Abstract
Background
Mupirocin is widely used for nasal decolonization of Staphylococcus aureus to prevent subsequent staphylococcal infection in patients and healthcare personnel. However, the prolonged and unrestricted use has led to the emergence of mupirocin-resistant (mupR) S. aureus. The aim of this systematic review was to investigate the prevalence, phenotypic and molecular characteristics, and geographic spread of mupR S. aureus in Africa.
Methods
We examined five electronic databases (EBSCOhost, Google Scholar, ISI Web of Science, MEDLINE, and Scopus) for relevant English articles on screening for mupR S. aureus from various samples in Africa. In addition, we performed random effects meta-analysis of proportions to determine the pooled prevalence of mupR S. aureus in Africa. The search was conducted until 3 August 2016.
Results
We identified 43 eligible studies of which 11 (26%) were obtained only through Google Scholar. Most of the eligible studies (28/43; 65%) were conducted in Nigeria (10/43; 23%), Egypt (7/43; 16%), South Africa (6/43; 14%) and Tunisia (5/43; 12%). Overall, screening for mupR S. aureus was described in only 12 of 54 (22%) African countries. The disk diffusion method was the widely used technique (67%; 29/43) for the detection of mupR S. aureus in Africa. The mupA-positive S. aureus isolates were identified in five studies conducted in Egypt (n = 2), South Africa (n = 2), and Nigeria (n = 1). Low-level resistance (LmupR) and high-level resistance (HmupR) were both reported in six human studies from South Africa (n = 3), Egypt (n = 2) and Libya (n = 1). Data on mupR-MRSA was available in 11 studies from five countries, including Egypt, Ghana, Libya, Nigeria and South Africa. The pooled prevalence (based on 11 human studies) of mupR S. aureus in Africa was 14% (95% CI =6.8 to 23.2%). The proportion of mupA-positive S. aureus in Africa ranged between 0.5 and 8%. Furthermore, the frequency of S. aureus isolates that exhibited LmupR, HmupR and mupR-MRSA in Africa were 4 and 47%, 0.5 and 38%, 5 and 50%, respectively.
Conclusions
The prevalence of mupR S. aureus in Africa (14%) is worrisome and there is a need for data on administration and use of mupirocin. The disk diffusion method which is widely utilized in Africa could be an important method for the screening and identification of mupR S. aureus. Moreover, we advocate for surveillance studies with appropriate guidelines for screening mupR S. aureus in Africa.
Similar content being viewed by others
Background
Staphylococcus aureus is a well-recognized human pathogen that is implicated in a wide array of superficial, invasive and toxigenic infections [1]. Meta-analyses of published studies have provided evidence that S. aureus nasal carriage is an important risk factor for subsequent infection among patients with surgical site infections and atopic dermatitis [2, 3]. Other high-risk groups include patients colonized with methicillin-resistant Staphylococcus aureus (MRSA) undergoing dialysis, and patients admitted in the intensive care unit [4, 5]. Consequently, infection prevention strategies such as nasal decolonization are employed to minimize the occurrence of staphylococcal infection and reduce the risk of transmission in healthcare settings [6, 7]. Mupirocin (2%) nasal ointment alone or in combination with 4% chlorhexidine (CHG) based body wash is considered as the main decolonization strategy for S. aureus carriage [8, 9]. Mupirocin is a naturally occurring antibiotic produced by Pseudomonas fluorescens that interferes with protein synthesis by competitive inhibition of the bacterial isoleucyl-tRNA synthetase (IRS) [10, 11]. It gained prominence in the mid-1990s for the eradication of S. aureus nasal carriage due to its effectiveness, safety and cost [12].
Mupirocin-resistant (mupR) S. aureus was first reported in the United Kingdom in 1987 [13]. Since then, it has been reported in several countries worldwide [14–17]. The emergence of mupR S. aureus has been associated with unrestricted policies and use of mupirocin for long periods in health care settings [8, 18]. Decolonization failure in patients with S. aureus carriage is associated with high-level mupirocin resistance (HmupR - minimum inhibitory concentration [MIC]: ≥512 μg/ml), while that of low-level mupirocin resistance (LmupR – MIC: 8-64 μg/ml) is still unclear [7, 19]. LmupR is mediated through point mutation (largely V588F and V631F) in the native isoleucyl-tRNA synthetase (ileS) gene [20]. In contrast, HmupR is mainly attributed to the acquisition of plasmids with the mupA (or ileS2) gene encoding an additional IRS with no affinity for mupirocin [11, 21]. Another determinant for HmupR is the acquisition of a plasmid-mediated mupB gene [22].
There is no data summarizing reports on screening, prevalence, characterization, and geographic spread of mupR S. aureus in Africa. This systematic review evaluated published articles that assessed for mupirocin resistance in African S. aureus isolates. The findings from this systematic review highlight the need to develop an early warning system, including harmonized strategies for the prompt screening and identification of mupR S. aureus in Africa.
Methods
Literature search strategy
The relevant English articles from human and animal investigations were retrieved by three authors (YA, SA, and AS) from five electronic databases (EBSCOhost, Google Scholar, ISI Web of Science, MEDLINE, and Scopus). The search terms for each database are reported in Table 1. The literature search was concluded on 3 August 2016.
