Abstract
Background
In recent years, the world has seen a surge in Enterobacteriaceae resistant to broad-spectrum beta-lactam antibiotics due to the production of extended-spectrum beta-lactamases (ESBLs) or plasmid-mediated AmpC (pAmpC) enzymes. Data on the epidemiology of cephalosporin-resistant Enterobacteriaceae in Sub-Saharan Africa are still limited.
Methods
Two hundred seventy-five non-repetitive stool samples were collected from Mozambican university students of both sexes. Samples were cultured on MacConkey agar with and without ceftriaxone (1 mg/L) for selection of third-generation cephalosporin-resistant isolates, which were subjected to antimicrobial susceptibility testing by disc diffusion, characterization of resistance genes by PCR and ERIC-PCR analysis for strain clonality.
Results
Among the 275 students, 55 (20%) carried a total of 56 E. coli (n = 35) and Klebsiella spp. (n = 21) isolates resistant to ceftriaxone and phenotypically positive for ESBL- and/or pAmpC-production. Forty-three percent of the isolates (24/56) contained only ESBL genes, 11% (6/56) only pAmpC genes, and 36% (20/56) both ESBL and pAmpC genes. The remaining six isolates were negative for the CTX-M/pAmpC genes included in the test panel. E. coli and Klebsiella spp. combined demonstrated 70% resistance to tetracycline and co-trimoxazole, 63% to ceftazidime and 34% to ciprofloxacin. In total, 89% of ESBL/pAmpC-positive isolates were defined as multi-resistant by being resistant to three or more antibiotic classes. ERIC-PCR fingerprinting demonstrated low similarity among isolates. None of the participants reported recent hospitalization and just 12.5% had taken antibiotics 3 months prior to the study.
Conclusion
This study demonstrated 20% colonization with multi-resistant E. coli and Klebsiella spp. among Mozambican students with a diversity of ESBL and pAmpC genes. Colonization was not related to prior hospitalization or antimicrobial consumption.
Similar content being viewed by others
Background
In recent years, the world has seen a surge in Enterobacteriaceae resistant to broad-spectrum beta-lactam antibiotics attributed to the production of extended-spectrum beta-lactamases (ESBLs) and/or plasmid-mediated AmpC (pAmpC) enzymes [1, 2]. ESBLs are, by the classical definition, inhibited by clavulanic acid [3] whereas pAmpC enzymes are not [4]. The plasmid location of ESBL and pAmpC genes facilitates their spread via horizontal gene transfer [5, 6].
Escherichia coli and Klebsiella pneumoniae are not only constituents of the commensal gut flora but also common opportunistic pathogens often implicated in urinary tract and bloodstream infections [7, 8]. They frequently harbor ESBL- and pAmpC-encoding genes. Broad-spectrum beta-lactamase production is associated with increased morbidity and mortality in both high- and low/middle-income countries [9, 10].
Data on the epidemiology of ESBL- and pAmpC-producing Enterobacteriaceae in Africa are still limited. The majority of publications report on the prevalence of ESBL-producing Enterobacteriaceae in clinical samples, and there are only few studies on colonization [11, 12]. Furthermore, there is a predominance of reports from Northern and Western Africa [13, 14] and only individual studies from Eastern and Southern Africa [15,16,17] (excluding South-Africa). The aim of this study was to determine the prevalence of fecal colonization with ESBL- and pAmpC-producing E. coli and Klebsiella spp. among healthy university students in Maputo, Mozambique, and to analyze the resistance phenotype, ESBL- and pAmpC resistance gene content and clonal relatedness of isolates.
Methods
Study sample
A random sample of 275 university students of both sexes living in three residences at the Eduardo Mondlane University in Maputo, Mozambique, provided non-repetitive stool specimens within a six-week period, from January to February 2016 upon informed, voluntary consent. Students with diarrheal disease were excluded from the study. Participants provided information about antibiotic use within 3 months and hospitalization within 6 months prior to the study. Stool samples were kept on ice and transported to the laboratory for immediate culture twice daily.
