stresses, hampering plant growth and productivity and frequently cause a series of morphological, physiological and biochemical changes (Mehrotra et al. 2014). In addition, heavy metal contamination of soil and water is a global problem giving rise to crop yield losses and having hazardous effects on human health when these metals enter the food chain (Vernay et al. 2007). Because plant vulnerability to such abiotic stresses is a serious threat to the entire plant ecosystem, molecular and genetic mechanisms of abiotic stress tolerance in plants are of great scientific interest (Alexandra et al. 2013). Plants respond and adapt to stress conditions with an array of biochemical and physiological changes, many of which are regulated by stress-responsive gene expression. The regulation of these abiotic stress responses to generate stress resistant plants is of great importance to plant growers (Sarika and Aaron 2014). One important strategy related to this goal is the development of stress-tolerant transgenic plants (Tuli et al. 2010). To develop such plants, stress-related plant genes must be identified, characterized and tested in plant systems for their potential use in commercially important crops (Kumar et al. 2013; Quaggiotti et al. 2007). Rehmannia glutinosa (Scrophulariaceae) is a well-known medicinal plant widely cultivated in Asian countries such as China, Korea, Japan, Vietnam and others. The tuberous roots are used to treat fever, nervous conditions, diabetes and hypertension, to strengthen liver function, and for hematopoietic, immuno-enhancing, and tonic purposes (Kim et al. 2012; Sun et al. 2010). Because root yield and quality are limited by various stresses. the incorporation of some stress-tolerant genes is needed to improve R. glutinosa resistance. The effects of several abiotic stresses, such as temperature, water, NaCl, Paraquat and choline chloride have been previously reported for R. glutinosa (Chung et al. 2006; Jia et al. 2009; Zhao et al. 2007; Piao et al. 2008). The NaCl- and waterlogging stress-intolerant Aux/IAA gene from R. glutinosa has also been recently described (Wang et al. 2013). Nevertheless, the effects of many other abiotic stresses and the functions of other stress-related genes remain unknown in R. glutinosa. In the present study, we therefore isolated the RghBGN gene from R. glutinosa. To elucidate the role of RghBGN in R. glutinosa growth, development, and stress responses, we investigated its expression patterns in the presence and absence of abiotic stresses and plant growth regulators.
Results
Cloning and sequencing of RghBNG
One 221 bp cDNA fragment (middle fragment) was amplified by RT-PCR with the primer pair P1, cloned and sequenced. Its 3′-end product and 5′-end product were generated by 3′-RACE and 5′-RACE, respectively. After both were sequenced, three fragments were aligned linked together to give a 548 bp full-length cDNA known as RghBNG (GenBank Accession No.JX290370) including a 5′-untranslated region of 68 bp and 3′- untranslated region of 199 bp. Its 282-bp open reading frame (ORF) was searched by ORF Finder (Figure 1). RghBNG gene was amplified by RT-PCR with primer pair P4.
Nucleotide sequence of the full-length cDNA and deduced amino acid sequence of RghBNG . Numbers on the left and right correspond to nucleotide and amino acid sequences, respectively. In the nucleotide sequence, the stop codon is boxed and the polyadenylated tail is indicated in bold italic letters. In the amino acid sequence, the B12D superfamily motif is shaded and in bold.