Eligible article identification
The identification of the eligible articles was conducted according to the guidelines for preferred reporting items for systematic reviews and meta-analyses (PRISMA) [23]. We defined an eligible article as a peer-reviewed publication that (i) included mupirocin in the antibiotic susceptibility testing of S. aureus isolates, and (ii) employed phenotypic ((disc diffusion, E-test, minimum inhibitory concentration (MIC), VITEK and other automated methods)), and/or molecular ((conventional or real-time polymerase chain reaction (PCR)) techniques. International multicentre studies that included African countries were also eligible for inclusion.
Data extraction and analysis
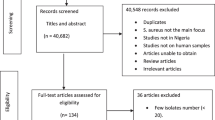
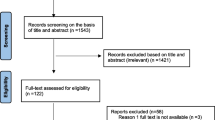
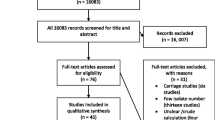
The relevant data were extracted from each of the eligible articles included in this systematic review. A study that analysed S. aureus isolates from another investigation but answered a different research question were both considered as one study (Table 2). We performed three levels of analysis (Fig. 1). First, to understand the characteristics and geographic spread of mupR S. aureus in Africa, studies that included mupirocin in the antibiotic susceptibility testing and employed phenotypic and/or molecular techniques were identified. Secondly, the prevalence of S. aureus with the mupA gene, isolates that expressed LmupR and HmupR, and mupR-MRSA in Africa were derived from each eligible study as follows:
Thirdly, to estimate the prevalence of mupR S. aureus in humans, studies that employed at least one of the screening methods with defined breakpoint for mupirocin resistance were included in the meta-analysis. The StatsDirect statistical software version 3.0.165 (England: StatsDirectLtd.2016) was utilized to assess the heterogeneity of the eligible studies included in the meta-analysis (Cochran Q-test) [24], and to ascertain the inconsistency across the studies (I2 statistic) [25]. The random effects model was used to determine the pooled prevalence of mupR S. aureus in Africa. The criterion for statistical significance for heterogeneity was set at alpha = 0.05. The risk of publication bias was assessed and visualized by a Funnel plot [26, 27].
Results
Eligible studies from electronic database search
We identified 43 reports (Table 1) of which 34 studies investigated only human samples. The remaining nine studies assessed samples from only animals (n = 5), human and environmental sources (n = 2), human and animal sources (n = 1), and cockroaches (n = 1). Most of the eligible studies (32/43; 74%) were obtained from EBSCOhost, ISI Web of Science, MEDLINE, and Scopus. The remaining studies (11/43; 26%) were obtained only through Google Scholar and consisted of studies conducted in Egypt [28–31], South Africa [32–34], Nigeria [35, 36], Ethiopia [37] and Kenya [38].
Screening and identification of mupR S. aureus in Africa
Only 12 of the 54 (22%) African countries reported data on screening for mupR S. aureus (Fig. 2). The first published article indicated that mupirocin had been in use in Africa, at least from the late 1980s [39]. Most of these studies (28/43; 65%) were conducted in Nigeria (10/43; 23%), Egypt (7/43; 16%), South Africa (6/43; 14%) and Tunisia (5/43; 12%) (Fig. 2). MupR S. aureus was mainly identified through the disk diffusion method (29/43; 67%). The guidelines by the Clinical and Laboratory Standards Institute (CLSI), previously known as National Committee for Clinical Laboratory Standards (NCCLS), were broadly used in Africa (Table 2). However, a number of studies [28, 29, 31, 33, 36, 40–46] utilized the disk diffusion method with CLSI guidelines that had no zone diameter breakpoint for mupirocin. Moreover, some studies [47–49] did not provide information on the year of publication of the CLSI guidelines. MupR S. aureus was reported in six African countries including South Africa [32–34, 46, 50, 51], Egypt [29–31, 52], Nigeria [36, 44, 53], Ghana [54, 55], Libya [56, 57] and Ethiopia [37] (Fig. 2; Table 2). The mupA-positive S. aureus was detected in five studies from Egypt [30, 52], South Africa [33, 50] and Nigeria [53]. LmupR and HmupR were both reported in six human studies conducted in South Africa [32, 33, 50], Egypt [30, 52] and Libya [57]. The mupR-MRSA isolates were identified in South Africa [32, 34, 50, 51], Egypt [30, 31, 52], Libya [56, 57], Ghana [55] and Nigeria [36] (Table 3). MupR-MRSA was not reported from MRSA isolates recovered from studies conducted in Egypt [28, 58], Tunisia [59, 60] and Algeria [47].
An assessment of data on mupR S. aureus at the regional level is described as follows (Fig. 3).
North Africa
Seventeen eligible studies were recorded from this region, including Egypt [28–31, 40, 52, 58], Tunisia [41–43, 59, 60], Libya [56, 57, 61], Algeria [47] and Morocco [62]. MupR S. aureus was reported in six studies conducted in two North African countries: Egypt [29–31, 52] and Libya [56, 57]. PCR detection of the mupA gene was performed in only two studies conducted in Egypt [30, 52]. In addition, one of the reports identified two mupA positive MRSA that exhibited LmupR [30]. MupR S. aureus was not detected in Tunisia [41–43, 59, 60], Algeria [47], and Morocco [62].
