Abstract
NZ2114 is a promising candidate for therapeutic application owing to its potent activity to Staphylococcus aureus. Our objective was to identify NZ2114 derivatives with improved activity through substitution of His16 and His18 with residues Arginine and Lysine. Eight mutants were designed and expressed in Pichia pastoris X-33 via pPICZαA. Five of them exhibited strong antimicrobial activity against S. aureus at low minimal inhibitory concentrations (MICs) of 0.057–0.454 μM. Among them, H1, H2, and H3 showed ideal pharmacodynamic effects on methicillin-resistant S. aureus ATCC43300. The total protein level of H1, H2, and H3 reached 1.70, 1.77 and 1.54 g/l at 120 h of induction in the 5-l fermenter, respectively. They killed over 99.9% of pathogens within 1.5 h at 2× and 4× MIC. The post antibiotic effect of H1, H2 and H3 to S. aureus ATCC43300 was 2.94, 1.75 and 1.55 h at 2× MIC, which was similar with their original peptide NZ2114 (1.43 h) and vancomycin (1.72 h). The fractional inhibitory concentration index (FICI) indicated indifferent effects between H1, H2, H3 and vancomycin, ampicillin, rifampicin. Additionally, they had low hemolysis and high stability in different environments (temperature, pH, proteases, and saline ions). All results indicate that H1, H2, and H3 can be produced in large-scale and have potential as therapeutic drugs against MRSA.
Similar content being viewed by others
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most common antimicrobial-resistant pathogens causing invasive infections (Dantes et al. 2013), which can produce a series of toxins and shows frequent and sometimes multi-drug resistance to antimicrobials (Stefani et al. 2012). According to the previous report, 25–35% of healthy human individuals carried S. aureus on the skin or mucous membranes (Wertheim et al. 2005). This means that up to two billion individuals may currently carried S. aureus worldwide, and conservative estimates based on Dutch and US prevalence data predicted that 2–53 million people carried MRSA (Grundmann et al. 2010). MRSA infections can affect more than 150,000 patients annually in the European Union (EU), resulting in extra in-hospital costs of EUR 380 million for EU healthcare systems (Köck et al. 2010). In another report, it is estimated that MRSA caused 171,200 healthcare-associated infections (HAIs) in Europe each year, which equals to 44% of all HAIs. MRSA also caused 5400 extra deaths and over a million extra days of hospitalisation associated with the infection (Gould et al. 2012).
The guidelines issued by Infectious Disease Society of America in 2011 recommend vancomycin, daptomycin, linezolid or clindamycin for MRSA infection (Liu et al. 2011). Although MRSA can be effectively treated by some antibiotics, sometimes their MIC values increased. Data from the tigecycline evaluation and surveillance trial showed that proportion of MRSA with MICs (vancomycin) ≥2 mg/l increased from 5.6% in 2004 to 11.1% in 2009 (P < 0.001) (Hawser et al. 2011).
Antimicrobial peptides (AMPs) are potent drugs known for their broad-spectrum of antimicrobial properties and particularly against antibiotic-resistant bacteria (Brogden and Brogden 2011). However, some flaws hamper the clinical usage of AMPs, such as toxicity to normal mammalian cells and the lack of a cost-effective way of commercial-scale production (Cao et al. 2015). Plectasin is a fungal defensin from Pseudoplectania nigrella and is especially active against Gram-positive bacteria such as S. aureus (MIC50: 16 μg/ml for methicillin-sensitive strains and 32 μg/ml for resistant strains) and Streptococcus pneumoniae (MIC50: 1 μg/ml for both penicillin-sensitive and resistant strains) by binding with the pyrophosphate moiety of lipid II, the essential precursor of the cell wall (Mygind et al. 2005; Ostergaard et al. 2009; Schneider et al. 2010). Peptide NZ2114 is a novel variant of plectasin (D9N, M13L, Q14R) that is significantly more potent than parental peptide (MIC50: 2 μg/ml for S. aureus and 0.25 μg/ml for S. pneumoniae) (Ostergaard et al. 2009; Zhang et al. 2014; Andes et al. 2009; Xiong et al. 2011). It also had long postantibiotic effect (PAE) (Zhang et al. 2014) and synergistic in combination with the conventionl antibiotics such as teicoplanin, moenomycin, and dalbavancin (Breidenstein et al. 2015). It had potent activities against S. aureus in rabbit meningitis, murine peritonitis, and thigh infection models (Ostergaard et al. 2009; Andes et al. 2009; Xiong et al. 2011). Additionally, NZ2114 showed low or no cell toxicities, long-lasting serum stability and in vivo half-life (Brinch et al. 2010).
However, two histidine (His) residues exist in the sequence of NZ2114, which have a pKa of approximately 6.0 and are largely unprotonated and uncharged at physiological conditions (Kashiwada et al. 2016). It was found that some AMPs rich in His exhibited higher activity at low pH, when the histidine was positively charged, as compared with neutral pH. For example, LAH4 weakly disrupted the membrane at physiological conditions. The cell lysis activities of peptides LL-1a and LL-1c decreased up to four times as the solution pHs at 7.4 (Mason et al. 2006). The arginine residues were previously suggested to be important for the antimicrobial activity of AMPs (Tu et al. 2009) and also to potentiate the internalization of peptides (Hansen et al. 2008). Arginine residues having a more dispersed positive charge on their side chain guanidinium group, have been reported to enhance the electrostatic interactions between peptides and anionic lipids (Strandberg et al. 2002). Veiga et al. (2012) demonstrated that the self-assembling β-hairpin peptides, having a high content of arginine, were extremely effective to killing both gram-positive and gram-negative bacteria, including multi-drug resistant strains. Silva et al. (2014) found that arginine residues were crucial for the display of antimycobacterial activity. On the other side, the positive charge of lysine residues were considered to be critical for interacting with the anionic interface as a result of initial electrostatic interaction (Koba et al. 2009). It showed that four positively charged lysine residues concentrated on the hydrophilic face of HPA3NT3-analog peptides, improved the amphipathic structure, and resulted in decrease of hemolysis (Gopal et al. 2009). The large loss of antimicrobial activity was also found when the C-terminal arginine and lysine residues of CL(14–25) was replaced with alanine residues (Taniguchi et al. 2014).
Therefore, to enhance the antimicrobial activity of NZ2114, replacement of the histidine at the 16th and 18th positions by arginine and lysine was performed. Eight mutants were designed and the antibacterial activities were evaluated. The peptides with excellent antibacterial property were chosen to test their pharmacodynamics, post antibiotic effect, synergy, hemolytic activity, and stability against S. aureus ATCC43300 (MRSA).
