Abstract
Background
Over the past two decades, several studies have focused on the association between a common polymorphism (rs1800795) from interleukin-6 (IL-6) gene and Diabetes Mellitus (DM) risk. However, the results remain ambiguous and indefinite.
Methods
A comprehensive analysis was performed to explore this relationship. A search was conducted in the PubMed, Embase, Chinese (CNKI and Wanfang), and GWAS Catalog databases, covering all publications until February 10, 2022. Odds ratios (OR) with 95% confidence intervals (CI) were used to evaluate the strength of the association. Publication bias was assessed using both Begg and Egger tests.
Results
Overall, 34 case–control studies with 7257 T2DM patients and 15,598 controls, and 12 case–control studies (10,264 T1DM patients and 9031 health controls) were included in the analysis. A significantly lower association was observed between the rs1800795 polymorphism and T2DM risk in Asians, mixed population, and hospital-based (HB) subgroups (C-allele vs. G-allele: OR = 0.76, 95% CI 0.58–0.99, P = 0.039 for Asians; CG vs. GG: OR = 0.74, 95% CI 0.58–0.94, P = 0.014 for mixed population; CC vs. GG: OR = 0.61, 95% CI 0.41–0.90, P = 0.014 for HB). However, increased associations were found from total, mixed population, and HB subgroups between rs1800795 polymorphism and T1DM susceptibility (CG vs. GG: OR = 1.32, 95% CI 1.01–1.74, P = 0.043 for total population, CC vs. GG: OR = 2.45, 95% CI 1.18–5.07, P = 0.016 for mixed individuals; C-allele vs. G-allele: OR = 1.29, 95% CI 1.07–1.56, P = 0.0009 for HB subgroup).
Conclusions
In summary, there is definite evidence to confirm that IL-6 rs1800795 polymorphism is associated with susceptibility to decreased T2DM and increased T1DM.
Similar content being viewed by others
Background
Diabetes mellitus (DM) is a chronic medical condition in which the body either produces too little insulin from pancreatic islets or lacks effective access to insulin [1]. Type 1 DM (T1DM) is most often diagnosed in children and adolescents with respect to islet function development. Type 2 DM (T2DM) is caused by insulin resistance, and the body cannot use insulin effectively and may gradually lose its production capacity [2,3,4]. To the best of our knowledge, age, obesity, and family history are the major risk factors of developing DM [5]. However, the exact pathogenesis of DM is not fully understood. Past genome-wide association studies (GWAS) have identified over 100 genetic sites, which suggests that there are significant associations between different sites and susceptibility to DM, indicating that genetic factors may be crucial for its occurrence and development [6, 7].
Interleukin-6 (IL-6), a classic proinflammatory cytokine, plays a prominent role in the inflammatory response and is associated with insulin resistance and T2DM [8]. In addition, chronic low-grade inflammation and activation of the innate immune system are closely associated with the pathogenesis of T1DM and its complications. Inflammatory cytokines such as IL-6 are determinants of these pathogenic processes [9, 10].
The IL-6 gene is located on chromosome 7p21. The gene, which includes seven exons, covers approximately 12.8 kb of genomic DNA [11]. A common single nucleotide polymorphism (SNP) in the IL-6 promoter in T2DM has been named rs1800795 (also named –174G/C) [12]. The rs1800795 polymorphism functionally affects IL-6 promoter activity, indicating that the carried CC genotype individual is associated with lower plasma levels of IL-6 compared with individuals with the GG genotype [13]. In addition, the G-allele in homozygotes (GG genotype) was associated with higher concentrations of IL-6, increasing the immune response [14, 15], demonstrating that this polymorphism is functional, or that it defined a difference in IL-6 expression levels according to the genotype of the polymorphism.
Several epidemiological studies have observed associations between genetic variants of IL-6 and the risk of DM. For instance, Saxena et al. observed that the rs1800795 polymorphism showed a highly significant association with T2DM [16]. In contrast, Dhamodharan et al. determined that the C allele conferred significant protection against T2DM [17]. In addition, Fathy et al. [18] demonstrated a lack of significant association between rs1800795 polymorphism and T2DM. For T1DM, an increased association was observed between T1DM and the polymorphism by Cooper et al. [19]. However, Tsiavou et al. observed no significant differences [20]. Two meta-analyses (Yin and Xu et al.) showed that rs1800795 is not associated with T1DM risk [21, 22]. On the other hand, Huth and Xia et al. performed a meta-analysis and concluded that this polymorphism could be associated with a decreased risk of T2DM [23, 24]. In the last 10 years, some larger and more comprehensive studies have been conducted on this association. Therefore, it is necessary to perform an updated meta-analysis to understand the associations between rs1800795 polymorphism and T1DM/T2DM [12, 15,16,17,18,19,20, 25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60].