Identification and susceptibility testing
All samples were cultured on MacConkey agar with and without ceftriaxone (1 mg/L) for selection of third-generation cephalosporin-resistant isolates. Lactose-positive isolates growing on the 1 mg/L ceftriaxone agar were identified to the species level using API20E (bioMérieux, South Africa). A total of 56 E. coli (n = 35) and Klebsiella spp. (n = 21) isolates were identified by this procedure. Two isolates were recovered from the same sample. These putative ESBL- and/or pAmpC-producers constituted the study sample and were subjected to antimicrobial susceptibility testing by disc diffusion to the following antibiotics: cefoxitin, ceftazidime, ceftriaxone, imipenem, amikacin, gentamicin, ciprofloxacin and co-trimoxazole. The results were interpreted according to the CLSI breakpoints to determine their susceptibility profile [18]. Multi-resistance was defined as resistance to three or more antibiotic classes. E. coli ATCC 25922 (wild-type) and K. pneumoniae ATCC 700603 (ESBL positive) were used as negative and positive quality control strains for antimicrobial susceptibility testing and ESBL screening, respectively.
Phenotypic and genotypic characterization of beta-lactamases
ESBL production was confirmed by double disc synergy using ROSCO discs (Rosco Diagnostic, Taastrup, Denmark) containing ceftazidime, ceftriaxone and amoxicillin/clavulanic acid. pAmpC production was determined using ROSCO discs containing cefotaxime, cefotaxime+boronic acid, ceftazidime and ceftazidime+boronic acid [18]. Isolates with reduced susceptibility to carbapenems were subjected to the Carba NP test as described previously [19]. E. coli ATCC 25922 and K. pneumoniae ATCC 700603 were used as negative and positive quality control strains, respectively.
For DNA extraction, 18–24 h colonies grown on Muller Hinton agar were inoculated into Luria-Bertani (LB) broth and incubated at 37 °C with shaking [20]. After 20 h of incubation, DNA was extracted using the ZR Fungal/Bacterial DNA MiniPrep kit (Zymo Research, Lithuania). PCR amplifications for detection of blaTEM, blaSHV, blaCTX, blaCMY, blaDHA, blaFOX and blaMOX (Table 1) were performed in a ThermalCycler T100™ (Bio-Rad, USA) with a final volume of 50 μL (25 μL of Master mix, 15 μL of water, 4 μL of each primer with a final concentration of 0.8 μM (Inqaba Biotechnology Industries, South Africa) and 2 μL of template DNA), with an initial denaturation temperature of 98 °C for 10 s, extension at 72 °C for 15 s and a final extension at 72 °C for 1 min. The annealing temperatures were as follows: blaTEM 60 °C, blaSHV 56 °C, blaCTX 57 °C, blaCMY 57 °C, and blaDHA, blaFOX and blaMOX 50 °C. PCR products were loaded on a 1.5% (w/v) agarose gel and visualized by UV transillumination (Bio-Rad ChemiDoc™MP System) after staining in 0.1 mg/mL Gel Red for 15 min. A random sample of 19 CTX-M PCR amplicons were sent to Inqaba Biotec (South Africa) for DNA sequencing. TEM and SHV amplicons were not sequenced.
Genomic DNA isolation and ERIC-PCR analysis
Genomic DNA was isolated and purified using the GeneJet Genomic DNA Purification Kit (Thermo Scientific, USA). Antibiotic-susceptible E. coli ATCC 25922 and beta-lactam resistant SHV-18 K. pneumoniae ATCC 700603 were used as quality controls. The total PCR reaction volume was 10 μL, which contained 100 ng of template DNA, 50 pmol of each primer, 2.8 μL nuclease-free water and 5 μL of DreamTaq PCR Mastermix (2X) (Thermo Scientific). The primers ERIC 1 and ERIC 2 [21] were used. PCR conditions were as follows: 94 °C for 3 min, 30 cycles of 30 s of denaturation at 94 °C, 1 min of annealing at 50 °C, 8 min of extension at 65 °C, and a final elongation of 16 min at 65 °C, in an Applied Biosystems 2720 thermal cycler. The ERIC-PCR products were loaded onto 1% (w/v) agarose gels and subjected to electrophoresis at 80 V using 1 × TAE buffer. Amplification products were visualized by UV transillumination (Syngene, UK) after staining in 0.1 mg/mL ethidium bromide for 15 min. Genotypic variation was analyzed using the Gel Compare II version 6.0 software package (Applied Maths, Belgium) by Jacquard index and Unweighted Pair Group Method with Arithmetic Mean (UPGMA) cluster analysis to produce a dendrogram.
Statistical analyses
Groups were compared by the Fischer Exact Test using the Epi Info StatCalc software version 7.2.2.6 (CDC, Atlanta, USA) with statistical significance defined as p < 0.05.
