Abstract
Background
Wolbachia bacteria are widely distributed throughout terrestrial arthropod species. These bacteria can manipulate reproduction and influence the vector competence of their hosts. Recently, Wolbachia have been integrated into vector control programmes for mosquito management. A number of supergroups and strains exist for Wolbachia, and they have yet to be characterized for many mosquito species. In this study, we examined Wolbachia prevalence and their phylogenetic relationship to other Wolbachia, using mosquitoes collected in Merced County in the Central Valley of California.
Methods
Adult mosquitoes were collected from 85 sites in Merced County, California in 2017 and 2018. Traditional and quantitative PCR were used to investigate the presence or absence and the density of Wolbachia, using Wolbachia-specific 16S rRNA and Wolbachia-surface protein (wsp) genes. The supergroup of Wolbachia was determined, and Multilocus Sequence Typing (MLST) by sequencing five housekeeping genes (coxA, gatB, ftsZ, hcpA and fbpA) was also used to determine Wolbachia supergroup as well as strain.
Results
Over 7100 mosquitoes of 12 species were collected: Aedes melanimon, Ae. nigromaculis, Ae. vexans, Ae. aegypti, Culex pipiens, Cx. stigmatosoma, Cx. tarsalis, Anopheles franciscanus, An. freeborni, An. punctipennis, Culiseta incidens and Cs. inornata. Eight showed evidence of Wolbachia. To our knowledge, this study is the first to report detection of Wolbachia in five of these species (Ae. melanimon, Cx. stigmatosoma, Cx. tarsalis, Cs. incidens and Cs. inornata). Culex pipiens and Cx. stigmatosoma had a high frequency and density of Wolbachia infection, which grouped into supergroup B; Cs. inornata clustered with supergroup A. MLST comparisons identified Cx. pipiens and Cx. stigmatosoma as wPip strain type 9 supergroup B. Six species had moderate to low (< 14%) frequencies of Wolbachia. Four species were negative, Ae. nigromaculis, An. franciscanus, An. freeborni and Ae. aegypti.
Conclusions
New records of Wolbachia detection were found in mosquitoes from Merced County, California. Culex stigmatosoma and Cs. inornata were new records for Wolbachia supergroup B and A, respectively. Other species with Wolbachia occurred with low frequency and low density. Detection of Wolbachia in mosquitoes can be used to inform potential vector control applications. Future study of Wolbachia within Cx. stigmatosoma and Cs. inornata in California and through the range of these species could further explore Wolbachia infection in these two species.

Similar content being viewed by others
Background
Wolbachia pipientis are a monophyletic group of obligate intracellular bacteria that belong to the order Rickettsiales. These endosymbionts were first discovered in the Culex pipiens mosquito [1, 2], and are now estimated to infect between 40–52% of arthropod species [3, 4]. Wolbachia routinely infect their host’s reproductive tissues, and they are capable of surviving in a variety of invertebrates [5,6,7]. Wolbachia are known to be transmitted vertically through maternal inheritance and have also been shown to transmit horizontally between species, genera, and orders [8,9,10,11]. Wolbachia infections can have a diverse range of effects depending on the host species, from commensal, mutualistic, to parasitic interactions [5].
In recent years, Wolbachia have been implemented for population control of vector species [12, 13]. This is largely a result of the reproductive alterations that Wolbachia induce within their hosts in a strain-specific manner [5, 14]. Such reproductive manipulations include termination of male offspring, feminization of genetic males, parthenogenesis, and cytoplasmic incompatibility [5, 15]. Cytoplasmic incompatibility is the only known phenotype to be expressed within mosquito species [16]; when infected males mate with uninfected females, viable offspring are not produced. Furthermore, Wolbachia have been shown to modulate host fitness and vector potential. For instance, studies have shown a protective effect of Wolbachia against infection with pathogenic RNA viruses [12, 14, 17,18,19,20,21]. In addition, Wolbachia-infections have shown other complex host-specific manipulations: they can have increased or decreased rates of reproductive phenotypes; reduced host life-span and egg viability [6, 22]; impact larval survival [23]; decreased female mosquito biting ability [24]; decreased relative abundance of resident bacteria [25]; and in some cases, increased viral susceptibility and host mortality [26, 27]. Artificial infection of this endosymbiont into arthropod vectors has been shown to impact transmission of vector-borne diseases including lymphatic filariasis, West Nile virus, chikungunya, dengue, Zika, and avian malaria [14, 19, 20, 26].