Materials and methods
Document retrieval and data extraction
We used online databases, including PubMed, Embase, CNKI, Wanfang, and GWAS Catalog (https://www.ebi.ac.uk/gwas/) until on Feb 10, 2022, with keywords including ‘Interleukin-6/IL-6’, ‘polymorphism/variant’, and ‘Diabetes Mellitus/DM/TIDM/T2DM’. Two researchers (Zhiying Cheng, Chunmin Zhang) evaluated the articles to identify the stages through the abstract and then the full article. Systematic analysis/meta-analysis, case studies, other polymorphisms, insufficient data for each genotype, and duplications were identified and removed from further analysis. In addition, our meta-analysis was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) (Additional file 1: Table S1) and Meta-analysis of Observational Studies in Epidemiology. This study was registered at PROSPERO (number 329822; https://www.crd.york.ac.uk/prospero/). Eligible studies were selected based on the following criteria: @) studies assessing the association between TIDM or T2DMAdditional file: As per journal requirements, every additional file must have a corresponding caption. In this regard, please be informed that the caption was taken from the Additional file 1 itself. Please advise if action taken appropriate and amend if necessary. and rs1800795 variants; @) case/control studies; and @) age-and sex-matched control subjects. The exclusion criteria were: @) not case/control studies; @) insufficient genotype frequency; @) duplicate studies; and @) significantly biased articles. Information including the name of the first author, year of publication, origin, race, DM type, genotype methods, and Hardy–Weinberg equilibrium (HWE) was collected.
Quality assessment
Quality was assessed using the Newcastle–Ottawa Scale (NOS) for cross-sectional study quality assessment. The methodological quality of each study (sampling strategy, response rate, and representativeness), comparability, and outcomes were assessed using the NOS tool. Studies with a score of more than 7 out of 10 were considered suitable. This cutoff point was determined after reviewing relevant meta-analyses from the literature [61,62,63].
Statistical analyses
The correlation between IL-6 rs1800795 polymorphism and the risk of TIDM/T2DM was measured using 95% confidence interval (CI) and odds-ratio (OR) according to the genotype frequencies of the case and control groups. Ethnic groups were divided into African, mixed, Caucasian, and Asian groups. Population-based (PB) and hospital-based (HB) control subgroups were also identified.
The statistical significance of the results was calculated using the Z-test. In these studies, the heterogeneity hypothesis was assessed using the Q-test based on the chi-squared test [64]. If significant heterogeneity (< 0.1) was detected, the random effects model was used, else the fixed effects model was selected [65, 66]. For IL-6 rs1800795, we studied the relationship between variation and the risk of T2DM in the C-allele vs. G-allele, CG vs. GG, and CC + CG vs. GG models; and C-allele vs. G-allele, CC vs. GG, CC vs. CG + GG, CG vs. GG, and CC + CG vs. GG models for T1DM risk. The asymmetry of the funnel plot was evaluated using Begg’s test, and publication bias was evaluated using Egger’s test. Statistical significance was set at P < 0.05 [67]. Pearson’s chi-squared test was used in the control group (P < 0.05), and the χ2 test was used to evaluate the deviation of rs1800795 polymorphism from the expected frequency of HWE [68]. All statistical tests were conducted using Stata (version 11.0; StataCorp LP, College Station, Texas, USA). The power of our meta-analysis was calculated online using the website http://www.power-analysis.com/.
Gene interaction network analysis of the IL-6 gene
To fully understand the role of IL-6 and its potential functional partners in DM, we used the STRING online server (http://string-db.org/) to construct an IL-6 gene–gene interaction network.
Results
Study selection and characteristics
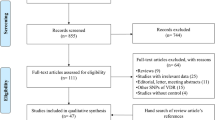
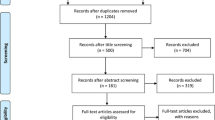
A total of 1356 articles were identified from the four main databases (PubMed, Embase, CNKI, and Wanfang). 1260 papers were excluded after reading the abstract, and 96 articles were used for a complete evaluation. Among them, 50 articles were excluded for the following reasons: systematic analysis/meta-analysis (10), only case studies (9), other polymorphisms in the IL-6 gene (15), insufficient data for each genotype (8), and duplication (8) (Fig. 1). Thus, 46 papers [13,14,15,16,17,18] accounting for a total of 17,521 DM patients and 24,629 healthy controls were included in our meta-analysis (34 case–control studies including 7257 T2DM patients and 15,598 controls, and 12 case–control studies including 17,521 T1DM and 9031 controls) [12, 15,16,17,18,19,20, 25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60] (Table 1). We checked the minor allele frequency (MAF) reported for the five main populations worldwide in the 1000 Genomes Browser (https://www.ncbi.nlm.nih.gov/snp/rs1800795#frequency_tab) (Fig. 2A). In addition, the C-allele frequency was significantly lower in both cases and controls (Fig. 2B) (Table 2). The relationship between this polymorphism and several organs is shown in Fig. 2C (https://www.gtexportal.org/home/). The distribution of genotypes in controls was not consistent with the HWE in T2DM (9 case–control studies) [15, 26, 32, 38, 41, 42, 51, 53, 60] and T1DM (2 case–control studies) [44, 48] (Table 1). Genotyping of the SNPs of IL-6 gene rs1800795 polymorphism was conducted using the genotyping methods listed in Table 1.