West Africa
S. aureus resistance to mupirocin was investigated in Nigeria [35, 36, 44, 48, 49, 53, 63–66] and Ghana [54, 55, 67, 68]. Only two studies from Ghana reported on mupR S. aureus [54, 55]. In Nigeria, three studies (including two from only human sources and one from both animal and human samples, respectively) reported on S. aureus isolates that demonstrated HmupR [36, 44, 53].
Central Africa
MupR S. aureus was not detected in studies conducted in Gabon [69], and São Tomé and Príncipe [70].
East Africa
In this review, we identified four eligible studies conducted in Kenya [38, 71, 72] and Ethiopia [37]. A report on the role of cockroaches as potential vectors of foodborne pathogens in Ethiopia identified 17 mupR S. aureus isolates [37]. All the S. aureus isolates (one animal and two human studies) from Kenya were susceptible to mupirocin [38, 71, 72].
Southern Africa
The six studies reported in this geographical area were from South Africa and consisted of two single centre studies [34, 46] and four multicenter studies [32, 33, 50, 51]. MupR S. aureus was identified in all the reports, while mupA-positive S. aureus isolates were noted in only two studies [33, 50].
Prevalence of mupR S. aureus in Africa
The random-effects pooled prevalence of mupR S. aureus in Africa is 14% (95% CI =6.8 to 23.2%). This was calculated based on 11 heterogeneous human studies (Figs. 4 and 5) conducted in South Africa [32, 33, 50, 51], Ghana [54, 55], Egypt [30, 52], Libya [56, 57] and Nigeria [53]. In Africa, the proportion of S. aureus isolates with the mupA gene, and those that expressed LmupR and HmupR ranged between 0.5 and 8%, 4 and 47%, 0.5 and 38%, respectively. The frequency of mupR-MRSA isolates ranged between 5 and 50% (Table 3).
Bias assessment (Funnel) plot for studies assessing rates of mupirocin-resistant Staphylococcus aureus in Africa. Random effects (DerSimonian-Laird). Pooled proportion = 0.139303 (95% CI = 0.067511 to 0.23165). Bias indicators, Begg-Mazumdar: Kendall’s tau = 0.2 P = 0.4454, Egger: bias = 4.771137 (95% CI = −2.517874 to 12.060148) P = 0.1728, Harbord: bias = 2.014783 (92.5% CI = −5.90181 to 9.931377) P = 0.6208
Association of MupR S. aureus with mupirocin use in Africa
There is no data on the use of mupirocin as an agent for S. aureus decolonization and its association with mupR S. aureus in Africa.
MupR S. aureus and biofilm production
A report from Egypt noted that mupR-MRSA were moderate to strong biofilm producers [52].
MupR S. aureus and co-resistance to other antibiotics
In this systematic review, two studies (conducted in Egypt and South Africa) showed that mupR S. aureus was associated with multi-drug resistance [30, 33].
Molecular characterization of mupR S. aureus in Africa
Only three studies provided molecular data on mupR S. aureus in Africa [45, 54, 55]. A report provided evidence of a 35 kb (non-conjugative) and 41.1 kb (conjugative) plasmid encoding mupA in S. aureus isolates from Nigeria and South Africa [45]. It also described an MRSA clone that demonstrated LmupR in South Africa. LmupR was also identified among MRSA isolates assigned with ST36, ST88, and ST789 in Ghana [55]. A cross-sectional S. aureus study identified a methicillin susceptible S. aureus (MSSA) strain with HmupR from a 51-year-old hospital staff in Ghana [54]. Molecular characterization indicated that the strain (spa type t4805) was PVL-positive.
Discussion
This is the first systematic review on mupR S. aureus in Africa and clearly showed the paucity of data on the continent. Nevertheless, this study indicated a high prevalence ((14% (95% CI =6.8 to 23.2)) of mupR S. aureus in Africa. These observations support the need for mupR S. aureus surveillance data to provide information on its epidemiology and clinical significance in Africa. It is noteworthy that Google Scholar was valuable in the identification of several eligible studies [28–38]. We observed that 26% (11/43) of the eligible studies were identified from African journals which were not indexed in commonly used electronic databases. Google Scholar has been considered as a useful supplement with other electronic databases for systematic review search [73] including recent meta-analyses of published studies on S. aureus in Africa [74, 75].
The phenotypic methods for the screening and identification of mupR S. aureus include disc diffusion (two-disc strategy: 5 μg and 200 μg), agar dilution, broth micro-dilution and E-test [19]. In this study, the disk diffusion method and the CLSI (formerly NCCLS) guidelines were strategies mainly applied to detect mupR S. aureus in Africa. However, we observed certain inconsistencies [28, 29, 31, 33, 36, 40–49]. For instance, a number of studies [28, 29, 31, 33, 36, 40–42, 44–46] applied the disk diffusion method with the CLSI guidelines that had no breakpoint values for mupirocin. The 2017 CLSI guidelines recommend the use of the 200 μg disk to differentiate between HmupR and the absence of HmupR (i.e. no zone = HmupR; any zone = absence of HmupR) [76]. The 200 μg disk with a different breakpoint (Susceptible ≥30 mm, Resistance < 18 mm) is also endorsed for the differentiation between HmupR and the absence of HmupR in the latest versions (accessed 28th May, 2018) of the European Committee for Antimicrobial Susceptibility Testing (EUCAST) and Comité de l’antibiogramme de la Société Française de Microbiologie (CA-SFM) [77, 78]. The breakpoint values for the detection of LmupR and differentiation from HmupR are not provided in these documents (CA-SFM, CLSI, and EUCAST). Despite this limitation, the disk diffusion method in conjunction with any of these guidelines could at least be valuable for the preliminary screening and identification of HmupR S. aureus in Africa. MRSA decolonization failure is of clinical significance as it is often attributed to persistence or re-colonization associated with isolates exhibiting HmupR, while that of LmupR is not clear [7, 19, 79]. In this review, the prevalence of S. aureus that exhibited LmupR, HmupR and mupR-MRSA in Africa was predicated on a range of methods using different guidelines. We suggest that surveillance data from Africa is established on harmonized guidelines to enhance quality assurance and comparison at the continental and global level.