Materials and methods
Strains, plasmid and reagents
Escherichia coli DH5α, P. pastoris X-33 and pPICZαA vector were purchased from Invitrogen (Beijing, China). The test strains for the antimicrobial activity assays and their sources are listed in Additional file 1: Table S1. Vancomycin, ampicillin, rifampicin and ciprofloxacin were purchased from the China Institute of Veterinary Drug Control. NZ2114 was prepared in our laboratory (Zhang et al. 2014). Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (NEB, Beijing, China). The kits for plasmid extraction and DNA purification were purchased from Tiangen (Beijing, China). Other chemical reagents were analytical grade.
Peptide design
The main structure of NZ2114 was kept unchangable to maintain the antimicrobial activity. Additionally, the histidine residues in position 16 and 18 were mutated into arginine or lysine. As results, eight derived peptides which H16 and H18 were replaced by arginine or lysine were generated. The amino acid sequences and properties of the parental peptide and designed peptides were calculated by bioinformatics programs, including antimicrobial peptide calculator and predictor (http://aps.unmc.edu/AP/prediction/prediction_main.php) and ProtParam (ExPASy Proteomics Server: http://www.expasy.org/tools/protparam.html). All of the parameters are summarized in Table 1.
Construction of recombinant plasmid pPICH1–pPICH8
The codon-optimized gene sequences of H1–H8 (Additional file 1: Table S2) were designed by the Reverse Translate Tool (www.bioinformatics.org/sms2/rev_trans.html), according to the preferential codon usage of P. pastoris (www.kazusa.or.jp/codon/). To ensure the integrity of the sequences in the expression process, the expression cassette included an XhoI recognition site, a P. pastoris Kex2 protease cleavage site, the H1–H8 genes, two stop codons, and an XbaI recognition site. The DNA sequences and pPICZαA vector were digested by XhoI and XbaI, gel-purified, and ligated together by T4 DNA ligase. The recombinant plasmids pPICH1–pPICH8 were transformed into E. coli DH5α, and positive cells were selected. The H1–H8 gene sequences were confirmed by DNA sequencing using the following two primers.
Primer 5′AOX1: 5′-GACTGGTTCCAATTGACAAGC-3′
Primer 3′AOX1: 5′-GCAAATGGCATTCTGACATCC-3′
F1: 5′-CCGCTCGAGAAGAGAGGTTT-3′
R1: 5′-GCTCTAGATTATTAGTAACAC-3′
Transformation and selection of positive transformants
The pPICH1–pPICH8 were linearized with PmeI and then transformed into the competent P. pastoris X-33 cells by electroporation following the Invitrogen’s instructions. The pPICZαA vector was also linearized and transformed into P. pastoris X-33 cells as a negative control. All zeocin-resistant colonies were selected in YPDS plates (10 g/l yeast extract, 20 g/l peptone, 20 g/l glucose, 182.2 g/l sorbitol, 20 g/l agar, and 100 μg/ml zeocin).
Expression of H1–H8 in P. pastoris in 48-well plates
A single colony of positive P. pastoris transformants was cultured at 29 °C (250 rpm) in 48-well plates containing 500 µl BMGY medium (10 g/l yeast extract, 20 g/l peptone, 10 ml/l glycerol, 13.4 g/l yeast nitrogen base, 400 µg/l biotin, and 100 ml/l 1 M potassium phosphate, pH 6.0). After 24 h, methanol (100%) was added each well to a final concentration of 0.5% (v/v), and the temperature was adjusted to 28 °C. Then methanol was repeatedly added every 24 h during the 96 h induction time. The supernatant was collected by centrifugation at 10,000×g for 10 min and stored at −20 °C. The expression conditions of H1–H8 were determined by the inhibition zone assay against S. aureus ATCC25923 and Tricine-sodium dodecyl sulfate polyacrylamide gel electrophoresis (Tricine-SDS-PAGE) (Zhang et al. 2014; Schägger 2006). Then the positive transformants with high inhibitory effect were selected to express in shake flasks (1-l shake flask containing 200-ml BMGY medium) using the same method.
Purification and identification of H1, H2, H3, H6, H8
The supernatant of 1-l shake flasks of H1, H2, H3, H6, and H8 was applied onto an SP Sepharose FF cation-exchange column (GE Healthcare, UK) pre-equilibrated with binding buffer (20 mM sodium phosphate buffer, pH 6.7). H1, H2, H3, H6, and H8 were eluted from the column with elution buffer (20 mM sodium phosphate buffer, 600 mM NaCl, pH 6.7) at a rate of 6 ml/min, and the eluent of corresponding elution peaks were collected. The eluent were analyzed by Tricine-SDS-PAGE and confirmed by MALDI-TOF MS at the Laboratory of Proteomics, Institute of Biophysics, Chinese Academy of Sciences according to the previously reported method (Zhang et al. 2014).
Antimicrobial activity assays of H1, H2, H3, H6, and H8 in vitro
The peptide solutions were diluted twofold with the range of final concentrations were 0.015–32 µg/ml (0.003–7.273 µM) for purified H1, H2, H3, H6, and H8. The test strains were grown in MHB medium at 37 °C to an OD600 of 0.4 and diluted to 1 × 105 CFU/ml. The 10 µl peptide and 90 µl cell suspension were added into each well. All assays were performed in triplicate. The antimicrobial activity of NZ2114 and vancomycin were tested as positive controls. The plates were incubated at 37 °C for 18–24 h. MIC was defined as the lowest peptide concentration of ones at which there was no visible growth (Tian et al. 2009).
Expression of H1, H2, H3 in P. pastoris in high-density cultivation in fermentors
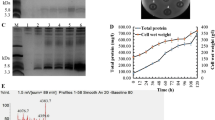
According to the results of the antimicrobial activity assays, H1, H2, and H3 were chosen to be expressed in the 5-l fermentor. A single colony of H1, H2, H3 was incubated in shaking flasks with 10 ml YPD medium at 30 °C (250 rpm). Overnight cultures were inoculated into 200 ml YPD medium and cultivated at 30 °C (250 rpm) to an OD600 of 5.0 and then transferred into a 5-l fermentor (BIOSTAT®B plus, Sartorius Stedim Biotech) containing 2 l basal salts medium (50 g/l NH4H2PO4, 20 g/l K2SO4, 15 g/l MgSO4·7H2O, 6 g/l KH2PO4, 0.4 g/l CaSO4, 1.5 g/l KOH, 45 g/l glucose, and 4.8‰ PMT1). The pH was controlled at 5.5 using H3PO4 and NH4OH, and the temperature was maintained at 29 °C. When the glucose was exhausted, methanol was supplied from 1 to 7 ml/l/h during the first 6 h. Then methanol was supplied to maintain a relative dissolved oxygen (DO) content between 20 and 40% under the speed of 6–8 ml/l/h (Bai et al. 2010). The fermentation liquid was collected every 24 h to quantify the cell wet weight and the total protein level which was assayed by a Bradford protein assay kit (Tiangen Biotech, Beijing, China). The expression of H1, H2, and H3 was determined by Tricine-SDS-PAGE. The supernatant was purified in the same way as the 1-l shake flasks.