IL-6 rs1800795 polymorphism and T2DM risk
The results of the meta-analysis suggested no associations between IL-6 rs1800795 polymorphism and T2DM risk (Table 3). If studies that were not consistent with HWE were excluded, no significant results were detected in any of the three models. Analysis of ethnicity subgroups showed a statistically significant association in Asians (ORC-allele vs. G-allele = 0.76, 95% CI 0.58–0.99, P = 0.039, random effect model; ORCC vs. GG = 0.45, 95% CI 0.24–0.85, P = 0.014, random effect model, ORCC vs. CG+GG = 0.48, 95% CI 0.27–0.86, P = 0.014, random effect model, Fig. 3) and mixed populations (ORCG vs. GG = 0.74, 95% CI 0.58–0.94, P = 0.014, fixed effect model, Fig. 4). Surprisingly, a marginal and poorly significant difference was found in the HB sources of the control subgroup (ORCC vs. GG = 0.61, 95% CI 0.41–0.90, P < 0.011, random effect model, ORCC vs. CG+GG = 0.64, 95% CI 0.46–0.90, P = 0.011, random effect model, Fig. 5). Furthermore, if studies that were not consistent with HWE were included, no significant association was found between Asians and HB subgroups (Table 3).
Forest plot of T2DM risk associated with IL-6 rs1800795 polymorphism (C-allele vs. G-allele) in the subgroup of Asian subgroup. The squares and horizontal lines correspond to the study-specific OR and 95% CI. The area of the squares reflects the weight (inverse of the variance). The diamond represents the summary OR and 95% CI
IL-6 rs1800795 polymorphism and T1DM risk.
There was a significant positive association between rs1800795 polymorphism and T1DM susceptibility in the total analysis (ORCC vs. GG = 1.32, 95% CI 1.01–1.74, P = 0.043, random effect model, Fig. 6) (Table 3). Additionally, a risk association was observed between this polymorphism in the mixed population (ORC-allele vs. G-allele = 1.39, 95% CI 1.10–1.77, P = 0.006, fixed effect model, ORCC vs. GG = 2.45, 95% CI 1.18–5.07, P = 0.016, fixed effect model, ORCC+CG vs. GG = 1.43, 95% CI 1.07–1.90, P = 0.015, fixed effect model, ORCC vs. CG+GG = 2.20, 95% CI 1.08–4.48, P = 0.031, fixed effect model, Fig. 7). Similar relationships were observed for the sources of the HB subgroup (ORC-allele vs. G-allele = 1.29, 95% CI 1.07–1.56, P = 0.009, fixed effect model, ORCG vs. GG = 1.47, 95% CI 1.11–1.94, P = 0.008, fixed effect model, Fig. 8). Furthermore, when we excluded studies that were not consistent with HWE, the results remain the same as above (Table 3).
Publication bias and sensitive analysis
Begg’s and Egger’s tests were performed to assess publication bias, which was not found for T2DM or T1DM analyses (T2DM: tC-allele vs. G-allele = − 1.32, P = 0.195 for Egger’s test, z = 1.02, P = 0.306 for Begg’s test, Fig. 9a, b; T1DM: tC-allele vs. G-allele = 1.82, P = 0.099 for Egger’s test, z = 1.17, P = 0.244 for Begg’s test, Fig. 10a,b, Table 4). To delete studies that may influence the power and stability of the whole study, we applied a sensitivity analysis, and no sensitive case–control studies were found (Figs. 9c, 10c, Table 4).
Gene–gene network diagram and interactions
Our analysis using the STRING online server indicated that IL-6 interacts with several genes. The ten most significant genes from the network of gene–gene interactions are shown in Fig. 11. These ten genes are: interleukin-6 receptor (IL6R); interleukin-6 receptor subunit beta (IL6ST); interleukin-1 beta (IL1B); interleukin-8 (CXCL8); growth-regulated alpha protein (CXCL1); C-X-C motif chemokine 2 (CXCL2); C–C motif chemokine 2 (CCL2); interleukin-17A (IL17A); tumor necrosis factor (TNF); and interleukin-1 alpha (IL1A).
Discussion
Diabetes has reached pandemic dimensions, and is becoming relevant in both developed and developing countries, affecting over 400 million people worldwide [69]. To date, several studies have focused on the relationship between IL-6 rs1800795 polymorphism and DM risk [26, 29, 30, 38]. A few meta-analysis-based studies have also indicated similar associations [21,22,23,24]. However, there is a lack of robust conclusions. Therefore, it is necessary to recombine previously published studies to perform a comprehensive meta-analysis to understand the above-mentioned association in further detail. To the best of our knowledge, meta-analysis is a powerful method when the results are based on a large number of samples and are inconsistent, including different ethnicities or countries [24]. The conclusion obtained from the meta-analysis is more robust than that of a single study [24]. To investigate the association between IL-6 rs1800795 and DM, our comprehensive study included 42,150 individuals. Our results indicate that IL-6 rs1800795 acts as a protective factor in T2DM. In other words, individuals carrying the C-allele may have a decreased association with T2DM, particularly among Asians, mixed populations, and HB source studies. However, IL-6 rs1800795 was found to be a risk factor for T1DM, and there was a significantly increased association between this polymorphism and T1DM risk in four genetic models in mixed-population and HB source studies.