We noted a prevalence of mupR-MRSA ranging between 5 and 50% in Africa (Table 3). This is of serious concern. Specifically, the relationship between mupirocin resistance and MRSA has important consequences on infection control measures and effectiveness of decolonization strategies [8]. MupR-MRSA could limit the choices available for the control and prevention of healthcare-associated MRSA infections (7, 8). Therefore, surveillance studies are important to investigate the emergence and spread of mupirocin resistance in hospital settings in Africa. This is important among patients at high risk of MRSA infections, including patients in the dermatology, dialysis and the Intensive Care Units. In addition, there is the need for more data on the molecular characterization of mupR S. aureus in Africa [45, 54, 55]. For instance, whole genome sequencing (WGS) will assist in understanding the transmission dynamics of mupR S. aureus in Africa. Moreover, WGS data will allow comprehensive investigation of the genetic basis for LmupR mutation (which is largely due to V588F and V631F in the native gene (ileS)) and mupB-positive S. aureus in Africa.
Language bias was the main limitation of this systematic review as we did not include studies published in French, Portuguese, Arabic and Spanish.
Conclusions
This study showed the need for more epidemiological data to understand the transmission, burden and risk factors associated with mupR S. aureus in Africa. In addition, there is a need for data on administration and use of mupirocin in community and hospital setting in Africa. This is important in antibiotic stewardship to mitigate the emergence and spread of mupR S. aureus in Africa. Finally, this systematic review highlighted the need for harmonized guidelines to facilitate the comparison of data on mupR S. aureus from Africa.
Abbreviations
- HmupR:
-
High-level mupirocin resistance
- LmupR:
-
Low-level mupirocin resistance
- MIC:
-
Minimum inhibitory concentration
- MRSA:
-
Methicillin-resistant Staphylococcus aureus
- MSSA:
-
Methicillin-susceptible S. aureus
- mupR:
-
Mupirocin-resistant
- PCR:
-
Polymerase chain reaction
- PVL:
-
Panton Valentine Leucocidin
- S. aureus :
-
Staphylococcus aureus
- ST:
-
Sequence type
References
Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–32.
Levy PY, Ollivier M, Drancourt M, Raoult D, Argenson JN. Relation between nasal carriage of Staphylococcus aureus and surgical site infection in orthopedic surgery: the role of nasal contamination. A systematic literature review and meta-analysis. Orthop Traumatol Surg Res. 2013;99:645–51. https://doi.org/10.1016/j.otsr.2013.03.030.
Totté JE, van der Feltz WT, Hennekam M, van Belkum A, van Zuuren EJ, Pasmans SG. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: a systematic review and meta-analysis. Br J Dermatol. 2016;175:687–95. https://doi.org/10.1111/bjd.14566.
Zacharioudakis IM, Zervou FN, Ziakas PD, Mylonakis E. Meta-analysis of methicillin-resistant Staphylococcus aureus colonization and risk of infection in dialysis patients. J Am Soc Nephrol. 2014;25:2131–41. https://doi.org/10.1681/ASN.2013091028.
Ziakas PD, Anagnostou T, Mylonakis E. The prevalence and significance of methicillin-resistant Staphylococcus aureus colonization at admission in the general ICU setting: a meta-analysis of published studies. Crit Care Med. 2014;42:433–44. https://doi.org/10.1097/CCM.0b013e3182a66bb8.
Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–62. https://doi.org/10.1016/S1473-3099(05)70295-4.
Septimus EJ, Schweizer ML. Decolonization in prevention of health-care associated infections. Clin Microbiol Rev. 2016;29:201–22. https://doi.org/10.1128/CMR.00049-15.
Poovelikunnel T, Gethin G, Humphreys H. Mupirocin resistance: clinical implications and potential alternatives for the eradication of MRSA. J Antimicrob Chemother. 2015;70:2681–92. https://doi.org/10.1093/jac/dkv169.
Global Guidelines for the prevention of surgical site infection. World Health Organization, Geneva. 2016. http://www.who.int/gpsc/ssi-prevention-guidelines/en/ Accessed 15 June 2017.
Fuller AT, Mellows G, Woolford M, Banks GT, Barrow KD, Chain EB. Pseudomonic acid: an antibiotic produced by Pseudomonas fluorescens. Nature. 1971;234:416–7.
Gilbart J, Perry CR, Slocombe B. High-level mupirocin resistance in Staphylococcus aureus: evidence for two distinct isoleucyl-tRNA synthetases. Antimicrob Agents Chemother. 1993;37:32–8.
Perl TM, Golub JE. New approaches to reduce Staphylococcus aureus nosocomial infection rates: treating S. aureus nasal carriage. Ann Pharmacother. 1998;32:S7–16.