Bioinfomatics analyses of RghBNG gene
Bioinfomatics analyses indicated that RghBNG has no homology to any known gene in NCBI at the cDNA level. However, its open reading frame may encode a protein of 93 amino acid residues in length (Figure 1).The protein is composed of 20 amino acids, of which alanine accounts for 9.7%, both arginine and valine for 8.6%, each of the rest for 1.1%-7.5%, alkaline amino acids for14.0% and acidic amino acids for10.8%, in which hydrophilic amino acids and hydrophobic amino acids are evenly distributed, and there are no signal peptide but four phosphorylation sites including 1 Ser site, 1 Thr site and 2 Tyr sites. Its secondary structure is composed of four parts such as α-helixes (56.99%), random coils (27.96%), β-sheets (9.68%) and β-turns (5.38%). Its theoretical isoelectric point (pI), molecular weight and molar extinction coefficient are, 9.25, 10.52 and 56.74, respectively. Furthermore, it has one conserved functional domain of B12D superfamily (Figure 1). Moreover, similarity analyses by the NCBI-blastp showed that the RghBNG protein had a high degree of similarity (58%--86%) to some unknown functional proteins from 14 known species (Figure 2). Meanwhile, it also has another high degree of similarity (>72%) to B12D proteins such as 80% to Beta vulgaris (CAK22419.1), 79% to Camellia sinensis var. assamica (Mast.) Kitam (AEC10990.1), 79% to Arabidopsis thaliala (NP_190397.1), 74% to Ipomoea batatas (AAD22104.1), 73% to Castanea sativa (AAL17696.1) and 72% to Wolffia arrhiza (ADB08699.1).
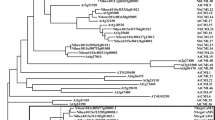
A molecular phylogenetic tree of RghBNG and related proteins generated by neighbor-joining method using MEGA 4.0.The unrooted phylogenetic tree was generated based on the alignment of amino acid sequences from 14 plant species: Populus trichocarpa (XP_002318014.1; 86% similarity to the RghBNG ), Glycine max (XP_003527039.1; 84%), Ricinus communis (XP_002515002.1; 82%), Lotus japonicus (AFK42697.1; 81%), Medicago truncatula (ACJ84101.1; 79%), Arabidopsis lyrata subsp (XP_002875880.1; 79%), Vitis vinifera (XP_002283743.1; 78%), Brachypodium distachyon (XP_003557311.1; 72%), Picea sitchensis (ABK24106.1; 70%), Oryza sativa Japonica Group (NP_001057280.1; 70%), Sorghum bicolor (XP_002438146.1; 70%), Hordeum vulgare subsp. Vulgare (BAK01669.1; 70%), Zea mays (ACN31786.1; 59%) and Physcomitrella patens subsp. Patens (XP_001779363.1; 58%). Numbers above nodes are bootstrap support percentages based on 1,000 replicates. The scale bar indicates evolutionary distance of amino acid substitutions per position. RghBNG from Rehmannia glutinosa is boxed.
To investigate the phylogenetic relationship of RghBNG with other known homologous proteins, the amino acid sequences of available unknown functional proteins from 14 known species were used to construct the phylogenetic tree (Figure 2). This analysis showed that RghBNG clusters with Glycine max, Medicago truncatula, Lotus japonicus and Vitis vinifera with a support value of 29, and that RghBNG protein is attributed to dicots (Figure 2), which is in accordance with APG classification system.
Expression pattern of RghBNG in different Rehmannia glutinosa tissues and stages
To determine the spatial expression pattern, cDNA was synthesized from three tissues of the plants at seedling stage, seven ones at full blooming stage and three ones at mature stage of Rehmannia glutinosa. It was found that RghBNG mRNA was expressed in all the tissues tested, with the strongest signal in root and the significant difference between root and leaf at seedling stage (α = 0.05); with the strongest signal in petal and the extremely significant one between petal and each of others, the one between receptacle or stamen and root or stem or leaf, the one between leaf and pistil at full blooming stage (α = 0.01); with the strongest one in root, and the extremely significant one between root and stem or leaf (α = 0.01), and the significant one between stem and leaf (α = 0.05) at mature stage (Figure 3). All qRT-PCR products of RghBNG gene was confirmed by sequencing.
Expression pattern of the Rehmannia glutinosa RghBNG gene in different tissues at three different developmental stages as determined by quantitative real-time PCR. Expression levels are normalized relative to those of TIP41, which was used as an internal control. Data are means ± SE of three replicates. a-d and A-D in relative (spatial) expression level indicate the significance at 0.05 and 0.01, respectively; a’-c’and A’-C’ in relative (temporal) expression level indicate the significance at 0.05 and 0.01, respectively.