Results
Setting and samples
Among the 275 samples collected in the study, 159 (58%) were retrieved from male students and 116 (42%) from females in an age range from 19 to 32 years old. The students lived in separate blocks and/or floors for male and female students. Each floor had one kitchen where students could prepare their own food. There were students from different courses including, but not limited to Engineering, Medicine, Political Science, Biology and Sociology. All participants declared a history of non-hospitalization 6 months prior to the study and 87.5% had not consumed any antibiotics for at least 3 months.
Frequency of E. coli and Klebsiella spp. ESBL colonization
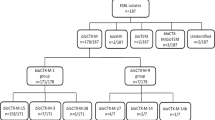
In 55 of 275 non-repetitive stool samples 35 E. coli and 21 Klebsiella spp. grew on MacConkey agar containing 1 mg/L ceftriaxone, with one sample providing two isolates. The overall prevalence of ceftriaxone-resistant E. coli and Klebsiella spp. colonization was thus 20% (55/275). There was no statistically significant difference in the rate of colonization between females (n = 28, 24%) and males (n = 28, 17%). All samples displayed growth on the ceftriaxone-free growth control agar.
Phenotypic analyses of antimicrobial susceptibility
All the 56 E. coli (n = 35) and Klebsiella spp. (n = 21) isolates were phenotypically confirmed as ESBL and/or pAmpC producers and showed high rates of resistance to tetracycline and co-trimoxazole (70%), ceftazidime (63%), cefoxitin (41%), and ciprofloxacin (34%). Two isolates displayed carbapenem zone diameters below the screening breakpoint for carbapenemase production, but were categorized as susceptible by the clinical breakpoint. They were subsequently negative in the Carba NP test and there were thus no carbapenemase producers (Table 2). Eighty-eight percent of the isolates were defined as multi-resistant. Antimicrobial susceptibility results are presented in the Additional file 1: Table S1.
ESBL gene identification by PCR and sequencing
The PCR results for CTX-M and pAmpC genes are summarized in Table 3. Among 56 isolates, 41% were positive for at least two genes, 32% were positive for at least three, and 23% were positive for at least four genes. Forty-three percent (24/56) of the isolates contained only CTX-M, 11% (6/56) only pAmpC, and 36% (20/56) both CTX-M and pAmpC sequences. Six isolates (11%) were negative for CTX-M and pAmpC genes but contained TEM and/or SHV sequences (data not shown). However, as these amplicons were not sequenced it cannot be concluded that they encode broad-spectrum beta-lactamases and the basis for ceftriaxone resistance in these isolates was consequently not determined. A total of 67 CTX-M/pAmpC genes were found in the 35 E. coli isolates and 41 in the 21 Klebsiella spp. isolates. CTX-M was the most common gene detected in both E. coli (n = 25) and Klebsiella spp. (n = 16) followed by DHA (n = 15 in E. coli and n = 9 in Klebsiella spp.). DNA sequencing of 19 randomly selected CTX-M amplicons from E. coli (n = 13) and Klebsiella spp. (n = 6) revealed a predominance of blaCTX-M-15 (n = 14) and occasional detection of blaCTX-M-55 (n = 3), blaCTX-M-3 (n = 1) and blaCTX-M-186 (n = 1). A full list of CTX-M and pAmpC gene content in individual strains is presented in the Additional file 1: Table S1.
ERIC-PCR results for E. coli
Distinct ERIC-PCR profiles were obtained for the 35 E. coli isolates (Fig. 1). The absence or presence of a band was noted in determining variation among isolates, and banding patterns comprised between 2 and 14 individual bands. Fragments of different molecular weights were observed in the ERIC-PCR fingerprints, ranging from 0.5–20 kb (Fig. 1). Amplification of different intensities was observed and visual analysis of the ERIC profiles included primary, secondary and tertiary amplification (Fig. 1). Primary amplification products refer to those products of high intensity, which appear extremely bright on the gels. Secondary amplification products are those products that are not as bright as the primary amplification products but more intense that the tertiary amplification products, while the tertiary amplification products are the minor amplification products of low intensity [22]. All isolates were typeable using this fingerprinting technique and band profiles were reproducibly obtained under similar experimental conditions on repeat amplification.