Rahman M, Noble WC, Cookson B. Mupirocin resistant Staphylococcus aureus. Lancet. 1987;330:387–8. https://doi.org/10.1016/S0140-6736(87)92398-1.
Hughes J, Stabler R, Gaunt M, Karadag T, Desai N, Betley J, Ioannou A, Aryee A, Hearn P, Marbach H, Patel A, Otter JA, Edgeworth JD, Tosas AO. Clonal variation in high- and low-level phenotypic and genotypic mupirocin resistance of MRSA isolates in south-East London. J Antimicrob Chemother. 2015;70:3191–9. https://doi.org/10.1093/jac/dkv248.
Boswihi SS, Udo EE, Al-Sweih N. Shifts in the clonal distribution of methicillin-resistant Staphylococcus aureus in Kuwait hospitals: 1992-2010. PLoS One. 2016;11:e0162744. https://doi.org/10.1371/journal.pone.0162744.
Hayden MK, Lolans K, Haffenreffer K, Avery TR, Kleinman K, Li H, Kaganov RE, Lankiewicz J, Moody J, Septimus E, Weinstein RA, Hickok J, Jernigan J, Perlin JB, Platt R, Huang SS. Chlorhexidine and mupirocin susceptibility of methicillin-resistant Staphylococcus aureus isolates in the REDUCE-MRSA trial. J Clin Microbiol. 2016;54:2735–42.
Gostev V, Kruglov A, Kalinogorskaya O, Dmitrenko O, Khokhlova O, Yamamoto T, Lobzin Y, Ryabchenko I, Sidorenko S. Molecular epidemiology and antibiotic resistance of methicillin-resistant Staphylococcus aureus circulating in the Russian Federation. Infect Genet Evol. 2017;53:189–94. https://doi.org/10.1016/j.meegid.2017.06.006.
Hetem DJ, Bonten MJ. Clinical relevance of mupirocin resistance in Staphylococcus aureus. J Hosp Infect. 2013;85:249–56. https://doi.org/10.1016/j.jhin.2013.09.006.
Swenson JM, Wong B, Simor AE, Thomson RB, Ferraro MJ, Hardy DJ, Hindler J, Jorgensen J, Reller LB, Traczewski M, McDougal LK, Patel JB. Multicenter study to determine disk diffusion and broth microdilution criteria for prediction of high- and low-level mupirocin resistance in Staphylococcus aureus. J Clin Microbiol. 2010;48:2469–75. https://doi.org/10.1128/JCM.00340-10.
Antonio M, McFerran N, Pallen MJ. Mutation affecting the Rossman fold of isoleucyl-tRNA synthetase are correlated with low-level mupirocin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:438–42. https://doi.org/10.1128/AAC.46.2.438-442.2002.
Hodgson JE, Curnock SP, Dyke KG, Morris R, Sylvester DR, Gross MS. Molecular characterization of the gene encoding high-level mupirocin resistance in Staphylococcus aureus J2870. Antimicrob Agents Chemother. 1994;38:1205–8. https://doi.org/10.1128/AAC.38.5.1205.
Seah C, Alexander DC, Louie L, Simor A, Low DE, Longtin J, Melano RG. MupB, a new high-level mupirocin resistance mechanism in Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56:1916–20. https://doi.org/10.1128/AAC.05325-11.
Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. https://doi.org/10.1371/journal.pmed.1000097.
Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–29.
Huggins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. https://doi.org/10.1136/bmj.327.7414.557.
Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. https://doi.org/10.1136/bmj.315.7109.629.
Sterne JA, Sutton AJ, Ioannidis JP, Terrin N, Jones DR, Lau J, Carpenter J, Rücker G, Harbord RM, Schmid CH, Tetzlaff J, Deeks JJ, Peters J, Macaskill P, Schwarzer G, Duval S, Altman DG, Moher D, Higgins JP. Recommendations for examining and interpreting funnel plots assymettry in meta-analysis of randomised control trials. BMJ. 2011;342:1–8. https://doi.org/10.1136/bmj.d4002.
Salama MF. Comparative molecular analysis of community or health care associated methicillin-resistant Staphylococcus aureus. Egypt J Med Microbiol. 2006;15:371–80.
Taher S, Roshdy H. Prevalence of Panton-Valentine Leukocidin genes among Staphylococcus aureus isolates in Mansoura University hospitals. Egypt J Med Microbiol. 2009;18:97–108.
Wali I, Ouda N, El-Seidi E. Mupirocin resistance among methicillin resistant Staphylococcus aureus isolates in an Egyptian hospital. Egypt J Med Lab Sci. 2011;20:1–11.
Melake N, Zakaria AS, Ibrahim NH, Salama M, Mahmoud AZ. Prevalence of agr specificity groups among in vitro biofilm forming methicillin resistant Staphylococcus aureus strains isolated from nasal carriers. Int J Microbiol Res. 2014;5:76–84. https://doi.org/10.5829/idosi.ijmr.2014.5.2.83184.
Marais E, Aithma N, Perovic O, Oosthuyen WF, Musenge E, Dusé AG. Antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolates from South Africa. S Afr Med J. 2009;99:170–3.
Wasserman E, Orth H, Senekal M, Harvey K. High prevalence of mupirocin resistance associated with resistance to other antimicrobial agents in Staphylococcus aureus isolated from patients in private health care, Western Cape. South Afr J Infect Dis. 2014;29:126–32.