The temporal expression profile of RghBNG RNAs during root, stem and leaf developments was determined by qRT-PCR. Their expression levels at three stages were summarized in Figure 3. It was found that RghBNG mRNA was expressed at all the stages tested, with the strongest signal at seedling stage, the extremely significant difference between blooming stage and seedling stage or mature stage (α = 0.01) and the significant one between seedling stage and mature stage (α = 0.05) for root; with the strongest signal at seedling stage and the extremely significant one between one and each of others among three ones (α = 0.01) for stem; with the strongest one at seedling stage, and the extremely significant one between seedling stage and blooming one or mature one (α = 0.01) for leaf. All qRT-PCR products of RghBNG gene was confirmed by sequencing.
Expression of the Rehmannia glutinosa RghBNG gene in roots of plants exposed to chromium, mercury, NaCl, gibberellic acid, 1-napthalene acetic acid (NAA) or 6-benzyladenine (6-BA). Following treatments, total RNAs were isolated, and RghBNG expression levels were monitored by quantitative real-time PCR. Error bars indicate standard deviation values of three independent analyses. a-c and A-C in relative expression level indicate the significance at 0.05 and 0.01, respectively.
Gene transcription regulation of the RghBNG gene
In order to investigate if the expression of the RghBNG gene is associated with the abiotic stress and plant growth regulators-induced defense mechanism in Rehmannia glutinosa, the expression pattern of the RghBNG was examined in young Rehmannia glutinosa plants subjected to six factors such as chromium, mercury, NaCl, Gibberellin, NAA and 6-BA treatments by qRT-PCR (Figures 4 and 5). For root as shown in Figure 4, compared to control, Chromium, exogenous Gibberellin and NaCl resulted in different increases in RghBNG transcript levels with the highest one induced by chromium, whose ones were extremely significant (α = 0.01), and that NAA and 6-BA down-regulated RghBNG transcript levels, whose ones were extremely significant (α = 0.01). However, mercury did not significantly decrease the expression level (α = 0.05). For leaf as shown in Figure 5, Chromium, NAA, Gibberellin and NaCl also resulted in different increases in RghBNG transcript levels with the highest one under chromium stress, whose ones were extremely significant (α = 0.01), while mercury and 6-BA decreased RghBNG transcript levels, whose ones was also extremely significant (α = 0.01) compared to control.
Expression of the Rehmannia glutinosa RghBNG gene in leaves of plants exposed to chromium, mercury, NaCl, gibberellic acid, 1-naphthalene acetic acid (NAA) or 6-benzyladenine (6-BA). Following treatments, total RNAs were isolated, and RghBNG expression levels were monitored by quantitative real-time PCR. Error bars indicate standard deviation values of three independent analyses. a-c and A-C in relative expression level indicate the significance at 0.05 and 0.01, respectively.
Discussion
Cloning, sequencing, and structural analysis of RghBNG
In this study, we cloned the full-length sequence of the RghBNG gene, deduced its amino acid sequence, and analyzed the structure of the encoded protein. The RghBNG gene was obtained from R. glutinosa using RT-PCR and RACE technology. No homology was found between the RghBNG cDNA sequence and that of any other plant species gene, but considerable similarity was uncovered between the amino acid sequence of the deduced RghBNG protein and proteins from other species, which could be divided into two groups: B12D proteins, and proteins of unknown function. In addition, RghBNG protein possesses the conserved structure domain of B12D protein family, B12D superfamily,(Walker et al. 1992, Figure 1), belonging to one kind of subunits of the NADH: ubiquinone oxidoreductase (Complex I) in mitochondrial membrane (Gabaldón et al. 2005; Zhang et al. 2012).