Dendrogram representing the genetic relatedness and cluster analysis of 35 E. coli strains isolated from stool samples of university students, based on ERIC-PCR fingerprinting patterns using Jacquard index and UPGMA algorithm. The scale at the top represents percentage similarity to E. coli ATCC 25922
The ERIC-PCR profiles allowed differentiation of the 35 E. coli isolates into 24 ERIC-PCR types which were grouped into 15 clusters (A-O), each of which were futher sub-divided into multiple sub-clusters (Fig. 1). CTX-M and pAmpC genes were amplified from isolates in different clusters. Isolates with similar profiles demonstrated varying beta-lactamase gene content.
ERIC-PCR results for Klebsiella spp.
Twenty-one Klebsiella spp. isolates were subjected to ERIC-PCR analysis in comparison to the SHV-18 containing K. pneumoniae ATCC 700603. Distinct profiles were obtained for all isolates tested using ERIC-PCR fingerprinting (Fig. 2). Banding patterns comprised between 2 and 16 individual bands. Fragments of different molecular weights were observed in the ERIC-PCR fingerprints, ranging from 0.5–20 kb (Fig. 2).
Dendrogram representing the genetic relatedness and cluster analysis of 21 Klebsiella spp. strains isolated from stool samples of university students, based on ERIC-PCR fingerprinting patterns using Jacquard index and UPGMA algorithm. The scale at the top represents percentage similarity to Klebsiella pneumoniae ATCC 700603
The ERIC-PCR profiles allowed differentiation of the 21 isolates into 17 ERIC-PCR types which were grouped into 12 clusters (A-L), each of which were further sub-divided into multiple sub-clusters (Fig. 2). CTX-M and pAmpC genes were identified in isolates from different clusters, predominantly clusters B-G. Isolates with similar profiles demonstrated varying beta-lactamase gene content.
Discussion
Antibiotic-resistant bacteria are an escalating cause of infections in Mozambique and worldwide, but information regarding the molecular epidemiology of ESBL- and pAmpC-producing Enterobacteriaceae in the country remains scarce. To our knowledge, this is the first study reporting on gastrointestinal colonization with ESBL- and pAmpC-producing Enterobacteriaceae among university students in Mozambique.
We systematically collected samples from healthy students to determine the prevalence of colonization by E. coli and Klebsiella spp. carrying ESBL and/or pAmpC, as well as their antibiotic susceptibility. In total, 20% of students (55/275) were colonized with ESBL and/or pAmpC positive E. coli and Klebsiella spp. Similar results were found in a study conducted in pre-school children attending childcare facilities in the Lao People’s Democratic Republic where the ESBL prevalence was 23% [23] while the prevalence amongst patients in intensive care in Korea was 28.2% with a higher frequency in E. coli (78%) compared with Klebsiella spp. (18%) [24]. A study recently conducted in Madagascar [18] in community settings demonstrated lower rates of colonization by ESBL-positive Gram-negative bacilli compared to this study, despite the fact that more species were included in addition to E. coli and Klebsiella spp. The present study showed a higher prevalence of ESBL and pAmpC colonization compared to a study conducted in rural communities in France where the frequency of ESBL colonization was 5.3% [25]. This large difference may be attributed to poorly controlled antibiotic consumption and sub-optimal hygiene conditions in developing countries [26]. In Mozambique, antibiotic therapy is mostly empirical because of scarce diagnostic microbiology facilities [27]. We found 36% co-existence of ESBL and pAmpC beta-lactamase genes which is high compared to reports from clinical isolates in Turkey (13.9%) [28].
The CTX-M positive isolates carried different genotypes; blaCTX-M-15 (n = 14), blaCTX-M-55 (n = 3), blaCTX-M-186 (n = 1) and blaCTX-M-3 (n = 1). These results are similar to findings in a study conducted in Tanzania [29] that found blaCTX-M-15 genes in 95% of the carriers, but differ from a study in Kenya [30] which demonstrated only 29% of the isolates carrying blaCTX-M-15 and 4% carrying blaCTX-M-3. No isolates harbored blaCTX-M-55 or blaCTX-M-186 in the Kenyan study. blaCTX-M-55 is endemic in many Asian countries, and the detection of this variant in Mozambique may reflect travel and trade with this part of the world [31].
In this study of healthy students, none of the participants had a history of hospitalization 6 months prior to the study, and 87.5% had not consumed any antibiotics at least 3 months prior to the study. This suggests that the high antibiotic resistance rates are not related to antibiotic consumption or hospitalization, and may indicate that the E. coli and Klebsiella ESBL- and pAmpC-producers isolated in this study were acquired in the community. In the university residences there is a mixture of students from different courses including medicine and health sciences, but the study was not powered to analyse differences between student groups (data not shown). ESBL- and/or pAmpC-colonized students working in health institutions may constitute a reservoir for further spread of multi-resistant microorganisms among patients.