Time-kill curve assay
Bacterial culture (S. aureus ATCC43300) was diluted to 1 × 105 CFU/ml and H1, H2, and H3 were added. The final concentration of H1, H2, and H3 was 1× , 2× and 4× MIC, respectively. The mixture was cultivated at 250 rpm, 37 °C. The 100-µl samples were taken from each flask at 0, 2, 4, 6, 8, 12 and 24 h of incubation, and serial dilutions of samples were plated to count visible colonies. Vancomycin was tested in the same way as a positive control and the culture without antimicrobial agent as a negative control (Xiong et al. 2011). All experiments were performed in triplicate.
The postantibiotic effect of H1, H2, H3 against S. aureus
The 100 µl peptides were added into tubes containing 900 µl bacterial cultures (1 × 108 CFU/ml) to make their final peptide concentration to 1× , and 2× MIC, respectively, and the mixture was cultured at 37 °C for 2 h. Tubes with 2× MIC vancomycin, 2× MIC NZ2114, and without antimicrobial agent were used as controls. After 2 h, the drug was removed by diluting 1:103 into the MHB and incubated at 37 °C, 250 rpm. The sample was taken to plate counting every 1 h until the bacterial cultures become turbid. The PAE was calculated by the following formula: PAE = T − C, where T is the time needed for the count of CFU in the test culture to increase 1 log10 (10-fold) above the 0 h and C is the time needed for the count of CFU in the untreated control culture to increase 1 log10 above the 0 h (Giguère et al. 2012).
Synergism assays of H1, H2, H3 with conventional antibiotics
The MIC values of four antibiotics (vancomycin, ampicillin, rifampicin and ciprofloxacin) to S. aureus ATCC43300 were tested as the MIC assay described above. The peptide solutions and antibiotics were diluted twofold with the final concentrations ranging from 1/16 to 8× MIC, and added into 96-well plates in a checkerboard fashion (White et al. 1996). The results of combination were evaluated by calculating the fractional inhibitory concentration index (FICI) of each combination. FIC of H1, H2, H3=MIC of H1, H2, H3, respectively, in combination with antibiotic/MIC of H1, H2, H3 alone; FIC of antibiotics=MIC of antibiotic in combination with peptide/MIC of antibiotic alone; FICI = FIC of H1, H2, H3+ antibiotic, respectively. The result of interaction between two antimicrobial drugs was determined according to: FICI ≤ 0.5 refers to synergy, 0.5 < FICI ≤ 1 refers to additivity, 1 < FICI ≤ 4 refers to indifference, and FICI > 4 is defined as antagonism (Tsuji and Rybak 2006).
Hemolytic assay
The mice blood cells were washed three times in stroke-physiological saline solution (0.9% NaCl) and centrifuged at 4 °C, 2000 rpm for 5 min. A 50-μl cell was diluted to 8% (v/v), added into 96-well plates, and mixed with 50-μl peptide to the final concentrations ranging from 0.23 to 23.09 μM (1–128 μg/ml). The plates were incubated at 37 °C for 1 h, and centrifuged at 4 °C, 5000 rpm for 5 min. The absorbance of supernatants was measured at 540 nm, and 0 and 100% hemolysis was measured by 0.9% NaCl and 0.1% Triton X-100, respectively. The hemolysis percentages were calculated by the following equation: [(Abs540 nm in H1, H2, H3 solution − Abs540 nm in 0.9% NaCl)/(Abs540 nm in 0.1% Triton X-100 − Abs540 nm in 0.9% NaCl)] ×100% (Jiao et al. 2015).
Circular dichroism (CD) of H1, H2, H3
CD spectroscopy analysis of H1, H2, H3 [5.68 µM (25 µg/ml) in ddH2O, 20 mM SDS, 50% TFE solution was carried out in a MOS-450 spectropolarimeter (Bio-Logic, Grenoble)]. The samples were loaded into a 1 mm cell path at room temperature and the spectra were recorded from 195 to 245 nm three times.
Effect of pH, temperature, NaCl concentration and proteinase on the activity of H1, H2, H3
The effect of pH, temperature, NaCl concentration and proteinase on the activity of peptides was evaluated. Peptides were diluted twofold, then adjusted to a pH range from 2.0 to 10.0 and incubated at 37 °C for 4 h. The thermal stability of purified peptides was determined after 1-h incubation of peptides at 4, 20, 40, 60, 80 and 100 °C in deionized water, respectively. Aliquots of peptides were incubated in various proteinases solutions [pepsin (pH 2.0), trypsin (pH 8.0), proteinase K (pH 7.0)] at a ratio of 1:10, w/w (proteinase: peptide) at 37 °C for 3 h. For the ion stability, peptides were incubated in 50, 100, 200, 300, 400, 500 mM sodium chloride solutions, respectively. Other methods and conditions were prepared as the MIC assay described above (Li et al. 2015; Qu et al. 2016).
Results
Peptides design
The net charge of derived peptides with one site mutants (H1, H2, H3 and H4) increased from +3 to +4, and the two sites mutants was +5 (H5, H6, H7 and H8). Due to the higher hydrophily of arginine and lysine, the grand average of hydropathicity (GRAVY) slightly decreased for one site mutants (from −0.672 to −0.690 for H1 and H3, and to −0.705 for H2 and H4, respectively), and it further decreased for two sites mutants (−0.708 for H5, −0.722 for H6 and H7, and −0.738 for H8, respectively). The related high hydrophilicity may contribute to the surface contact of pathogens and peptides. The Boman index indicates the binding activity of drugs to protein, the large value may induce high negative side effects. Generally, it was considered that the value was acceptable as 1–3 kcal/mol (Weistroffer 2007). The Boman index of derived peptides was all between 1.54 and 2.03 kcal/mol, indicating they may have little negative side effects as novel antimicrobial agents (Table 1).