Therefore, IL-6 rs1800795 polymorphism may have different effects in different types of DM, and also have different influences on different ethnicities, such as Asians and mixed populations. This could be due to the following: the pathogenic mechanisms of T2DM and T1DM are different, with differences in several significantly expressed genes. Further studies should focus on the functions and mechanisms of mutation or wild-type IL-6 rs1800795 polymorphism to define the dissimilarity between T2DM and T1DM. On the other hand, the same gene may have different effects, even opposite, and the IL-6 gene may behave differently for T2DM and T1DM. Therefore, rs1800795 polymorphism affecting the expression of IL-6 may also differ in its roles in T2DM and T1DM. Different races have heterogeneity, and the same gene may also have different roles in different ethnicities [70, 71]. Third, heterogeneity in the selection strategy may exist, which may have affected our results. To evaluate the stability and validity of the current study, we performed a power analysis. The power in T2DM was 1 and that in T1DM was 0.166, indicating that the conclusions from T2DM were more powerful and persuasive than those for T1DM. This suggests that more studies on rs1800795 and T1DM risk should be conducted in future to obtain a robust conclusion.
The development and outcome of DM are complex and multifactorial. Focusing only on each gene or polymorphism provides a limited understanding of the same. Hence, we attempted to detect other potential genes related to DM using the online STRING server. The other ten most probable genes were obtained from the network. Among them, six genes belonged to the interleukin family and three were in the front. Four genes were related to the chemokine (C–X–C motif) ligand family. For example, the first related gene is IL-6R, which is the receptor of the IL-6 gene. Qi et al. reported that the IL6R rs8192284 variant was significantly associated with plasma CRP level and could predict diabetes risk [72]. Jiao et al. performed a meta-analysis and suggested that the IL-1B (-511) T-allele polymorphism is associated with a decreased T2DM risk in East Asians [73]. Silva et al. concluded that functional CXCL8 rs4073, rs2227307, and rs2227306 SNPs are relevant genetic factors for T2DM [74]. Trapali et al. indicated that the TNF-α308G/A polymorphism is significantly associated with T2DM susceptibility [75]. In summary, there is a need toexplore these partners of the IL-6 gene and gene–gene interactions in the development and treatment of DM.
Although we performed a comprehensive meta-analysis, this study has several limitations. First, studies from mixed populations and Africans are limited, which leads to missing or insufficient results and may influence the conclusion. Second, one single gene or one polymorphism may not have the power to result in the development of DM, which is a complex process including gene–gene or gene-environment interactions, and further studies should pay close attention to the same. Third, four databases were included, and some valuable studies from other databases or languages could not be identified, which should have an impact on the current conclusions. Finally, most of the studies were selected using the PCR–RFLP technique in current publications, and the authors may apply to duplicate selected samples for the second time at least 10% of the total samples to confirm the genotypes detected by PCR–RFLP, as real-time PCR is a reference method which can verify the genotyping in PCR–RFLP technique to avoid false positives.
Conclusions
In summary, our meta-analysis provided evidence that the IL-6 rs1800795 polymorphism was associated with significantly increased T1DM risk in a mixed population. In contrast, a decreased association was found in T2DM susceptibility in Asians. Consequently, further well-designed large-scale studies, particularly those related to gene–gene and gene-environment interactions, are warranted.
Availability of data and materials
All data generated or analyzed in this study are included in this published article.
Abbreviations
- DM:
-
Diabetes mellitus
- GWAS:
-
Genome-wide association studies
- IL-6:
-
Interleukin-6
- SNP:
-
Single nucleotide polymorphism
- HB:
-
Hospital-based
- PB:
-
Population-based
- SOC:
-
Source of control
- PCR–RFLP:
-
Polymerase chain reaction followed by restriction fragment length polymorphism
- PCR-SSP:
-
Polymerase chain reaction followed with sequence specific primers
- MALDI-TOF:
-
A chip-based matrix-assisted laser-desorption/ionization time-of-flight
References
Edgerton DS, Kraft G, Smith M, Farmer B, Williams PE, Coate KC, Printz RL, O’Brien RM, Cherrington AD. Insulin’s direct hepatic effect explains the inhibition of glucose production caused by insulin secretion. JCI insight. 2017;2(6): e91863.
Association AD. Diagnosis and classification of diabetes mellitus. Diabetes care. 2010;33(1):S62-69.
Diamant AL, Babey SH, Hastert TA, Brown ER. Diabetes: the growing epidemic. Policy Brief. 2007;9:1–12.
Marciano L, Camerini AL, Schulz PJ. The role of health literacy in diabetes knowledge, self-care, and glycemic control: a meta-analysis. J Gen Intern Med. 2019;34(6):1007–17.