The roles of other known B12D protein genes
The B12D gene was first isolated from barley (Aalen et al. 1994) and was initially characterized as a transcript related to barley aleurone and embryo development (Steinum et al. 1998). Several B12D protein-like protein genes have been subsequently cloned and their roles have been studied. For example, B12D protein has been shown to be localized in peroxisomes of Arabidopsis and rice (Kaur and Hu 2011), and GA3 up-regulates B12D protein gene expression in imbibed barley embryoless grains (Aalen et al. 1994). However, abscisic acid (ABA) suppresses B12D expression in imbibed barley embryoless grains (Steinum et al. 1998), while ABA and 20% mannitol inhibit its expression in barley embryos (Aalen et al. 1994). In another study, B12D protein was specifically expressed by the induction of ABA (Aalen and Steinum 2001). A B12D-like protein of unknown function was found to be constitutively expressed in Arabidopsis seedlings, roots, leaves, inflorescences, flowers and siliques at different developmental stages (Zhu et al. 2001). A soybean B12D protein gene (Glyma11g37370.1) has been observed to increase the female index of soybean cyst nematode (Matthews et al. 2013). The C832 (DT045059) gene, which has been found to be differentially expressed between wheat near-isogenic lines Chisholm-T and Chisholm-S under Al stress, shows similarity with known putative B12D protein genes in GenBank and TIGR databases (Guo et al. 2007). Plant-specific B12D proteins of unknown function are strongly down-regulated in root tissues of wheat genotypes with distinct responses to water deficit (Ergen et al. 2009). A B12D protein encoded by TDF41 (NP_001057280) is induced by sorbitol, and indirectly promotes plantlet regeneration frequency of rice calli (Feng et al. 2011). An abiotic stress-regulated cDNA clone (CD725202) from Pennisetum encodes proteins involved in cellular homeostasis, including a B12D-like protein that protects plants from stress-induced damages and facilitates the establishment of cellular homeostasis (Mishra et al. 2007). A cDNA sequence (CdR311) with identity to the mRNA sequence of a Zea mays B12D-like protein (AY104072) is up-regulated in roots of Bermuda grass growing under petroleum stress, but its biological activity remains unknown (Peña-Castro et al. 2006), B12D proteins may be involved in general functions such as signal transduction, disease resistance, cell growth, and differentiation (Vinod et al. 2010). A B12D protein cDNA (BP947551) showed 100% transcript variation in NaCl-treated leaves and roots of Burma mangrove, making it a perfect candidate as a stress-responsive protein (Miyama et al. 2006). Another B12D-like protein gene (BP945437) has been discovered to be highly repressed in salt-treated Burma mangrove roots (Miyama and Hanagata 2007). Castanea sativa B12D-like protein mRNA is differentially expressed upon wounding and infection of chestnut with chestnut blight fungus (Schafleitner and Wilhelm 2002). B12D proteins also have been inferred to participate in ATP synthesis and energy metabolism (Gabaldón et al. 2005; Zhang et al. 2012).
Spatial and temporal expression of RghBNG
The roles of RghBNG in plant development were examined by characterizing its spatial and temporal expression patterns during R. glutinosa growth and development by qRT-PCR. RghBNG mRNA transcripts were detected in all tested tissues and organs, but the transcript levels varied. The highest levels were clearly detected in petals at the full-blooming stage and roots at both seedling and mature stages (Figure 3), demonstrating that RghBNG may play an important role in R. glutinosa growth and development.
Transcriptional expression of RghBNG in response to abiotic stresses and plant growth regulators
Plants are frequently exposed to many forms of stress. Plant adaptation to stress has been suggested to be mediated by both preexisting and induced defense mechanisms. Plant signaling pathways are driven by plant growth regulators and reactive oxygen species. Phytohormones generate a signal transduction network that leads to a cascade of events responsible for plant adaptation to external conditions (Al-Momany and Abu-Romman 2014). To assign possible gene physiological functions, detailed expression analysis of the target gene is an important and necessary step (Pérez-Torres et al. 2009). In our study, RghBNG responses to abiotic stresses and plant growth regulators were investigated by qRT-PCR. The RghBNG gene was induced by GA3 and NaCl, in agreement with previous studies of B12D proteins (Steinum et al. 1998; Aalen et al. 1994; Aalen and Steinum 2001; Miyama et al. 2006; Zhang et al. 2012). Furthermore, our results revealed that the RghBNG gene significantly responded to abiotic stress induced by chromium and plant growth regulators such as NAA and 6-BA (Figures 4 and 5). These findings reveal that RghBNG proteins possessed some known roles for B12D proteins such as GA3 and NaCl induction and novel roles for B12D proteins (one group) or the proteins of unknown function (the other group) mentioned above, for example, responses to Cr, Hg, NAA and 6-BA.