To control the rapid dissemination of resistant Enterobacteriaceae among students and, consequently, the general population, it is necessary to educate students about the importance of personal hygiene. It is also necessary to perform further studies to determine the prevalence of colonization in different groups of the population. The prevalence in different studies may vary depending on socio-economic status of individuals involved [32], and this makes it difficult to estimate the prevalence in the general population in Mozambique. One may suppose that transmission between students is facilitated because of the conditions they live under with many individuals sharing the same bathroom and kitchen. However, the diversity of strains demonstrated using ERIC-PCR indicates that there is at present no widespread clonal outbreak, although there could be horizontal transfer of plasmids or other mobile genetic elements. One may speculate that students are exposed to ESBL and/or pAmpC isolates from some external source like dissemination in the food supply. Very little is known about the occurrence of ESBL and pAmpC strains in the food supply in Mozambique, but it is reported from other countries that ESBL and pAmpC E. coli and Klebsiella spp. may disseminate in food animals and environmental sources [33,34,35,36,37].
Conclusions
This study demonstrated a 20% prevalence of colonization by multi-drug resistant, non-clonally-related ESBL- and/or pAmpC positive E. coli and Klebsiella spp. isolates among male and female university students in Mozambique. Colonization of ESBL/pAmpC in the population of study was not related to prior antimicrobial consumption or hospitalization. Similar studies should be done to further explore the epidemiology of multi-drug resistant Enterobacteriaceae in the population as a whole.
Abbreviations
- ATCC:
-
American Type Culture Collection
- DNA:
-
Deoxyribonucleic acid
- ERIC-PCR:
-
Enterobacterial repetitive intergenic consensus PCR
- ESBL:
-
Extended-spectrum beta-lactamases
- pAmpC:
-
Plasmid-mediated AmpC beta-lactamase
- PCR:
-
Polymerase chain reaction
- TAE:
-
Buffer containing Tris base, acetic acid and EDTA (Ethylenediaminetetraacetic acid)
- UPGMA:
-
Unweighted pair group method with arithmetic mean
References
Al-Jasser AM. Extended-spectrum beta-lactamases (ESBL’s): global problem. Kuwait Med J. 2006;38:171–85.
Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8(3):159–66.
Rawat D, Nair D. Extended-spectrum beta-lactamases in gram-negative bacteria. J Glob Infect Dis. 2010;2(3):263–74.
Villa L, Pezzella C, Tosini F, Visca P, Petrucca A, Carattoli A. Multiple-antibiotic resistance mediated by structurally related IncL/M plasmids carrying an extended spectrum beta-lactamase gene and class 1 integron. Antimicrob Agents Chemother. 2000;44(10):2911–4.
Coudron PE, Hanson ND, Climo MW. Occurrence of extended-spectrum and AmpC beta-lactamases in bloodstream isolates of Klebsiella pneumoniae: isolates harbor plasmid-mediated FOX-5 and ACT-1 AmpC beta-lactamases. J Clin Microbiol. 2003;41(2):772–7.
Dolejska M, Villa L, Hasman H, Hansen L, Carattoli A. Characterization of IncN plasmids carrying blaCTX-M-1 and qnr genes in Escherichia coli and Salmonella from animals, the environment and humans. J Antimicrob Chemother. 2013;68(2):333–9.
Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657–86.
Kothari C, Gaind R, Singh LC, Sinha A, Kumari V, Arya S, Chellani H, Saxena S, Deb M. Community acquisition of beta-lactamase producing Enterobacteriaceae in neonatal gut. BMC Microbiol. 2013;13:136.
de Kraker ME, Davey PG, Grundmann H, BURDEN study group. Mortality and hospital stay associated with resistant Staphylococcus aureus and Escherichia coli bacteremia: estimating the burden of antibiotic resistance in Europe. PLoS Med. 2011;8(10):e1001104.
Blomberg B, Jureen R, Manji KP, Tamim BS, Mwakagile DS, Urassa WK, Fataki M, Msangi V, Tellevik MG, Maselle SY, Langeland N. High rate of fatal cases of pediatric septicemia caused by gram-negative bacteria with extended-spectrum beta-lactamases in Dar es Salaam, Tanzania. J Clin Microbiol. 2005;43(2):745–9.