Khan RMM, Chua ZJY, Tan JC, Yang Y, Liao Z, Zhao Y. From pre-diabetes to diabetes: diagnosis, treatments and translational research. Medicina (Kaunas, Lithuania). 2019;55(9):546.
Gaulton KJ. Mechanisms of type 2 diabetes risk loci. Curr DiabRep. 2017;17(9):72.
Papazafiropoulou AK, Papanas N, Melidonis A, Maltezos E. Family history of type 2 diabetes: does having a diabetic parent increase the risk? Curr Diabetes Rev. 2017;13(1):19–25.
Pickup JC, Mattock MB, Chusney GD, Burt D. NIDDM as a disease of the innate immune system: association of acute-phase reactants and interleukin-6 with metabolic syndrome X. Diabetologia. 1997;40(11):1286–92.
Campbell IL, Kay TW, Oxbrow L, Harrison LC. Essential role for interferon-gamma and interleukin-6 in autoimmune insulin-dependent diabetes in NOD/Wehi mice. J Clin Investig. 1991;87(2):739–42.
Foulis AK, Farquharson MA, Meager A. Immunoreactive alpha-interferon in insulin-secreting beta cells in type 1 diabetes mellitus. Lancet (London, England). 1987;2(8573):1423–7.
Bowcock AM, Kidd JR, Lathrop GM, Daneshvar L, May LT, Ray A, Sehgal PB, Kidd KK, Cavalli-Sforza LL. The human “interferon-beta 2/hepatocyte stimulating factor/interleukin-6” gene: DNA polymorphism studies and localization to chromosome 7p21. Genomics. 1988;3(1):8–16.
Vozarova B, Fernandez-Real JM, Knowler WC, Gallart L, Hanson RL, Gruber JD, Ricart W, Vendrell J, Richart C, Tataranni PA, et al. The interleukin-6 (-174) G/C promoter polymorphism is associated with type-2 diabetes mellitus in Native Americans and Caucasians. Hum Genet. 2003;112(4):409–13.
Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, Woo P. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Investig. 1998;102(7):1369–76.
Larcombe LA, Orr PH, Lodge AM, Brown JS, Dembinski IJ, Milligan LC, Larcombe EA, Martin BD, Nickerson PW. Functional gene polymorphisms in canadian aboriginal populations with high rates of tuberculosis. J Infect Dis. 2008;198(8):1175–9.
Nadeem A, Mumtaz S, Naveed AK, Mansoor Q, Aslam M, Siddiqui A, Ismail M. Association of IL-6 C-174G (rs 1800795) single nucleotide polymorphism with type 2 diabetes mellitus in Pakistani population. J Pak Med Assoc. 2017;67(3):428–33.
Saxena M, Agrawal CG, Srivastava N, Banerjee M. Interleukin-6 (IL-6)-597 A/G (rs1800797) & -174 G/C (rs1800795) gene polymorphisms in type 2 diabetes. Indian J Med Res. 2014;140(1):60–8.
Dhamodharan U, Viswanathan V, Krishnamoorthy E, Rajaram R, Aravindhan V. Genetic association of IL-6, TNF-alpha and SDF-1 polymorphisms with serum cytokine levels in diabetic foot ulcer. Gene. 2015;565(1):62–7.
Fathy SA, Mohamed MR, Ali MAM, El-Helaly AE, Alattar AT. Influence of IL-6, IL-10, IFN-gamma and TNF-alpha genetic variants on susceptibility to diabetic kidney disease in type 2 diabetes mellitus patients. Biomarkers. 2019;24(1):43–55.
Cooper JD, Smyth DJ, Bailey R, Payne F, Downes K, Godfrey LM, Masters J, Zeitels LR, Vella A, Walker NM, et al. The candidate genes TAF5L, TCF7, PDCD1, IL6 and ICAM1 cannot be excluded from having effects in type 1 diabetes. BMC Med Genet. 2007;8:71.
Tsiavou A, Hatziagelaki E, Chaidaroglou A, Manginas A, Koniavitou K, Degiannis D, Raptis SA. TNF-alpha, TGF-beta1, IL-10, IL-6, gene polymorphisms in latent autoimmune diabetes of adults (LADA) and type 2 diabetes mellitus. J Clin Immunol. 2004;24(6):591–9.
Xu WD, Zhou M, Peng H, Pan HF, Ye DQ. Lack of association of IL-6 polymorphism with rheumatoid arthritis/type 1 diabetes: a meta-analysis. Joint Bone Spine. 2013;80(5):477–81.
Yin YW, Sun QQ, Zhang BB, Hu AM, Wang Q, Liu HL, Hou ZZ, Zeng YH, Xu RJ, Shi LB. The lack of association between interleukin-6 gene -174 G/C polymorphism and the risk of type 1 diabetes mellitus: a meta-analysis of 18,152 subjects. Gene. 2013;515(2):461–5.
Huth C, Heid IM, Vollmert C, Gieger C, Grallert H, Wolford JK, Langer B, Thorand B, Klopp N, Hamid YH, et al. IL6 gene promoter polymorphisms and type 2 diabetes: joint analysis of individual participants’ data from 21 studies. Diabetes. 2006;55(10):2915–21.