Conclusions
A full-length cDNA of the RghBNG gene from R. glutinosa was cloned and characterized, and its spatial and temporal expression profiles and abiotic stress and plant growth regulator-responsive patterns were determined. This study has provided essential molecular data that can be used in follow-up studies to elucidate the biological roles of RghBNG in growth, development, and stress and plant growth regulator-responses of R. glutinosa.
Materials and methods
Plant materials and treatment conditions
Sterile 25-day-old plants of R. glutinosa f. hueichingensis ‘Wen85-5′ were grown in hormone-free Murashige-Skoog (MS) media or transplanted into flower pots in a greenhouse to obtain full-blooming plants and mature plants (Zhou et al. 2009). Leaf sample were collected for gene cloning. For expression pattern analysis, samples of fresh leaves, stems, roots, petals, receptacles, stamens and pistils were collected from R. glutinosa plants during corresponding vegetative growth, full-blooming and early senescence phases. To investigate gene expression regulation under abiotic and hormonal stresses, 1.5-cm-long shoots with two leaves from 25-day-old plants, five shoots per treatment, were inserted and maintained for 20 days in a 450-mL glass bottle containing MS medium either lacking (control) or supplemented with one of the following impact factors: 200 μM NAA, 500 μM 6-BA, 150 μM K2Cr2O7 or 200 μM HgCl2. At the end of the treatment period, five control plants each were sprayed every other hour for 48 h either with a solution of 100 μM GA3 or 250 mM NaCl, and leaf and root samples were then collected. The experiment was repeated three times in a greenhouse. All samples were immediately frozen in liquid nitrogen after collection and stored at −80°C until further analysis.
Total RNA extraction and first-strand cDNA preparation
Total RNA was extracted from leaf samples using an RNA kit (Lifefeng Biotechnology Co., Shanghai, China). RNA concentration and purity were estimated spectrophotometrically based on absorbance at 260 and 280 nm. First-strand cDNA was synthesized from 1 μg total RNA, which was reversely transcribed using M-MLV reverse transcriptase with oligo (dT)15 as a primer in a 10-μL reaction volume (Takara, Japan).
Cloning of RghBNG cDNA
The middle region of the RghBNG gene was amplified using first-strand cDNA as a template with primer pair P1 (Table 1). The amplification reaction was performed in a total volume of 25 μL using a CodeDR011 PCR amplification kit (Takara). Amplification conditions were as follows: 94°C for 5 min followed by 30 cycles of 94°C for 30 s, 46°C for 1 min and 72°C for 1 min, with a final extension of 72°C for 10 min. RghBNG 5′ and 3′ ends were cloned with primer pairs P2-P2′ and P3′-P3 using 5′-and 3′-Full RACE kits (Takara) according to the manufacturer’s instructions. The full-length cDNA of the RghBNG gene was obtained by aligning the three cDNA fragments using DNAMAN4.0 software. The ORF was determined with ORF Finder and cloned by RT-PCR with primer pair P4 (Table 1), which was designed based on the ORF nucleotide sequence. All amplicons were separated, excised, purified, ligated, transformed and sequenced following Al-Momany and Abu-Romman (2014), except that we replaced the T vector and host used in that study with pMD19-T vector and Escherichia coli DH5α.
Bioinformatics analyses
The RghBNG nucleotide sequence was aligned using BLAST on the NCBI website. The ExPASy proteomics server (http://www.expasy.ch/tools/protparam.html) was used to determine the amino acid sequence, molecular weight, isoelectric point and other physicochemical properties of the RghBNG protein. The Expert Protein Analysis System proteomics server (http://www.expasy.org/) and the Predict Protein server (http://www.Predictprotein.org/) were used to predict the structure and function of the encoded protein. Phylogenetic trees were constructed by the neighbor-joining method with 1,000 bootstrap replicates using MEGA 4.0 software.