Expression of H1–H8 in Pichia pastoris in 48-well plates
Thirty-six positive transformants of each peptide were screened by inhibition zone against S. aureus ATCC25923. The transformants showed various antimicrobial activity except H4 (date not shown). According to the diameter of inhibition zone, one transformant of each peptide was selected to analyze the expression condition by Tricine-SDS–PAGE. As shown in Additional file 1: Figure S1, there were no visible bands in the lane of H4, H5, H7, while H1, H2, H3, H6, and H8 displayed obviously bands, so they were chosen to the following assays.
Purification and identification of H1, H2, H3, H6, and H8
The supernatants of H1, H2, H3, H6, and H8 in 1-l shake flasks were purified. H1, H2, H3, H6, and H8 were detected by Tricine-SDS-PAGE from the eluent containing 20 mM sodium phosphate buffer, 600 mM NaCl, pH 6.7 (Fig. 1a). Target bands around 4 kDa were detected, and there were no other bands (Fig. 1a). MALDI-TOF MS analysis indicated that only a target peak of 4428.97, 4428.41, 4401.63, 4421.98, and 4392.18 Da from purified peptides (Fig. 1b–f) were detected, which were consistent with their theoretical value of 4428, 4428, 4402, 4421, and 4393 Da, respectively.
Purification and identification of H1, H2, H3, H6, and H8. a Tricine-SDS-PAGE analysis of H1, H2, H3, H6, and H8 in fermentation supernatants of 1-l shake flasks. Lane M a total of 6 μl of protein molecular weight marker. Lane 1–5, 10 μl of purified H1, H2, H3, H6, and H8, respectively. b–f MALDI-TOF MS analysis of the purified H1, H2, H3, H6, and H8 fermentation supernatants in 1-l shake flasks
Antimicrobial activity assays of H1, H2, H3, H6, and H8 in vitro
H1, H2, H3, H6, and H8 showed a potent antimicrobial activity against Gram-positive bacteria, including three S. aureus strains (MIC: 0.057–0.454 μM), three Streptococcus suis strains (MIC: 0.007–0.057 μM), and two S. pneumonia strains (MIC: 0.227–1.818 μM) (Table 2). Due to the higher charge and hydrophily of arginine and lysine compared to histidine, their activity (MIC: 0.057–0.454 μM) against S. aureus ATCC43300 (MRSA) was improved two to sixteen times than NZ2114 (0.909 μM) and two to twelve times than vancomycin (0.714 μM). In addition, the activity of H1, H2, and H3 against Methicillin-susceptible Staphylococcus aureus (MSSA ATCC25923, and ATCC6538) was improved twice than NZ2114. Meanwhile, the too high charge and hydrophily did not result in the improved activity of H6 and H8 against both MRSA (ATCC43300) and MSSA (ATCC6538). All of them could not inhibit Gram-negative pathogens (Table 2). Due to the excellent activity of H1, H2, and H3 against S. aureus, they were selected for the further pharmacodynamics study.
Expression of H1, H2, H3 in P. pastoris in high-density cultivation in fermenters
H1, H2, and H3 were cultured and induced in 5-l fermenters. Target peptides in the supernatant were detected after 24 h of induction by Tricine-SDS-PAGE, and the concentration increased with induction time (Fig. 2a, c, e). The cell wet weight of H1, H2, and H3 increased during the induction time and it up to 542.68, 408.97, and 488.98 g/l at 120 h of induction, respectively, and their total protein level reached 1.70, 1.77, and 1.54 g/l, respectively (Fig. 2b, d, f).
High-density cultivation of H1, H2, H3 in the fermentor level. a, c, e Tricine-SDS-PAGE analysis of H1, H2, H3 fermentation supernatants at different induction times, respectively. Lane 1–5 a total of 5-μl fermentation supernatants taken at 24, 48, 72, 96, 120 h. Lane M 6 μl protein molecular weight marker. b, d, f Time curve of the cell wet weight and total secreted protein levels of H1, H2, H3 during induction (three duplicate observations were made; bars represent standard error of mean)
Time-killing curve assay
In vitro time killing curves were performed to evaluate the pharmacodynamic properties and bactericidal ability. In the absence of antimicrobial agent, the bacterial counts (log10 CFU/ml) reached to 15.17 for S. aures ATCCs43300 at 24 h (Fig. 3). The time killing curves of H1, H2, and H3 had a significant dose-dependent. The bacterial counts decreased to 2 log10 CFU/ml (a 99.9% reduction) in 6, 1.5 and 1 h with 1× , 2× , and 4× MIC of H1, respectively. However, after 8 and 12 h, 1× and 2× MIC of H1 had a regrowth and reached to 12.80 and 3.40 log10 CFU/ml at 24 h, respectively (Fig. 3a). Meanwhile, the bacterial counts decreased to 2 log10 CFU/ml in 1.5, 1, and 0.5 h with 1× , 2× , and 4× MIC of H2, respectively, and maintained to 24 h without regrowth of pathogens (Fig. 3b). The H3 showed a similar trend to H1, but the bacterial counts of 1× MIC of H3 regrowed after 8 h of incubation (Fig. 3c). The efficacy of 1× MIC of H1 and H3 was equivalent to 2× MIC of vancomycin, and all other concentrations of peptides were better than that with vancomycin.
Time-kill curves of H1 (a), H2 (b) and H3 (c). CK: S. aureus ATCC43300 (MRSA) were incubated in the presence of medium alone; Van-2MIC: S. aureus were incubated in the presence of the vancomycin(Van) at 2× MIC; H1-MIC, H1-2MIC, H1-4MIC, H2-MIC, H2-2MIC, H2-4MIC, H3-MIC, H3-2MIC, H3-4MIC: S. aureus were incubated in the presence of H1, H2, H3 at 1× , 2× , 4× MIC, respectively; Three duplicate observation were made; Bars represent the standard error of the mean
The postantibiotic effect (PAE) of H1, H2, H3 against S. aureus
The PAE of H1 to ATCC43300 was 2.94 h at 2× MIC, which was much longer compared with the 1.43 h for original peptide NZ2114 and 1.72 h for vancomycin at 2× MIC (Table 3). The PAE of H2 and H3 at 1× MIC were 0.63 and 0.55 h, respectively. They had similar values of 1.75 and 1.55 h with NZ2114 and vancomycin at 2× MIC (Table 3).