Storberg V. ESBL-producing Enterobacteriaceae in Africa - a non-systematic literature review of research published 2008-2012. Infect Ecol Epidemiol. 2014;4:20342.
Tansarli GS, Poulikakos P, Kapaskelis A, Falagas ME. Proportion of extended-spectrum β-lactamase (ESBL)-producing isolates among Enterobacteriaceae in Africa: evaluation of the evidence--systematic review. J Antimicrob Chemother. 2014;69(5):1177–84.
Sangare SA, Rondinaud E, Maataoui N, Maiga AI, Guindo I, Maiga A, Camara N, Dicko OA, Dao S, Diallo S, Bougoudogo F, Andremont A, Maiga II, Armand-Lefevre L. Very high prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in bacteriemic patients hospitalized in teaching hospitals in Bamako, Mali. PLoS One. 2017;12(2):e0172652.
Isendahl J, Turlej-Rogacka A, Manjuba C, Rodrigues A, Giske CG, Nauclér P. Fecal carriage of ESBL-producing E. coli and K. pneumoniae in children in Guinea-Bissau: a hospital-based cross-sectional study. PLoS One. 2012;7(12):e51981.
Nelson E, Kayega J, Seni J, Mushi MF, Kidenya BR, Hokororo A, Zuechner A, Kihunrwa A, Mshana SE. Evaluation of existence and transmission of extended spectrum beta lactamase producing bacteria from post-delivery women to neonates at Bugando medical center, Mwanza-Tanzania. BMC Res Notes. 2014;7:279.
Moremi N, Claus H, Vogel U, Mshana SE. Faecal carriage of CTX-M extended-spectrum beta-lactamase-producing Enterobacteriaceae among street children dwelling in Mwanza city, Tanzania. PLoS One. 2017;12(9):e0184592.
Herindrainy P, Randrianirina F, Ratovoson R, Ratsima Hariniana E, Buisson Y, Genel N, Decré D, Arlet G, Talarmin A, Richard V. Rectal carriage of extended-spectrum beta-lactamase producing gram-negative bacilli in community settings in Madagascar. PLoS One. 2011;6(7):e22738.
Clinical and Laboratory Standards Institute (CLSI). Performance Standards for disk Susceptibility Testing. Approved standard. 12th ed; 2015.
Vasoo S, Cunningham SA, Kohner PC, Simner PJ, Mandrekar JN, Lolans K, Hayden MK, Patel R. Comparison of a novel, rapid chromogenic biochemical assay, the Carba NP test, with the modified Hodge test for detection of carbapenemase-producing gram-negative bacilli. J Clin Microbiol. 2013;51(9):3097–101.
Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153–62.
Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acid Res. 1991;19:6823–31.
Bassam BJ, Caetano-Anollés G, Gresshoff PM. DNA amplification fingerprinting of bacteria. Appl Microbiol Biotechnol. 1992;38(1):70–6.
Stoesser N, Xayaheuang S, Vongsouvath M, Phommasone K, Elliott I, Del Ojo Elias C, Crook DW, Newton PN, Buisson Y, Lee SJ, Dance DA. Colonization with Enterobacteriaceae producing ESBLs in children attending pre-school childcare facilities in the Lao People’s Democratic Republic. J Antimicrob Chemother. 2015;70(6):1893–7.
Kim J, Lee JY, Kim SI, Song W, Kim JS, Jung S, Yu JK, Park KG, Park YJ. Rates of fecal transmission of extended-spectrum beta-lactamase producing and carbapenem-resistant Enterobacteriaceae among patients in intensive care units in Korea. Ann Lab Med. 2014;34(1):20–5.
Woerther PL, Angebault C, Jacquier H, Clermont O, El Mniai A, Moreau B, Djossou F, Peroz G, Catzeflis F, Denamur E, Andremont A. Characterization of fecal extended-spectrum beta-lactamase producing Escherichia coli in a remote community during a long time period. Antimicrob Agents Chemother. 2013;57(10):5060–6.
Woerther PL, Burdet C, Chachaty E, Andremont A. Trends in human fecal carriage of extended-spectrum beta-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev. 2013;26(4):744–58.