Analyses of RghBNG gene expression and transcriptional regulation
We used qRT-PCR to monitor RghBNG transcript levels in tissues and organs of R. glutinosa plants at three developmental phases as well as leaves and roots subjected to abiotic and hormonal stress treatments. PCR experiments were carried out using the first-strand cDNAs as templates with primer pair p5 (Table 1) as described above. As a control, TIP41 was also amplified by RT-PCR using primer pair P6 (Table 1). qRT-PCR was performed on an ABI 7500 Real-Time PCR instrument (Cwbiotech, Beijing, China) in 20-μL reaction volumes comprising 1 μL of cDNA template, 10 μL of 2× UltraSYBR mixture (with ROX), 1 μL of each primer (5 μM), and 8 μL ddH2O. PCR conditions were as follows: 95°C for 10 min followed by 40 cycles of 95°C for 15 s, 60°C for 1 min and 72°C for 5 min. The fluorescent product was detected in the last step of each cycle. Amplicons were diluted 16-fold and used to generate a calibration curve for determination of the PCR amplification efficiency of each gene. After acquiring Ct values of RghBNG and TIP41, relative mRNA expression (relative quantity) of target genes was determined with the formula described in Liu et al. (2012). Results were presented as means ± standard error (SE) of data from triplicate replicates. Data were additionally analyzed by one-way analysis of variance and the least significant difference test in SPSS 13.0.
Abbreviations
- RghBNG :
-
Rehmann glutinosa. f. hueichingensis binary novel gene
- NAA:
-
1-Naphthaleneacetic acid
- 6-BA:
-
6-Benzylaminopurine
- RT-PCR:
-
Reverse transcription PCR
- qRT-PCR:
-
Quantitative real time PCR
- RACE:
-
Rapid amplification of cDNA ends
- GA3:
-
Gibberellic acid
- MS:
-
Murashige and Skoog medium
References
Aalen RB, Steinum SZ (2001) Stability of barley aleurone transcripts: Dependence on protein synthesis, influence of the starchy endosperm and destabilization by GA3. Physiol Plantarum 112:403–413
Aalen RB, Opsahl-Ferstad HG, Linnestad C, Olsen OA (1994) Transcripts encoding an oleosin and a dormancy related protein are present in both the aleurone layer and the embryo of developing barley (Plordeum vitlgare L.) seeds. Plant J 5:385–396
Alexandra SD, Konstantin VK, Valeriya SK (2013) Expression of calcium-dependent protein kinase (CDPK) genes under abiotic stress conditions in wild-growing grapevine Vitis amurensis. J Plant Physiol 170:1491–1500
Al-Momany B and Abu-Romman S (2014) Cloning and molecular characterization of a flavin-dependent oxidoreductase gene from barley. J Appl Genetics. doi:10.1007/s13353-014-0227-8
Bhatnagar-Mathur P, Vadez V, Sharma KK (2008) Transgenic approaches for abiotic stress tolerance in plants: retrospect and prospects. Plant Cell Rep 27:411–424
Chung IM, Kim JJ, Lim JD, Yu CY, Kim SH, Hahn SJ (2006) Comparison of resveratrol, SOD activity, phenolic compounds and free amino acids in Rehmannia glutinosa under temperature and water stress. Environ Exp Bot 56(1):44–53
Ergen NZ, Thimmapuram J, Bohnert HJ, Budak H (2009) Transcriptome pathways unique to dehydration tolerant relatives of modern wheat. Funct Integr Genomics 9:377–396
Feng XL, Zhao PK, Hao JQ, Hu JQ, Kang DM, Wang H (2011) Effects of sorbitol on expression of genes involved in regeneration of upland rice (Oryza sativa L.). Plant Cell Tissue Organ Cult 106:455–463
Gabaldón T, Rainey D, Huynen MA (2005) Tracing the evolution of a large protein complex in the eukaryotes, NADH: ubiquinone oxidoreductase (Complex I). J Mol Biol 348(4):857–870