Synergism assays of H1, H2, H3 with conventional antibiotics
The synergism effect of H1, H2 and H3 with traditional antibiotics which have different action mechanisms (vancomycin and ampicillin on cell wall, rifampicin on RNA, ciprofloxacin on DNA) was evaluated (Table 4). The MIC values of H1, H2, H3, vancomycin, ampicillin, rifampicin, and ciprofloxacin against ATCC43300 were 0.056, 0.113, 0.056, 0.714, 5.724, 0.019, and 1.510 μM, respectively. When they were used with combination together, almost MIC values were not changed and some MIC values changed to 1/4 or 1/2× MIC. However, the MIC value of ampicillin increased to 2× MIC with H2 and H3. Meanwhile, MICs of H2 and rifampicin increased twice when combined used with ciprofloxacin and H3. The effect of interaction between H1, H2, H3 and antibiotics all showed as indifference with FICI from 1.25 to 3 (Table 4). These results were very different from previous results of original peptide NZ2114 (FICI: 0.125 for vancomycin and ampicillin) (Zhang et al. 2014).
Hemolytic assay
Different concentrations of H1, H2, H3, and NZ2114 [0.23–23.09 μM (1–128 μg/ml)] were tested to observe their lysis activity on mice red blood cells (RBCs) in hemolysis assays (Additional file 1: Figure S2). The hemolytic activity of original peptide NZ2114 maintained 1.00% at its concertration from 0.23 to 23.09 μM. Due to the higher charge and hydrophilicity, derived peptides had higher hemolysis compared with NZ2114 but the value was very low in the MICs. H2 and H3 had little or no hemolytic activity at the concentration from 0.23 to 7.27 μM (1–32 μg/ml). H3 had a hemolysis with 6.15% at 14.55 μM (64 μg/ml), while H2 only had 1.51% hemolytic activity at 23.09 μM (128 μg/ml). H1 had higher hemolysis than H2 and H3, which reached 5.99% at 7.27 μM (32 μg/ml).
CD spectra of H1, H2, H3
As shown in Fig. 4 and Table 5, NZ2114 had the high α-helix content of 74.1 and 71.7% in ddH2O and 50% TFE, and the content of β-inverse parallel was very low (2.9 and 1.3%). There was lower α-helix (24.8%) and higher antiparallel (18.9%) content in 20 mM SDS. The secondary structures of H1, H2 and H3 were different with NZ2114. The proportion of α-helix was 55.6, 35.6, and 35.1% and the β-inverse parallel was 3.8, 7.7, and 9.8% for H1, H2, and H3 in ddH2O, respectively. And similar trends were observed in 20 mM SDS (51.6, 26.8, 35.2% for α-helix; 5.3, 10.3, and 10.5% for β-inverse parallel). However, there was much higher α-helix content (82.9, 52.6, and 63.8% for H1, H2, and H3) and lower β-antiparallel proportion (0.4, 3.0, and 2.5% for H1, H2, and H3) in 50% TFE.
Effect of pH, temperature, NaCl concentration and proteinase on the activity of H1, H2, H3
The MIC values of H1 decreased twice (0.057 μM to 0.028 μM) in pH 10 than other pHs and it increased twice (0.057–0.114 μM) in pH 6 than others for H3. Additionally, the MIC of H2 maintained 0.114 μM in all five gradients (Additional file 1: Table S3). Three peptides all showed a high thermal stability, the MIC values of H1 and H3 increased twice only at 100 °C (0.057–0.114 μM), and H2 increased twice in 80 and 100 °C (0.114–0.227 μM) (Additional file 1: Table S4). As shown in Additional file 1: Table S5, the MIC values of H1 (0.057 μM) were not affected in the presence of different NaCl concentrations, but H2 and H3 increased twice at 200 mM (0.114–0.227 μM) and 400 mM (0.057–0.114 μM), respectively. The three peptides showed their own stability to different proteinases (Additional file 1: Table S6). The MIC value of H1 increased twice in pepsin (0.057–0.114 μM), and H2 and H3 increased twice in trypsin (0.227–0.454 μM) and proteinase K (0.057–0.114 μM), respectively.
Discussion
Currently, 2764 AMPs are registered in the antimicrobial peptide database (APD) (http://aps.unmc.edu/AP/main.php). However, only few AMPs entered into clinical trials. The use of AMPs was primarily hampered by their low oral or intravenous stability, high toxicity, serum binding activity, low activity in physiological condition and so on (Mohammad et al. 2015). Plectasin and its derived peptide NZ2114 had potent activity to S. aureus, which are the idea candidates for traditional drugs (Zasloff 2016). However, two histidine residues exist in the sequence of NZ2114, which are uncharged in the physiological condition (Kashiwada et al. 2016). To further improve the antimicrobial activity and properties of NZ2114, new derived peptides are needed.
Although AMPs vary widely in length, structure, and source, they have some important common traits, such as positive charges, presumed to be important for interaction with the negatively charged surface of pathogens, and amphipathicity, which enables better combine with the hydrophilic surface and interact with the hydrophobic part of the microbial membrane (Silva et al. 2014). The arginine and lysine are the key residues for AMPs which are stable charged and hydrophilic in physiological condition. Many studies showed that AMPs having appropriate proportion of arginine and lysine had an improved amphipathicity and higher activity (Veiga et al. 2012; Silva et al. 2014; Gopal et al. 2009; Taniguchi et al. 2014). As results, eight derived peptides (H1–H8) which H16 and H18 were replaced by arginine or lysine were generated.
All derived peptides were tried to express via P. pastoris but H4, H5 and H7 were not expressed. The antimicrobial activity of H1, H2, H3, H6 and H8 were assayed and H1, H2, and H3 had higher activity compared with original peptide NZ2114. However, the activity of mutants H6 and H8 which had more net positive charges did not significantly increase (Table 2), which indicated that the electrostatic interaction and cell-penetrating was not the all bactericidal mechanisms of mutants.
Unlike some AMPs with a wide antimicrobial spectrum, H1, H2, and H3 showed a narrow spectrum and they mainly killed the Gram-positive bacterium, such as S. aureus, and Streptococci, and showed a strong antimicrobial activity (0.007–0.454 μM; Table 2), which was very stronger than the activities of plectasin and NZ2114 (Hara et al. 2008; Zhang et al. 2014). Especially, H1, H2, and H3 showed higher antimicrobial activity against MRSA (S. aureus ATCC43300) with MIC of 0.057, 0.114, and 0.057 μM than NZ2114 (0.909 μM) and vancomycin (0.714 μM). Their characteristics of narrow-spectrum antibiotic and low MIC values are very attractive for developing them as candidate agent against MRSA infection.