Swe Swe K, Naidoo N, Jaglal P. Molecular epidemiology of a suspected methicillin-resistant Staphylococcus aureus outbreak in a renal unit of a central academic hospital in KwaZulu-Natal, South Africa. South Afr J Infect Dis. 2015;30:6–10.
Bamaiyi PH, Aniesona AT. Prevalence and antimicrobial susceptibility patterns of bovine and ovine Staphylococcus aureus isolates in Maiduguri, Nigeria. Adv Anim Vet Sci. 2013;1:59–64.
Mai-siyama IB, Okon KO, Adamu NB, Askira UM, Isyaka TM, Adamu SG, Mohammed A. Methicillin-resistant Staphylococcus aureus (MRSA) colonization rate among ruminant animals slaughtered for human consumption and contact persons in Maiduguri, Nigeria. Afr J Microbiol Res. 2014;8:2643–9. https://doi.org/10.5897/AJMR2014.6855.
Tachbele E, Erku W, Gebre-Michael T, Ashenafi M. Cockroach-associated food-borne bacterial pathogens from some hospitals and restaurants in Addis Ababa, Ethiopia: Distribution and antibiograms. JRTPH. 2006;5:34–41.
Njage PMK, Dolci S, Jans C, Wangoh J, Lacroix C, Meile L. Phenotypic and genotypic antibiotic resistance patterns of Staphylococcus aureus from raw and spontaneously fermented camel milk. BJAST. 2013;3(3):87–98.
Rode H, Hanslo D, de Wet PM, Millar AJW, Cywes S. Efficacy of mupirocin in methicillin-resistant Staphylococcus aureus burn wound infection. Antimicrob Agents Chemother. 1989;33:1358–61.
Salem-Bekhit M. Phenotypic and genotypic characterization of nosocomial isolates of Staphylococcus aureus with reference to methicillin resistance. Trop J Pharm Res. 2014;13:1239–46. https://doi.org/10.4314/tjpr.v13i8.7.
Ben Slama K, Gharsa H, Klibi N, Jouini A, Lozano C, Gómez-Sanz E, Zarazaga M, Boudabous A, Torres C. Nasal carriage of Staphylococcus aureus in healthy humans with different levels of contact with animals in Tunisia: genetic lineages, methicillin resistance, and virulence factors. Eur J Clin Microbiol Infect Dis. 2011;30:499–508. https://doi.org/10.1007/s10096-010-1109-6.
Gharsa H, Slama KB, Lozano C, Gomez-Sanz E, Klibi N, Sallem RB, Gomez P, Zarazaga M, Boudabous A, Torres C. Prevalence, antibiotic resistance, virulence traits and genetic lineages of Staphylococcus aureus in healthy sheep in Tunisia. Vet Microbiol. 2012;156:367–73. https://doi.org/10.1016/j.vetmic.2011.11.009.
Gharsa H, Sallem RB, Slama KB, Gomez-Sanz E, Lazano C, Jouini A, Klibi N, Zarazaga M, Boudabous A, Torres C. High diversity of genetic lineages and virulence genes in nasal Staphylococcus aureus isolates from donkeys destined to food consumption in Tunisia with predominance of the ruminant associated CC133 lineage. BMC Vet Res. 2012;8:203. https://doi.org/10.1186/1746-6148-8-203.
Olonitola OS, Inabo HI, Olayinka BO, Bugo ID. Nasal carriage of methicillin-resistant Staphylococcus aureus by primary school pupils in a university staff school, Zaria, Nigeria. Int J Bio. Chem Sci. 2007;1:71–5. https://doi.org/10.4314/ijbcs.v1i1.39701.
Shittu AO, Udo EE, Lin J. Phenotypic and molecular characterization of Staphylococcus aureus isolates expressing low- and high-level mupirocin resistance in Nigeria and South Africa. BMC Infect Dis. 2009;9:10. https://doi.org/10.1186/1471-2334-9-10.
Zinn CS, Westh H, Rosdahl VT. SARISA study group. An international multicenter study of antimicrobial resistance and typing of hospital Staphylococcus aureus isolates from 21 laboratories in 19 countries or states. Microb Drug Resist. 2004;10:160–8. https://doi.org/10.1089/1076629041310055.
Ouchenane Z, Smati F, Rolain J-M, Raoult D. Molecular characterization of methicillin-resistant Staphylococcus aureus isolates in Algeria. Pathol Biol (Paris). 2011;59:e129–32. https://doi.org/10.1016/j.patbio.2009.11.004.
Okon KO, Basset P, Uba A, Lin J, Oyawoye B, Shittu AO, Blanc DS. Co-occurrence of predominant Panton-Valentine Leukocidin-positive sequence type (ST) 152 and multidrug-resistant ST 241 Staphylococcus aureus clones in Nigerian hospitals. J Clin Microbiol. 2009;47:3000–3. https://doi.org/10.1128/JCM.01119-09.
Raji A, Ojemhen O, Umejiburu U, Ogunleye A, Blanc D, Basset P. High genetic diversity of Staphylococcus aureus in a tertiary care hospital in Southwest Nigeria. Diagn Microbiol Infect Dis. 2013;77:367–9. https://doi.org/10.1016/j.diagmicrobio.2013.08.030.