Wolbachia offers a potential effective alternative to traditional chemical pesticide applications for the control of disease vectors, for example through cytoplasmic incompatibility. Cytoplasmic incompatibility was first proposed as a method of biological control for Culex pipiens fatigans in 1967, although initially it was not attributed to Wolbachia [28]. Since then, the use of Wolbachia-mediated incompatible technique strategies have been studied for pest control of a number of insects including Aedes aegypti, Ae. albopictus, Ae. polynesiensis, Ceratitits capitata, Rhagoletis cerasi, Glossina morsitans, Culex pipiens and Cx. quinquefasciatus [28,29,30,31,32,33,34]. This method of control aims to reduce vector populations through the introduction of Wolbachia-infected ‘sterile’ males, which compete with uninfected males for mates at the release site. Aedes aegypti do not naturally harbor Wolbachia; when Wolbachia have been detected in Ae. aegypti [35,36,37,38,39], the range of detected strain types suggest they may be due to environmental contamination [40].
Naturally uninfected arthropod species like Ae. aegypti can be amenable to Wolbachia-infection through microinjection of the endosymbiont from another insect species into developing embryos [41, 42]. Currently, eight novel strains (wAlbA, wAlbB, wAu, wMel, wMelCS, wMelPop-CLA, wPip and wRi) have been transinfected into Ae. aegypti to be evaluated for vector control applications [42,43,44,45,46]. Aedes aegypti is widespread in tropical and subtropical regions globally [47, 48], and since its detection in California in 2011 it has become widespread in southern California and the Central Valley [49, 50]. One example of where Wolbachia-infected Ae. aegypti males have been used to reduce mosquito populations through cytoplasmic incompatibility is through the DeBug Fresno California programme, which released male Ae. aegypti with the wAlbB strain of Wolbachia to reduce local Ae. aegypti populations [34]. A second method of using Wolbachia-infections for mosquito control relies on the introduction of Wolbachia-infected male and female mosquitoes to replace uninfected mosquito populations [12, 51, 52]. Aedes aegypti populations (each with a unique Wolbachia strain, wMel or wAlbB), have been introduced into regions of Australia and Malaysia, respectively [12, 52, 53]. Both strains were shown to reduce the incidence of dengue virus infections [53, 54].
Each Wolbachia strain has particular biological characteristics when moved into another vector, and identification of strains is key. Supergroups are used to differentiate major phylogenetic subdivisions within Wolbachia pipientis [55]. The 16S rRNA gene and the Wolbachia-surface protein (wsp) have been used to characterize supergroups [55, 56]. Within supergroups, Wolbachia strains are identified and can be characterized by multilocus sequence typing (MLST) which relies on five conserved bacterial housekeeping genes (gatB, coxA, hcpA, ftsZ and fbpA). Strains are commonly characterized based on the host species in which they are first identified [55, 57, 58]. For instance, wPip is the strain of Wolbachia which was identified from the Culex pipiens mosquito species. Mosquitoes can be singly or superinfected with more than one Wolbachia strain, or infected with multiple variants of the same strain [31].
Merced County is located in the Central Valley of California and includes a diverse range of habitats and mosquito species. While previous studies have identified the presence or absence of Wolbachia within some mosquito species throughout California using traditional PCR [58], the current infection status for species in the Central Valley of California and Merced County is unknown. Wolbachia-infected mosquitoes as a method of mosquito control has great potential globally, and this vector-control method continues to be developed and refined.
The objectives of this study were to determine the presence or absence of Wolbachia in twelve mosquito species collected throughout Merced County, and to characterize the Wolbachia supergroup and strain for species with detections. Our study expands current knowledge of Wolbachia presence in mosquitoes in Merced and in the Central Valley of California, and would aid in the design of future Wolbachia-based mosquito control applications.
Methods
Mosquito collections
Adult mosquitoes were collected weekly from June to September, in both 2017 and 2018, using Encephalitis Vector Survey (EVS) traps (Bioquip, Rancho Dominguez, CA, USA) in Merced County. Traps sites were selected for habitats known to harbor the different species of mosquitoes. The EVS traps contained (1–2 kg) of dry ice (carbon dioxide) per container as an attractant for host-seeking female mosquitoes. Traps were hung on trees or fences in close proximity to a water source. The GPS coordinates of the site were recorded using a Garmin etrex High Sensitivity GPS unit (Garmin Ltd., Olathe, KS, USA). Traps were placed during the early afternoon and retrieved the following morning. Samples were transported on ice to a – 20 °C laboratory freezer. Adults were identified on a cold plate using a taxonomic key specific to Californian mosquitoes [59] and stored in 1.5 ml Eppendorf tubes until DNA extraction.