The lack of economic feasibility to manufacture AMPs at large-scale is another roadblock in the clinical implementation of AMPs (Findlay et al. 2010). Majority of directly expressed AMPs, such as LL-37 (Hong et al. 2007), CecropinAD (Jin et al. 2009), and N2 (Yang et al. 2016), showed unsatisfactory yields. In our previous works, Agplectasin (Mao et al. 2013), NZ2114 (Zhang et al. 2014), and MP1106 (Cao et al. 2015) were expressed in P. pastoris X-33 in high level. Similarly, H1, H2, and H3 were expressed in P. pastoris X-33 with high yields, their total protein level in 5-l fermentation reached 1.70, 1.77, and 1.54 g/l, respectively (Fig. 2b, d, f). However, because the fermentation was performed in summer, the temperature cannot be controlled at 29 °C as previous operation. To maintain the dissolved oxygen content, we had to reduce the flow rate, which lead to the partial losing of yield. If the induction temperature can maintain at 29 °C, at which this key temperature is very important for its high expression in yeast (Li et al. 2001, 2007). the production of H1, H2, H3 may be further improved like their original peptide NZ2114 (2390 mg/l in 29 °C and 2310 mg/l in 25 °C) (Zhang et al. 2014).
The PAE is a very important pharmacodynamics parameter in choosing of antibiotic dosage regimens in clinical use (Pankuch and Appelbaum 2009a). Obviously observed, the PAEs of H1, H2, and H3 increased with the concentration from 1× MIC to 2× MIC (Table 3). They showed similar values to vancomycin (2× MIC: 1.72 h) and NZ2114 (2× MIC: 1.43 h). H1 also had comparative value (2× MIC: 2.94 h) to some conventional antibiotics, for instance, daptomycin (2.0 h), tigecycline (3.2 h), and arbekacin (3.0–3.2 h), respectively (Pankuch and Appelbaum 2009b; Pankuch et al. 2003; Watanabe et al. 1997). Their appropriate PAE is critical to lengthen the interval of administration, reduce the daily dosages, and thus potentially reduce potential drug resistance.
To combat antibiotic resistance, combination antibiotic therapy is practiced in the clinical use due to its advantages such as wider coverage, higher activity, bactericidal synergy and the inhibition on toxin production (Leibovici et al. 2010; Müller et al. 2013). Vancomycin, one of the most effective antibiotics against MRSA, is often combined used with rifampicin, gentamicin, dalfopristin, and β-lactams to slow the development of resistance and enhance the antibacterial activity. The synergistic effect of H1, H2, and H3 is very different from their parent peptide NZ2114 (Zhang et al. 2014). The FICI of NZ2114 combined with ampicillin, and vancomycin to S. aureus ATCC43300 was 0.125, showing additivity effect (Zhang et al. 2014). However, the indifference effects (1.25 ≤ FICI ≤ 3) were observed for all combinations between H1, H2, H3 and four traditional antibiotics to S. aureus ATCC43300 (Table 4), which might result from the changed antimicrobial property and mechanism and its details should be studied in our next research.
The plectasin and NZ2114 showed no hemolysis in rabbit and human RBCs (Yang et al. 2011; Zhang et al. 2014). In this work, H1, H2, and H3 showed low hemolysis in mice RBCs. Although the values were higher than NZ2114 due to the high charge and hydrophilicity, they were still very low in the range of MICs. In addition, NZ2114 showed the highest activity in pH value of 8.0 and above 80% of initial activity was retained over a range of temperatures from 20 to 80 °C but maintained 20% activity at 100 °C (Zhang et al. 2014). Owing to contribution from the three pairs of disulfide bond into the stability of structure, H1, H2, and H3 all had high stability with minor differences. H1 was not sensitive to NaCl concentration, but sensitive to alkaline and high temperature environments. H2 was sensitive to high temperature and NaCl concentration, but not sensitive to different pHs environment. H3 was sensitive to high temperature and high NaCl concentration. Generally, no toxicity to erythrocytes and high stability of pH, temperature, proteases, and saline ions of H1, H2 and H3 meet the key requirements of new antimicrobial agents.
In summary, series of novel AMPs were designed and successfully expressed in P. pastoris. Among them, H1, H2, and H3 had high yields (1.70, 1.77 and 1.54 g/l) in 5-l fermentor level. H1, H2, and H3 also showed strong antimicrobial activity against S. aureus. They killed MRSA strain ATCC43300 in a short time with low concentrations and had long post antibiotic effect. Meanwhile, H1, H2, and H3 exhibited indifference effects when they were combined with conventional antibiotics. Furthermore, they had low toxicity to mice erythrocytes and high stability. All results indicate that H1, H2, and H3 have potential as candidates for the therapeutic agents with the better properties than their native peptide NZ2114.