Shittu AO, Lin J. Antimicrobial susceptibility patterns and characterization of clinical isolates of Staphylococcus aureus in KwaZulu-Natal province, South Africa. BMC Infect Dis. 2006;6:125. https://doi.org/10.1186/1471-2334-6-125.
Perovic O, Iyaloo S, Kularatne R, Lowman W, Bosman N, Wadula J, Seetharam S, Duse A, Mbelle N, Bamford C, Dawood H, Mahabeer Y, Bhola P, Abrahams S, Singh-Moodley A. Prevalence and trends of Staphylococcus aureus bacteraemia in hospitalized patients in South Africa, 2010-2012: laboratory-based surveillance mapping of antimicrobial resistance and molecular epidemiology. PLoS One. 2015;10:e0145429. https://doi.org/10.1371/journal.pone.0145429.
Barakat GI, Nabil YM. Correlation of mupirocin resistance with biofilm production in methicillin-resistant Staphylococcus aureus from surgical site infections in a tertiary Centre, Egypt. J Glob Antimicrob Resist. 2016;4:16–20. https://doi.org/10.1016/j.jgar.2015.11.010.
Shittu A, Lin J, Kolawole D. Antimicrobial susceptibility patterns of Staphylococcus aureus and characterization of MRSA in southwestern Nigeria. Wounds. 2006;18:77–84.
Egyir B, Guardabassi L, Nielsen SS, Larsen J, Addo KK, Newman MJ, Larsen AR. Prevalence of nasal carriage and diversity of Staphylococcus aureus among inpatients and hospital staff at Korle Bu teaching hospital, Ghana. J Glob Antimicrob Resist. 2013;1:189–93. https://doi.org/10.1016/j.jgar.2013.05.006.
Egyir B, Guardabassi L, Monecke S, Addo KK, Newman MJ, Larsen AR. Methicillin-resistant Staphylococcus aureus strains from Ghana include USA300. J Glob Antimicrob Resist. 2015;3:26–30. https://doi.org/10.1016/j.jgar.2014.11.006.
Ahmed MO, Abuzweda AR, Alghazali MH, Elramalli AK, Amri SG, Aghila ES, Abouzeed YM. Misidentification of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals in Tripoli, Libya. Libyan J Med. 2010;5:5230. https://doi.org/10.3402/ljm.v5i0.5230.
Ahmed MO, Elramalli AK, Amri SG, Abuzweda AR, Abouzeed YM. Isolation and screening of methicillin-resistant Staphylococcus aureus from health care workers in Libyan hospitals. EMHJ. 2012;18:37–42.
Enany S, Yaoita E, Yoshida Y, Enany M, Yamamoto T. Molecular characterization of Panton-Valentine Leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus isolates in Egypt. Microbiol Res. 2010;165:152–62. https://doi.org/10.1016/j.micres.2009.03.005.
Ben Nejma MB, Mastouri M, Jrad BBH, Nour M. Characterization of ST80 Panton-Valentine Leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus clone in Tunisia. Diagn Microbiol Infect Dis. 2013;77:20–4. https://doi.org/10.1016/j.diagmicrobio.2008.02.010.
Ben Nejma MB, Merghni A, Mastouri M. Genotyping of methicillin resistant Staphylococcus aureus strains isolated from hospitalized children. Int J Pediatr. 2014;2014:314316. https://doi.org/10.1155/2014/314316.
Ferghani NEL. An open study of mupirocin in Libyan patients with skin infections. J Int Med Res. 1995;23:508–17. https://doi.org/10.1177/030006059502300615.
Souly K, Ait el Kadi M, Lhmadi K, Biougnach H, Boughaidi A, Zouhdi M, Benasila S, Elyoussefi Z, Bouattar T, Zbiti N, Skalli Z, Rhou H, Ouzeddoun N, Bayahia R, Benamar L. Epidemiology and prevention of Staphylococcus aureus nasal carriage in hemodialyzed patients. Med Mal Infect. 2011;41:469–74. https://doi.org/10.1016/j.medmal.2011.05.005.
Shittu AO, Okon K, Adesida S, Oyedara O, Witte W, Strommenger B, Layer F, Nübel U. Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol. 2011;11:92. https://doi.org/10.1186/1471-2180-11-92.
Shittu A, Oyedara O, Abegunrin F, Okon K, Raji A, Taiwo S, Ogunsola F, Onyedibe K, Elisha G. Characterization of methicillin-susceptible and -resistant staphylococci in the clinical setting: a multicentre study in Nigeria. BMC Infect Dis. 2012;12:286. https://doi.org/10.1186/1471-2334-12-286.
Ayepola OO, Olasupo NA, Egwari LO, Becker K, Schaumburg F. Molecular characterization and antimicrobial susceptibility of Staphylococcus aureus isolates from clinical infection and asymptomatic carriers in Southwest Nigeria. PLoS One. 2015;10:e0137531. https://doi.org/10.1371/journal.pone.0137531.
Akobi B, Aboderin O, Sasaki T, Shittu A. Characterization of Staphylococcus aureus isolates from faecal samples of the Straw-Coloured Fruit Bat (Eidolon helvum) in Obafemi Awolowo University (OAU), Nigeria. BMC Microbiol. 2012;12:279. https://doi.org/10.1186/1471-2180-12-279.