Xia J, Sun RL. Association between interleukin-6 rs1800795 polymorphism and the decreased risk of type 2 diabetes mellitus: an updated meta-analysis. Int J Clin Exp Med. 2019;12(1):86–97.
Campos LP, Graciolo V, Sousa MM, Martins BR, Souza SW, Alberton D, Picheth G, Rego FGM. Polymorphisms rs1800795 of interleukin-6 and rs2228145 of interleukin-6 receptor genes in Euro- Brazilians with adult-onset type 1 diabetes mellitus. Genet Mol Res. 2019;18(3):gmr18260.
Helaly MA, Hatata EZ, Elmagd MA, Ibrahem EF, Alsajd A, El-Aal IA, Settin A. Association of IL-10 and IL-6 gene polymorphisms with type 2 diabetes mellitus among Egyptian patients. Eur J Gen Med. 2013;10(3):158–62.
Chen XQ, Xu YC. Interleukin-6 gene polymorphism and its expression related to the susceptivity of type two Diabetes. Master's Degree Thesis. 2002.
Kong JH, Yu M. Study of association of interleukin-6 gene polymorphisms with type 2 diabetes mellitus. Master's Degree Thesis. 2010.
Bouhaha R, Baroudi T, Ennafaa H, Vaillant E, Abid H, Sassi R, Vatin V, Froguel P, Gaaied AB, Meyre D, et al. Study of TNFalpha -308G/A and IL6 -174G/C polymorphisms in type 2 diabetes and obesity risk in the Tunisian population. Clin Biochem. 2010;43(6):549–52.
Buraczynska M, Zukowski P, Drop B, Baranowicz-Gaszczyk I, Ksiazek A. Effect of G(-174)C polymorphism in interleukin-6 gene on cardiovascular disease in type 2 diabetes patients. Cytokine. 2016;79:7–11.
Danielsson P, Truedsson L, Eriksson KF, Norgren L. Inflammatory markers and IL-6 polymorphism in peripheral arterial disease with and without diabetes mellitus. Vasc Med (London, England). 2005;10(3):191–8.
Erdogan M, Kulaksizoglu M, Solmaz S, Berdeli A. The relationship of Interleukin-6 -174 G>C gene polymorphism in type 2 diabetic patients with and without diabetic foot ulcers in Turkish population. Foot (Edinb). 2017;30:27–31.
Eze IC, Imboden M, Kumar A, Adam M, von Eckardstein A, Stolz D, Gerbase MW, Kunzli N, Turk A, Schindler C, et al. A common functional variant on the pro-inflammatory Interleukin-6 gene may modify the association between long-term PM10 exposure and diabetes. Environ Health. 2016;15:39.
Ghavimi R, Sharifi M, Mohaghegh MA, Mohammadian H, Khadempar S, Rezaei H. Lack of association between rs1800795 (-174 G/C) polymorphism in the promoter region of interleukin-6 gene and susceptibility to type 2 diabetes in Isfahan population. Adv Biomed Res. 2016;5:18.
Hamid YH, Rose CS, Urhammer SA, Glumer C, Nolsoe R, Kristiansen OP, Mandrup-Poulsen T, Borch-Johnsen K, Jorgensen T, Hansen T, et al. Variations of the interleukin-6 promoter are associated with features of the metabolic syndrome in Caucasian Danes. Diabetologia. 2005;48(2):251–60.
Jahromi MM, Millward BA, Demaine AG. A polymorphism in the promoter region of the gene for interleukin-6 is associated with susceptibility to type 1 diabetes mellitus. J interferon Cytokine Res. 2000;20(10):885–8.
Javor J, Ferencik S, Bucova M, Stuchlikova M, Martinka E, Barak L, Strbova L, Grosse-Wilde H, Buc M. Polymorphisms in the genes encoding TGF-beta1, TNF-alpha, and IL-6 show association with type 1 diabetes mellitus in the Slovak population. Arch Immunol Ther Exp. 2010;58(5):385–93.
Karadeniz M, Erdogan M, Berdeli A, Yilmaz C. Association of interleukin-6 -174 G>C promoter polymorphism with increased risk of type 2 diabetes mellitus patients with diabetic nephropathy in Turkey. Genet Test Mol Biomarkers. 2014;18(1):62–5.
Kavitha L, Vijayshree Priyadharshini J, Sivapathasundharam B. Association among interleukin-6 gene polymorphisms, type 2 diabetes mellitus, and chronic periodontitis: a pilot study. J Invest Clin Dent. 2017;8(3): e12230.
Lara-Gomez RE, Moreno-Cortes ML, Muniz-Salazar R, Zenteno-Cuevas R. Association of polymorphisms at -174 in IL-6, and -308 and -238 in TNF-alpha, in the development of tuberculosis and type 2 diabetes mellitus in the Mexican population. Gene. 2019;702:1–7.