Abbreviations
- MIC:
-
minimal inhibitory concentration
- RBCs:
-
red blood cells
- AMP:
-
anitimicrobial peptide
- APD:
-
antimicrobial peptide database
References
Andes D, Craig W, Nielsen LA, Kristensen HH (2009) In vivo pharmacodynamic characterization of a novel plectasin antibiotic, NZ2114, in a murine infection model. Antimicrob Agents Chemother 53(2009):3003–3009
Bai XJ, Teng D, Tian ZG, Zhu Y, Yang YL, Wang JH (2010) Contribution of bovine lactoferrin inter-lobe region to iron binding stability and antimicrobial activity against Staphylococcus aureus. Biometals 23(3):431–439
Breidenstein EBM, Courvalin P, Meziane-Cherif D (2015) Antimicrobial activity of plectasin NZ2114 in combination with cell wall targeting antibiotics against vana-type Enterococcus faecalis. Microb Drug Resist 21(2015):373–379
Brinch KS, Tulkens PM, Bambeke F, Frimodt-Moller N, Hoiby N, Kristensen HH (2010) Intracellular activity of the peptide antibiotic NZ2114: studies with Staphylococcus aureus and human THP-1 monocytes, and comparison with daptomycin and vancomycin. J Antimicrob Chemother 65(2010):1720–1724
Brogden NK, Brogden KA (2011) Will new generations of modified antimicrobial peptides improve their potential as pharmaceuticals? Int J Antimicrob Agents 38(3):217–225
Cao XT, Zhang Y, Mao RY, Teng D, Wang XM, Wang JH (2015) Design and recombination expression of a novel plectasin-derived peptide MP1106 and its properties against Staphylococcus aureus. Appl Microbiol Biotechnol 99(6):2649–2662
Dantes R, Mu Y, Belflower R, Aragon D, Dumyati G, Harrison LH, Lessa FC, Lynfield R, Nadle J, Petit S, Ray SM, Schaffner W, Townes J, Fridkin S (2013) National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern Med 173(21):1970–1978
Findlay B, Zhanel GG, Schweizer F (2010) Cationic amphiphiles, a new generation of antimicrobials inspired by the natural antimicrobial peptide scaffold. Antimicrob Agents Chemother 54(10):4049–4058
Giguère S, Lee EA, Guldbech KM, Berghaus LJ (2012) In vitro synergy, pharmacodynamics, and postantibiotic effect of 11 antimicrobial agents against Rhodococcus equi. Vet Microbiol 160(1):207–213
Gopal R, Park S, Ha K, Cho SJ, Kim SW, Song PI, Nah J, Park Y, Hahm K (2009) Effect of leucine and lysine substitution on the antimicrobial activity and evaluation of the mechanism of the HPA3NT3 analog peptide. J Pept Sci 15(9):589–594
Gould IM, David MZ, Esposito S, Garau J, Lina G, Mazzei T, Peters G (2012) New insights into meticillin-resistant Staphylococcus aureus (MRSA) pathogenesis, treatment and resistance. Int J Antimicrob Agents 39(2):96–104
Grundmann H, Aanensen DM, Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW, European Staphylococcal Reference Laboratory Working Group (2010) European staphylococcal reference laboratory working group, geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med 7(1):e1000215
Hansen M, Kilk K, Langel U (2008) Predicting cell-penetrating peptides. Adv Drug Deliv Rev 60:572–579
Hara S, Mukae H, Sakamoto N, Ishimoto H, Amenomori M, Fujita H, Ishimatsu Y, Yanagihara K, Kohno S (2008) Plectasin has antibacterial activity and no affect on cell viability or IL-8 production. Biochem Biophys Res Commun 374(4):709–713
Hawser SP, Bouchillon SK, Hoban DJ, Dowzicky M, Babinchak T (2011) Rising incidence of Staphylococcus aureus with reduced susceptibility to vancomycin and susceptibility to antibiotics: a global analysis 2004–2009. Int J Antimicrob Agents 37(3):219–224
Hong IP, Lee SJ, Kim YS, Choi SG (2007) Recombinant expression of human cathelicidin (hCAP18/LL-37) in Pichia pastoris. Biotechnol Lett 29(1):73–78
Jiao J, Mao RY, Wang X, Zhang Y, Teng D, Feng XJ, Wang JH (2015) GAP-initiated constitutive expression of a novel plectasin-derived peptide MP1106 by Pichia pastoris and its activity against Streptococcus suis. Process Biochem 50(2):253–261
Jin FL, Xu XX, Yu XQ, Ren SX (2009) Expression and characterization of antimicrobial peptide Cecropin AD in the methylotrophic yeast Pichia pastoris. Process Biochem 44(1):11–16
Kashiwada A, Mizuno M, Hashimoto J (2016) pH-Dependent membrane lysis by using melittin-inspired designed peptides. Org Biomol Chem 14(26):6281–62888
Koba H, Okuda K, Watanabe H, Tagami J, Senpuku H (2009) Role of lysine in interaction between surface protein peptides of Streptococcus gordonii and agglutinin peptide. Oral Microbiol Immunol 24:162–169
Köck R, Becker K, Cookson B, Gemert-Pijnen J, Harbarth S, Kluytmans J, Mielke M, Skov RL, Struelens MJ, Tacconelli E, Navarro Torné A, Witte W, Friedrich AW (2010) Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill 15:12–20
Leibovici L, Paul M, Andreassen S (2010) Balancing the benefits and costs of antibiotic drugs: the TREAT model. Clin Microbiol Infec 16(12):1736–1739
Li Z, Xiong F, Lin Q, d’Anjou M, Daugulis AJ, Yang DS, Hew CL (2001) Hew low-temperature increases the yield of biologically active herring antifreeze protein in Pichia pastoris. Protein Expr Purif 21(3):438–445
Li P, Anumanthan A, Gao XG, Ilangovan K, Suzara VV, Düzgüneş N, Renugopalakrishnan V (2007) Expression of recombinant proteins in Pichia pastoris. Appl Biochem Biotechnol 142(2):105–124
Li RF, Lu YL, Lu YB, Zhang HR, Huang L, Yin YL, Zhang L, Liu S, Lu ZF, Sun YN (2015) Antiproliferative effect and characterization of a novel antifungal peptide derived from human Chromogranin A. Exp Ther Med 10(6):2289–2294
Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray BE, Rybak MJ, Talan DA, Chambers HF (2011) Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin Infect Dis 52(3):e18–e55
Mao RY, Teng D, Wang XM, Xi D, Zhang Y, Hu XY, Yang YL, Wang JH (2013) Design, expression, and characterization of a novel targeted plectasin against methicillin-resistant Staphylococcus aureus. Appl Microbiol Biotechnol 97(9):3991–4002
Mason AJ, Gasnier C, Kichler A, Prevost G, Aunis D, Metz-Boutigue M, Bechinger B (2006) Enhanced membrane disruption and antibiotic action against pathogenic bacteria by designed histidine-rich peptides at acidic pH. Antimicrob Agents Chemother 50(10):3305–3311
Mohammad H, Thangamani S, Seleem MN (2015) Antimicrobial peptides and peptidomimetics-potent therapeutic allies for staphylococcal infections. Curr Pharm Design 21(16):2073–2088
Müller P, Alber DG, Turnbull L, Schlothauer RC, Carter DA, Whitchurch CB, Harry EJ (2013) Synergism between medihoney and rifampicin against methicillin-resistant Staphylococcus aureus (MRSA). PLoS ONE 8(2):e57679
Mygind PH, Fischer RL, Schnorr KM, Hansen MT, Sönksen CP, Ludvigsen S, Raventós D, Buskov S, Christensen B, Maria L, Taboureau O, Yaver D, Elvig-Jørgensen SG, Sørensen MV, Christensen BJ, Kjærulff S, Frimodt-Moller N, Lehrer RI, Zasloff M, Kristensen H (2005) Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature 437(7061):975–980