Egyir B, Guardabassi L, Esson J, Nielsen SS, Newman MJ, Addo KK, Larsen AR. Insights into nasal carriage of Staphylococcus aureus in an urban and a rural Community in Ghana. PLoS One. 2014;9:e96119. https://doi.org/10.1371/journal.pone.0096119.
Amissah NA, Glasner C, Ablordey A, Tetteh CS, Kotey NK, Prah I, van der Werf TS, Rossen JW, van Dijl JM, Stienstra Y. Genetic diversity of Staphylococcus aureus in Buruli ulcer. PLoS Negl Trop Dis. 2015;9:e0003421. https://doi.org/10.1371/journal.pntd.0003421.
Ngoa UA, Schaumburg F, Adegnika AA, Kösters K, Möller T, Fernandes JF, Alabi A, Issifou S, Becker K, Grobusch MP, Kremsner PG, Lell B. Epidemiology and population structure of Staphylococcus aureus in various population groups from a rural and semi-urban area in Gabon, Central Africa. Acta Trop. 2012;124:42–7. https://doi.org/10.1016/j.actatropica.2012.06.005.
Conceição T, Silva IS, de Lencastre H, Aires-de-Sousa M. Staphylococcus aureus nasal carriage among patients and health care workers in São Tomé and Príncipe. Microb Drug Resist. 2014;20:57–66. https://doi.org/10.1089/mdr.2013.0136.
Aiken AM, Mutuku IM, Sabat AJ, Akkerboom V, Mwangi J, Scott JAG, Morpeth SC, Friedrich AW, Grundmann H. Carriage of Staphylococcus aureus in Thika level 5 hospital, Kenya: a cross-sectional study. Antimicrob Resist Infect Control. 2014;3:22. https://doi.org/10.1186/2047-2994-3-22.
Omuse G, Kabera B, Revathi G. Low prevalence of methicillin resistant Staphylococcus aureus as determined by an automated identification system in two private hospitals in Nairobi, Kenya: a cross sectional study. BMC Infect Dis. 2014;14:669. https://doi.org/10.1186/s12879-014-0669-y.
Haddaway NR, Collins AM, Coughlin D, Kirk S. The role of Google scholar in evidence reviews and its applicability to Grey literature searching. PLoS One. 2015;10:e0138237. https://doi.org/10.1371/journal.pone.0138237.
Eshetie S, Tarekegn F, Moges F, Amsalu A, Birhan W, Huruy K. Methicillin resistant Staphylococcus aureus in Ethiopia: a meta-analysis. BMC Infect Dis. 2016;16:689. https://doi.org/10.1186/s12879-016-2014-0.
Deyno S, Fekadu S, Astatkie A. Resistance of Staphylococcus aureus to antimicrobial agents in Ethiopia: a meta-analysis. Antimicrob Resist Infect Control. 2017;6:85. https://doi.org/10.1186/s13756-017-0243-7.
Clinical and Laboratory Standard Institute (CLSI). Performance standards for antimicrobial susceptibility testing: 27th edition. CLSI supplement M100. Wayne CLSI. 2017.
The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 8.1, 2018. http://www.eucast.org. Accessed 28th May, 2018.
Comite de l’antibiogramme de la Societe Francaise de Microbiologie – recommendations 2018 v.1.0 mai. http://www.sfm-microbiologie.org. Accessed 28th May, 2018.
Hurdle JG, O’Neill AJ, Mody L, Chopra I, Bradley SF. In vivo transfer of high-level mupirocin resistance from Staphylococcus epidermidis to methicillin-resistant Staphylococcus aureus associated with failure of mupirocin prophylaxis. J Antimicrob Chemother. 2005;56:1166–8.
Shittu AO, Lin J, Morrison D, Kolawole DO. Isolation and molecular confirmation of a multiresistant catalase-negative Staphylococcus aureus in Nigeria. J Infect. 2003;46:203–4. https://doi.org/10.1053/jinf.2002.1106.
Acknowledgments
SMA was supported by the Organization for Women in Science in the Developing World (OWSD). AOS received funding through the Deutscher Akademischer Austausch Dienst (DAAD award) Staff Exchange Programme (2016). MK was a Wellcome Trust (UK) Fellow (102429/Z/13/Z). His research is currently supported by the Carnegie Corporation of New York (USA) early-career fellowship, the CIHR CTN International Fellowship (Canada), and the US National Institutes of Health (1R01HD093578-01). We appreciate the kind assistance of Oluwafemi Daramola in the preparation of the manuscript.
Funding
This review received support through the Deutscher Akademischer Austausch Dienst (DAAD award) Staff Exchange Programme (2016). However, the opinions expressed in this review are that of the authors.
Availability of data and materials
All supporting materials (Figures and Tables) are included in the manuscript.
Author information
Authors and Affiliations
Contributions
AOS conceived the project. YOA, SMA and AOS extracted the data and reviewed the articles. MOA and AOO wrote the initial draft of the manuscript. AOS, SMA, YOA, and MK wrote the subsequent draft. All the authors reviewed and agreed on the final version of the manuscript before submission for publication.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Shittu, A.O., Kaba, M., Abdulgader, S.M. et al. Mupirocin-resistant Staphylococcus aureus in Africa: a systematic review and meta-analysis. Antimicrob Resist Infect Control 7, 101 (2018). https://doi.org/10.1186/s13756-018-0382-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-018-0382-5