Mohlig M, Boeing H, Spranger J, Osterhoff M, Kroke A, Fisher E, Bergmann MM, Ristow M, Hoffmann K, Pfeiffer AF. Body mass index and C-174G interleukin-6 promoter polymorphism interact in predicting type 2 diabetes. J Clin Endocrinol Metab. 2004;89(4):1885–90.
Mukhopadhyaya PN, Acharya A, Chavan Y, Purohit SS, Mutha A. Metagenomic study of single-nucleotide polymorphism within candidate genes associated with type 2 diabetes in an Indian population. Genetics Mol Res. 2010;9(4):2060–8.
Mysliwiec M, Mysliwska J, Zorena K, Balcerska A, Malinowska E, Wisniewski P. Interleukin 6–174(G>C) gene polymorphism is related to celiac disease and autoimmune thyroiditis coincidence in diabetes type 1 children. Diabetes Res Clin Pract. 2008;82(1):108–12.
Mysliwska J, Zorena K, Mysliwiec M, Malinowska E, Raczynska K, Balcerska A. The -174GG interleukin-6 genotype is protective from retinopathy and nephropathy in juvenile onset type 1 diabetes mellitus. Pediatr Res. 2009;66(3):341–5.
Neelofar K, Ahmad J, Ahmad A, Alam K. Study of IL4-590C/T and IL6-174G/C gene polymorphisms in type 2 diabetic patients with chronic kidney disease in North Indian population. J Cell Biochem. 2017;118(7):1803–9.
Perez-Bravo F, Soto MF, Lopez AP, Eyzaguirre CF, Codner E. -174 G/C polymorphism of interleukin 6 gene in women with type 1 diabetes. Rev Med Chil. 2011;139(2):158–64.
Plataki MN, Zervou MI, Samonis G, Daraki V, Goulielmos GN, Kofteridis DP. Association of the interleukin-6 rs1800795 polymorphism with type 2 diabetes mellitus in the population of the Island of Crete, Greece. Genet Test Mol Biomarkers. 2018;22(7):448–52.
Settin A, Ismail A, El-Magd MA, El-Baz R, Kazamel A. Gene polymorphisms of TNF-alpha-308 (G/A), IL-10(-1082) (G/A), IL-6(-174) (G/C) and IL-1Ra (VNTR) in Egyptian cases with type 1 diabetes mellitus. Autoimmunity. 2009;42(1):50–5.
Siekiera U, Jarosz-Chobot P, Janusz J, Koehler B. Polymorphism of TNF-alpha (308 A/G), IL-10 (1082 A/G, 819 C/T 592 A/C), IL-6 (174 G/C), and IFN-gamma (874 A/T); genetically conditioned cytokine synthesis level in children with diabetes type 1. Endokrynologia, diabetologia i choroby przemiany materii wieku rozwojowego : organ Polskiego Towarzystwa Endokrynologow Dzieciecych. 2002;8(1):29–34.
Ururahy MA, de Souza KS, Oliveira YM, Loureiro MB, da Silva HP, Freire-Neto FP, Bezerra JF, Luchessi AD, Doi SQ, Hirata RD, et al. Association of polymorphisms in IL6 gene promoter region with type 1 diabetes and increased albumin-to-creatinine ratio. Diabetes Metab Res Rev. 2015;31(5):500–6.
Xiao LM, Yan YX, Xie CJ, Fan WH, Xuan DY, Wang CX, Chen L, Sun SY, Xie BY, Zhang JC. Association among interleukin-6 gene polymorphism, diabetes and periodontitis in a Chinese population. Oral Dis. 2009;15(8):547–53.
Zhang X, Ma L, Peng F, Wu Y, Chen Y, Yu L, Lei Z, Zhang C. The endothelial dysfunction in patients with type 2 diabetes mellitus is associated with IL-6 gene promoter polymorphism in Chinese population. Endocrine. 2011;40(1):124–9.
Ayelign B, Negash M, Andualem H, Wondemagegn T, Kassa E, Shibabaw T, Akalu Y, Molla MD. Association of IL-10 (- 1082 A/G) and IL-6 (- 174 G/C) gene polymorphism with type 2 diabetes mellitus in Ethiopia population. BMC Endocr Disord. 2021;21(1):70.
Cirelli T, Nepomuceno R, Rios ACS, Orrico SRP, Cirelli JA, Theodoro LH, Barros SP, Scarel-Caminaga RM. Genetic polymorphisms in the Interleukins IL1B, IL4, and IL6 are associated with concomitant periodontitis and type 2 diabetes mellitus in Brazilian patients. J Periodontal Res. 2020;55(6):918–30.
da Silva CB, Vieira DA, de Melo LF, Chagas AL, Gomes AD, de Faria Jr CL, Teixeira R, de Magalhães Queiroz DM, Rocha GA, Soares MM, et al. Interleukin-6-174G/C polymorphism is associated with a decreased risk of type 2 diabetes in patients with chronic hepatitis C virus. World J Hepatol. 2020;12(4):137–48.
Haghnazari L, Sabzi R. Relationship between TP53 and interleukin-6 gene variants and the risk of types 1 and 2 diabetes mellitus development in the Kermanshah province. J Med Life. 2021;14(1):37–44.