Ostergaard C, Sandvang D, Frimodt-Moller N, Kristensen HH (2009) High cerebrospinal fluid (CSF) penetration and potent bactericidal activity in CSF of NZ2114, a novel plectasin variant, during experimental pneumococcal meningitis. Antimicrob Agents Chemother 53(4):1581–1585
Pankuch GA, Appelbaum PC (2009a) Postantibiotic effect of ceftaroline against gram-positive organisms. Antimicrob Agents Chemother 53(10):4537–4539
Pankuch GA, Appelbaum PC (2009b) Postantibiotic effect of tigecycline against 14 gram-positive organisms. Antimicrob Agents Chemother 53(2):782–784
Pankuch GA, Jacobs MR, Appelbaum PC (2003) Postantibiotic effects of daptomycin against 14 staphylococcal and pneumococcal clinical isolates. Antimicrob Agents Chemother 47(9):3012–3014
Qu P, Gao W, Chen HX, Li D, Yang N, Zhu J, Feng XJ, Liu CL, Li ZQ (2016) The central hinge link truncation of the antimicrobial peptide fowlicidin-3 enhances its cell selectivity without antibacterial activity loss. Antimicrob Agents Chemother 60(5):2798–2806
Schägger H (2006) Tricine–SDS-PAGE. Nat Protoc 1:16–22
Schneider T, Kruse T, Wimmer R, Wiedemann I, Sass V, Pag U, Jansen A, Nielsen AK, Mygind PH, Raventos DS, Neve S, Ravn B, Bonvin AMJJ, Maria L, Andersen AS, Gammelgaard LK, Sahl H, Kristensen H (2010) Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science 328(5982):1168–1172
Silva T, Magalhaes B, Maia S, Gomes P, Nazmi K, Bolscher JGM, Rodrigues PN, Bastos M, Gomes MS (2014) Killing of Mycobacterium avium by Lactoferricin peptides: improved activity of arginine- and d-amino-acid-containing molecules. Antimicrob Agents Chemother 58(6):3461–3467
Stefani S, Chung DR, Lindsay JA, Friedrich AW, Kearns AM, Westh H, MacKenzie FM (2012) Meticillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. Int J Antimicrob Agents 39(4):273–282
Strandberg E, Morein S, Rijkers DTS, Liskamp R, Wel P, Killian JA (2002) Lipid dependence of membrane anchoring properties and snorkeling behavior of aromatic and charged residues in transmembrane peptides. Biochemistry 41(23):7190–7198
Taniguchi M, Takahashi N, Takayanagi T, Ikeda A, Ishiyama Y, Saitoh E, Kato T, Ochiai A, Tanaka T (2014) Effect of substituting arginine and lysine with alanine on antimicrobial activity and the mechanism of action of a cationic dodecapeptide (CL(14-25)), a partial sequence of cyanate lyase from rice. Biopolymers 102(1):58–68
Tian ZG, Dong TT, Yang YL, Teng D, Wang JH (2009) Expression of antimicrobial peptide LH multimers in Escherichia coli C43 (DE3). Appl Microbiol Biotechnol 83(1):143–149
Tsuji BT, Rybak MJ (2006) Etest synergy testing of clinical isolates of Staphylococcus aureus demonstrating heterogeneous resistance to vancomycin. Diagn Microbiol Infect Dis 54(1):73–77
Tu Z, Young A, Murphy C, Liang JF (2009) The pH sensitivity of histidine-containing lytic peptides. J Pept Sci 15(11):790–795
Veiga AS, Sinthuvanich C, Gaspar D, Franquelim HG, Castanho M, Schneider JP (2012) Arginine-rich self-assembling peptides as potent antibacterial gels. Biomaterials 33(35):8907–8916
Watanabe T, Ohashi K, Matsui K, Kubota T (1997) Comparative studies of the bactericidal, morphological and post-antibiotic effects of arbekacin and vancomycin against methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother 39(4):471–476
Weistroffer PL (2007) Cathelicidins: a history and current knowledge with experimental data on the antimicrobial and cytotoxic activities of SMAP29 and congeners. University of Iowa, Iowa. http://ir.uiowa.edu/etd/164
Wertheim HF, Melles DC, Vos MC, Leeuwen W, Belkum A, Verbrugh HA, Nouwen JL (2105) The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 5(12):751–762
White RL, Burgess DS, Manduru M, Bosso JA (1996) Comparison of three different in vitro methods of detecting synergy: time-kill, checkerboard, and E test. Antimicrob Agents Chemother 40(1):1914–1918
Xiong YQ, Hady WA, Deslandes A, Rey A, Fraisse L, Kristensen HH, Yeaman MR, Bayer AS (2011) Efficacy of NZ2114, a novel plectasin-derived cationic antimicrobial peptide antibiotic, in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 55(2011):5325–5330
Yang YL, Teng D, Zhang J, Tian ZG, Wang SR, Wang JH (2011) Characterization of recombinant plectasin: solubility, antimicrobial activity and factors that affect its activity. Process Biochem 46(5):1050–1055
Yang N, Wang XM, Teng D, Mao RY, Hao Y, Zong LF, Feng XJ, Wang JH (2016) Modification and characterization of a new recombinant marine antimicrobial peptide N2. Process Biochem 51(6):734–739
Zasloff M (2016) Antimicrobial peptides: role in human health and disease in antimicrobial peptides: do they have a future as therapeutics?. Springer International Publishing, Berlin, pp 147–154
Zhang Y, Teng D, Mao RY, Wang XM, Xi D, Hu XY, Wang JH (2014) High expression of a plectasin-derived peptide NZ2114 in Pichia pastoris and its pharmacodynamics, postantibiotic and synergy against Staphylococcus aureus. Appl Microbiol Biotechnol 98(2014):681–694
Authors’ contributions
RYM, DT, and JHW conceived and designed the research. HXC designed peptides and performed the in vitro pharmacodynamics. YH and XMW performed the hemolysis and stability (temperature, pH, proteases, and saline ions) assays. RYM, XJF and JHW wrote the manuscript. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank all the colleagues at Gene Engineering Laboratory, Feed Research Institute, Chinese Academy of Agricultural Sciences for their technical assistance throughout this study.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All datasets on which the conclusions of the manuscript were presented in the main paper.
Ethical approval
This article does not contain any studies with human and animal participants.
Funding
This study was supported by the National Natural Science Foundation of China (Nos. 31601968, 31672456, 31572444, 31572445, and 31372346), the Special Fund for Agro-scientific Research in the Public Interest in China (No. 201403047), and the AMP Direction of National Innovation Program of Agric Sci & Technol in Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-FRI-02). And all running of this experiment and paper was completed in the Gene Engineering Laboratory, Feed Research Institute, CAAS.
Author information
Authors and Affiliations
Corresponding authors
Additional file
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Chen, H., Mao, R., Teng, D. et al. Design and pharmacodynamics of recombinant NZ2114 histidine mutants with improved activity against methicillin-resistant Staphylococcus aureus . AMB Expr 7, 46 (2017). https://doi.org/10.1186/s13568-017-0345-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-017-0345-x