Mandomando I, Sigaúque B, Morais L, Espasa M, Vallès X, Sacarlal J, Macete E, Aide P, Quintò L, Nhampossa T, Machevo S, Bassat Q, Menéndez C, Ruiz J, Roca A, Alonso PL. Antimicrobial drug resistance trends of bacteremia isolates in a rural hospital in southern Mozambique. Am J Trop Med Hyg. 2010;83(1):152–7.
Korten V, Ulusoy S, Zarakolu P, Mete B, Turkish MYSTIC Study Group. Antibiotic resistance surveillance over a 4-year period (2000-2003) in Turkey: results of the MYSTIC program. Diagn Microbiol Infect Dis. 2007;59(4):453–7.
Tellevik MG, Blomberg B, Kommedal Ø, Maselle SY, Langeland N, Moyo SJ. High prevalence of faecal carriage of ESBL-producing Enterobacteriaceae among children in Dar es Salaam, Tanzania. PLoS One. 2016;11(12):e0168024.
Kiiru J, Kariuki S, Goddeeris BM, Butaye P. Analysis of beta-lactamase phenotypes and carriage of selected beta-lactamase genes among Escherichia coli strains obtained from Kenyan patients during an 18-year period. BMC Microbiol. 2012;12:155.
Zhang J, Zheng B, Zhao L, Wei Z, Ji J, Li L, Xiao Y. Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect Dis. 2014;14:659.
Schellenberg JA, Victora CG, Mushi A, de Savigny D, Schellenberg D, Mshinda H, Bryce J. Inequities among the very poor health care for children in rural southern Tanzania. Lancet. 2003;361(9357):561–6.
Tekiner IH, Özpınar H. Occurrence and characteristics of extended spectrum beta-lactamases-producing Enterobacteriaceae from foods of animal origin. Brazilian J Microbiol. 2016;47(2):444–51.
Abdallah HM, Reuland EA, Wintermans BB, Al Naiemi N, Koek A, Abdelwahab AM, Ammar AM, Mohamed AA, Vandenbroucke-Grauls CM. Extended-spectrum beta-lactamases and/or carbapenemases-producing Enterobacteriaceae isolated from retail chicken meat in Zagazig, Egypt. PLoS One. 2015;10(8):e0136052.
Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH. Extended-spectrum beta-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin Microbiol Infect. 2012;18(7):646–55.
Zurfluh K, Nüesch-Inderbinen M, Morach M, Zihler Berner A, Hächler H, Stephan R. Extended-spectrum beta-lactamase-producing Enterobacteriaceae isolated from vegetables imported from the Dominican Republic, India, Thailand, and Vietnam. Appl Environ Microbiol. 2015;81(9):3115–20.
Stefani S, Giovanelli I, Anacarso I, Condò C, Messi P, de Niederhäusern S, Bondi M, Iseppi R, Sabia C. Prevalence and characterization of extended-spectrum beta-lactamase-producing Enterobacteriaceae in food-producing animals in Northern Italy. New Microbiol. 2014;37(4):551–5.
Acknowledgements
The authors would like to acknowledge all participants in the study, as well as technical staff involved in the collection and analysis of samples.
Funding
The study was funded by the Norwegian Agency for Development Cooperation under the Norwegian Programme for Capacity Development in Higher Education and Research for Development (NORHED) Grant QZA 0484 RSA 13/0010 entitled “Antibiotic Stewardship and Conservancy in Africa”. The funding body had no role in the design of the study, collection, analysis, or interpretation of data, or in writing the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Author information
Authors and Affiliations
Contributions
The study was designed LMC, TFZ, AS, SYE and GSS. Collection of data and laboratory analyses were performed by LMC, JOS, UG and HYC. All authors contributed to the interpretation of the data and writing of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical approval was received from the Biomedical Research Ethics Committee of University of KwaZulu-Natal (BE214/16) and the Bioethical Council ISCISA-Mozambique (TFCMCSCLJ03/15). All participants signed an informed consent form. Students colonized with ESBL- and/or pAmpC-producing E. coli and Klebsiella spp. did not receive information about their test results during the study period, but got such information at the end of the study.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Table S1. Resistance genes identified and sensitivity results of E. coli and Klebsiella spp. (DOCX 38 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Chirindze, L.M., Zimba, T.F., Sekyere, J.O. et al. Faecal colonization of E. coli and Klebsiella spp. producing extended-spectrum beta-lactamases and plasmid-mediated AmpC in Mozambican university students. BMC Infect Dis 18, 244 (2018). https://doi.org/10.1186/s12879-018-3154-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-018-3154-1