Hameed I, Masoodi SR, Malik PA, Mir SA, Ghazanfar K, Ganai BA. Genetic variations in key inflammatory cytokines exacerbates the risk of diabetic nephropathy by influencing the gene expression. Gene. 2018;661:51–9.
Martínez-Ramírez OC, Salazar-Piña DA, de Lorena RM, Castro-Hernández C, Casas-Ávila L, Portillo-Jacobo JA, Rubio J. Association of NFκβ, TNFα, IL-6, IL-1β, and LPL polymorphisms with type 2 diabetes mellitus and biochemical parameters in a Mexican Population. Biochem Genet. 2021;59(4):940–65.
Rodrigues KF, Pietrani NT, Bosco AA, Campos FMF, Sandrim VC, Gomes KB. IL-6, TNF-α, and IL-10 levels/polymorphisms and their association with type 2 diabetes mellitus and obesity in Brazilian individuals. Arch Endocrinol Metab. 2017;61(5):438–46.
Saxena M, Srivastava N, Banerjee M. Cytokine gene variants as predictors of type 2 diabetes mellitus. Curr Diabetes Rev. 2018;14(3):307–19.
Lin Z, Sun Y, Xue H, Chen L, Yan C, Panayi AC, Mi B, Liu G. The effectiveness and safety of LMWH for preventing thrombosis in patients with spinal cord injury: a meta-analysis. J Orthop Surg Res. 2021;16(1):262.
Melisse B, de Beurs E, van Furth EF. Eating disorders in the Arab world: a literature review. J Eat Disord. 2020;8(1):59.
Shu X, Wu G, Zhang Y, Wang Y, Zheng Y, Guo Q, Ji R, Zhou Y. Diagnostic value of linked color imaging based on endoscopy for gastric intestinal metaplasia: a systematic review and meta-analysis. Ann Transl Med. 2021;9(6):506.
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–88.
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–48.
Hayashino Y, Noguchi Y, Fukui T. Systematic evaluation and comparison of statistical tests for publication bias. J Epidemiol. 2005;15(6):235–43.
Napolioni V. The relevance of checking population allele frequencies and Hardy-Weinberg Equilibrium in genetic association studies: the case of SLC6A4 5-HTTLPR polymorphism in a Chinese Han Irritable Bowel Syndrome association study. Immunol Lett. 2014;162(1):276–8.
(IDF). IDF: IDF Diabetes Altas, 8th ed. International Diabetes Federation 2017, Brussels, Belgium. ISBN 9782930229874.
Mi YY, Yu QQ, Xu B, Zhang LF, Min ZC, Hua LX, Feng NH, Yao Y. Interferon gamma +874 T/A polymorphism contributes to cancer susceptibility: a meta-analysis based on 17 case-control studies. Mol Biol Rep. 2011;38(7):4461–7.
Mi YY, Yu QQ, Yu ML, Xu B, Zhang LF, Cheng W, Zhang W, Hua LX, Feng NH. Review and pooled analysis of studies on -607(C/A) and -137(G/C) polymorphisms in IL-18 and cancer risk. Med Oncol (Northwood, London, England). 2011;28(4):1107–15.
Qi L, Rifai N, Hu FB. Interleukin-6 receptor gene, plasma C-reactive protein, and diabetes risk in women. Diabetes. 2009;58(1):275–8.
Silva BRD, Cirelli T, Nepomuceno R, Theodoro LH, Orrico SRP, Cirelli JA, Barros SP, Scarel-Caminaga RM. Functional haplotype in the Interleukin8 (CXCL8) gene is associated with type 2 diabetes mellitus and periodontitis in Brazilian population. Diabetes Metab Syndr. 2020;14(6):1665–72.
Jiao J, Wang Z, Guo Y, Liu J, Huang X, Ni X, Gao D, Sun L, Zhu X, Zhou Q, et al. Association between IL-1B (-511)/IL-1RN (VNTR) polymorphisms and type 2 diabetes: a systematic review and meta-analysis. PeerJ. 2021;9: e12384.
Trapali M, Houhoula D, Batrinou A, Kanellou A, Strati IF, Siatelis A, Halvatsiotis P. Association of TNF-α 308G/A and LEPR Gln223Arg polymorphisms with the risk of type 2 diabetes mellitus. Genes. 2021;13(1):59.
Acknowledgements
Not applicable.
Funding
None.
Author information
Authors and Affiliations
Contributions
CZ conceived of the study, CZ prepared the data, ZC were involved in the data analyses, ZC drafted the original manuscript. ZC and MY prepared the figures. All the authors agreed to the submission of the present work. All the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors proclaim that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
PRISMA 2019 checklist.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Cheng, Z., Zhang, C. & Mi, Y. IL-6 gene rs1800795 polymorphism and diabetes mellitus: a comprehensive analysis involving 42,150 participants from a meta-analysis. Diabetol Metab Syndr 14, 95 (2022). https://doi.org/10.1186/s13098-022-00851-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13098-022-00851-8