Guo PG, Bai GH, Carver B, Li RH, Bernardo A, Baum M (2007) Transcriptional analysis between two wheat near-isogenic lines contrasting in aluminum tolerance under aluminum stress. Mol Genet Genomics 277:1–12
Jia YY, He YJ, Liang ZS, Dong F, Fang L (2009) Effect of different moisture treatments on photosynthetic characteristics of Rehmannia glutinosa Libosch. J Northwest Sci-Tech Univ Agri For (Nat Sci Ed) 8:182–186 (in Chinese)
Kaur N, Hu JP (2011) Defining the plant peroxisomal proteome: from Arabidopsis to rice. Front Plant Sci 2:1–20
Kim YS, Ryuk JA, Ko BS (2012) Discrimination of Korean Rehmannia glutinosa from Chinese Rehmannia glutinosa using sequence-characterized amplified region marker. J Korean Soc Appl Biol Chem 55:1–6
Kumar S, Asif MH, Chakrabarty D, Tripathi RD, Dubey RS, Trivedi PK (2013) Expression of a rice Lambda class of glutathione S-transferase, OsGSTL2, in Arabidopsis provides tolerance to heavy metal and other abiotic stresses. J Hazard Mater 248–249:228–237
Liu HH, Wang JW, Si JM, Jia J, Li L, Han CC et al (2012) Molecular cloning and in silico analysis of the duck (Anas platyrhynchos) MEF2A gene cDNA and its expression profile in muscle tissues during fetal development. Genet Mol Biol 35:182–190
Matthews BF, Beard H, Macdonald MH, Kabir S, Youssef RM, Hosseini P et al. (2013) Engineered resistance and hypersusceptibility through functional metabolic studies of 100 genes in soybean to its major pathogen, the soybean cyst nematode. Planta 237:1337–1357
Mehrotra R, Bhalothia P, Bansal P, Basantani MK, Bharti V, Mehrotra S (2014) Abscisicacid and abiotic stress tolerance – Different tiers of regulation. J Plant Physiol 171:486–496
Mishra RN, Reddy PS, Nair S, Markandeya G, Reddy RR, Sopory SK et al. (2007) Isolation and characterization of expressed sequence tags (ESTs) from subtracted cDNA libraries of Pennisetum glaucum seedlings. Plant Mol Biol 64:713–732
Miyama M, Hanagata N (2007) Microarray analysis of 7029 gene expression patterns in burma mangrove under high-salinity stress. Plant Sci 172:948–957
Miyama M, Shimizu H, Sugiyama M, Hanagata N (2006) Sequencing and analysis of 14,842 expressed sequence tags of burma mangrove, Bruguiera gymnorrhiza. Plant Sci 171:234–241
Peña-Castro JM, Barrera-Figueroa BE, Ferna’ndez-Linares L, Ruiz-Medrano R, Xoconostle-Ca’zares B (2006) Isolation and identification of up-regulated genes in bermudagrass roots (Cynodon dactylon L.) grown under petroleum hydrocarbon stress. Plant Sci 170:724–731
Pérez-Torres E, ParedesM PV, Becerra V (2009) Gene expression analysis: a way to study tolerance to abiotic stresses in crops species. Chil J Agri Res 69(2):260–269
Piao RZ, Zhao HY, Jin YJ, Lin H (2008) Influences of paraquat stress on membrane lipid peroxidation and antioxidative activity in Rehmannia glutinosa and piemarker. Jiangsu Agril Sci 3:271–274 (in Chinese)
Quaggiotti S, Barcaccia G, Schiavon M, Nicolé S, Galla G, Rossignolo V et al (2007) Phytoremediation of chromium using Salixs pecies: Cloning ESTs and candidate genes involved in the Cr response. Gene 402:68–80
Sarika GS, Aaron MR (2014) Expression patterns and regulation of SlCRF3and SlCRF5 in response to cytokinin and abiotic stresses in tomato (Solanum lycopersicum). J Plant Physi 171:349–358
Schafleitner R, Wilhelm E (2002) Isolation of wound-responsive genes from chestnut (Castanea sativa) microstems by mRNA display and their differential expression upon wounding and infection with the chestnut blight fungus (Cryphonectria parasitica)”. Physiol Mol Plant Pathol 61:339–348
Steinum TM, Berner HS, Stacy RAP, Salehian Z, Aalen RB (1998) Differential regulation of the barley (Hordeum vulgare)transcripts B22E and B12D in mature aleurone layers. Physiologia plantarum 102:337–345
Sun P, Guo YH, Qi JJ, Zhou LL, Li XE (2010) Isolation and expression analysis of tuberous root development related genes in Rehmannia glutinosa. Mol Bio Rep 37:1069–1079
Tuli R, Chakrabarty D, Trivedi PK, Tripathi RD (2010) Recent advances in arsenic accumulation and metabolism in rice. Mol Breed 26:307–323
Vernay P, Gauthier-Moussard C, Hitmi A (2007) Interaction of bioaccumulation of heavy metal chromium with water relation, mineral nutrition and photosynthesis in developed leaves of Lolium perenne L. Chemosphere 68:1563–1575
Vinod MS, Sankararamasubramanian HM, Priyanka R, Ganesan G, Parida A (2010) Gene expression analysis of volatile-rich male flowers of dioecious Pandanus fascicularis using expressed sequence tags. J Plant Physiol 167:914–919
Walker JE, Arizmendi JM, Dupuis A, Ian MF, Moshe F, Simon MM et al (1992) Sequences of 20 subunits of NADH: ubiquinone oxidoreductase from bovine heart mitochondria. application of a novel strategy for sequencing proteins using the polymerase chain reaction. J Mol Biol 226:1051–1072
Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures towards genetic engineering for stress tolerance. Planta 218:1–14
Wang FQ, Tian YH, Li MJ, Yang JF, Zhang B, Lin WX et al (2013) Molecular cloning and expression analysis of an Aux/IAA gene (RgIAA1) from Rehmannia glutinosa. China J C Mater Med 38(23):10–15 (in Chinese)
Zhang YY, Huang N, Xiao XH, Huang L, Su WH, Xu LP, Que YX (2012) Cloning and expression analysis of B12D gene in sugarcane. http://www.paper.edu.cn
Zhao HJ, Tan JF, Qi CM (2007) Photosynthesis of Rehmannia glutinosa subjected to drought stress is enhanced by choline chloride through alleviating lipid peroxidation and increasing proline accumulation. Plant Growth Regul 51(3):255–262
Zhou YQ, Duan HY, Zhou CE, Li JJ, Gu FP, Wang F et al (2009) Hairy root induction and plant regeneration of Rehmann glutinosa Libosch. f. hueichingensis Hsiao via Agrobacterium rhizogenes-mediated transformation. Russ J Plant Physl 56(2):224–231
Zhu T, Budworth P, Han B, Brown D, Chang HS, Zou GZ et al. (2001) Toward elucidating the global gene expression patterns of developing Arabidopsis: Parallel analysis of 8300 genes by a high-density oligonucleotide probe array Plant Physiol. Biochem 39:221–242
Acknowledgments
The work was supported by Personnel Training United Fund of NSFC-Henan province (U1304304), National Training Programs of Innovation and Entrepreneurship for Undergraduates (201310476102), Program for Innovative Research Team (in Science and Technology) in University of Henan Province (No. 13IRTSTHN009) and Key Research Program of Science and technology of Education department of Henan province (No.14B180028).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Yanqing Zhou: AB and FG. Yonghua Zhang: AB. Yu Zhang: JY and MT. Jingyun Li; ES. Wanshen Wang: ES. Hongying Duan: FG. Jun Wei: AB. Juanjuan Chen: MT. All authors read and approved the final manuscript.
Yonghua Zhang and Jun Wei contributed equally to this work.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Zhou, Y., Zhang, Y., Wei, J. et al. Cloning and analysis of expression patterns and transcriptional regulation of RghBNG in response to plant growth regulators and abiotic stresses in Rehmannia glutinosa . SpringerPlus 4, 60 (2015). https://doi.org/10.1186/s40064-015-0830-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40064-015-0830-0