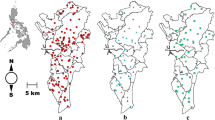
Aedes aegypti larvae were collected in addition to adults from several sites in Merced, California. Larvae were collected from water sources, transported to the lab, and reared at laboratory temperature in a BugDorm (MegaView Science, Taichung, Taiwan). Emerged adults were stored at – 20 °C and identification was confirmed using a taxonomic key. A map of trapping locations was constructed for 2017 and 2018 using qGIS v3.8.3-Zanzibar [60]. The Census TIGER/Line file for Merced County, California was retrieved (www.census.gov/cgi-bin/geo/shapefiles2010/main), and site location GPS coordinates were overlaid on the county map.
DNA extraction
The whole body of the mosquito was used for individual extractions of genomic DNA using the Qiagen DNeasy Blood and Tissue Kit (Qiagen Inc., Valencia, CA, USA), following the manufacturer’s protocol for tissue extraction with a 2 h incubation at 65 °C [61]. Extracted samples were stored at – 20 °C. The DNA quantity was measured using the Qubit® dsDNA HS Assay kit (Life Technologies, Carlsbad, CA, USA). The quantity of DNA in the samples averaged 10–15 ng/µl.
Screening samples for Wolbachia and relative Wolbachia density determination
Presence or absence of Wolbachia was determined by amplicon detection of the Wolbachia-specific 16S rRNA gene and the general Wolbachia surface protein (wsp) via qPCR in individual field-collected mosquitoes. For each mosquito species collected, a subset of individuals was screened for Wolbachia, and individuals tested came from multiple sites or collection dates (Additional file 1: Table S1). The primer combinations for the Wolbachia-16S rRNA gene and Wolbachia-surface-protein (wsp) used in our assays are detailed on Table 1. The qPCR cycling conditions were those recommended for the master mix and consisted of holding at 95 °C for 10 min and 40 cycles of 15 s at 95 °C and 1 min at 60 °C. A melt curve stage at the end of the reaction was included. Each sample was analyzed in duplicates (technical replicates), and a non-template control was included. The qPCR assays were run on Applied Biosystems 5700 Fast Real-time PCR (Applied Biosystems, Foster City, CA, USA).
The relative Wolbachia density was determined via qPCR for two species, Culex pipiens and Culex stigmatosoma. Relative density was determined by measuring the signal amplifications of the Wolbachia 16S rRNA or wsp gene and the respective reference gene for each mosquito species. The RpS3 gene was used as a reference gene and primers specific for this location (Table 1) were employed to compare Wolbachia densities in the collected samples. The RpS3 gene is known to be a single copy gene in mosquitoes [62] and is described to be highly conserved [63]. Culex pipiens is known to be naturally infected with Wolbachia was used as a control to compare the relative Wolbachia density to Cx. stigmatosoma. Samples were compared and the data was analyzed post-run using the \(\mathrm{\Delta \Delta }\) Ct method [64]. Data were evaluated using the GraphPad Prism 8.4.2 statistical software, comparing the two species using Studentʼs t-test.
Determination of Wolbachia supergroups
A subset of samples that screened positive for Wolbachia by qPCR were used to characterize the Wolbachia supergroup. Samples were run using the Wolbachia wsp supergroup A and wsp supergroup B primers [65] (Table 1). Polymerase chain reaction (PCR) was performed using a mixture of 2 µl of DNA, 1 µl of each forward and reverse primer at 10 µM concentration, 1 µl of Taq polymerase, 5 µl of buffer, 1 µl of dNTPs (2.5 µM) (Takara-Clonetech Bio, Mountain View, CA, USA) and 40 µl of sterile water to make the reaction volume of 51 µl. The temperature profile for wsp amplification was the following: initial denaturation for 3 min at 95 °C, followed by 35 cycles of 1 min at 94 °C, 1 min at 55 °C, and 1 min at 72 °C, a final elongation of 10 min at 72 °C and a final hold at 4 °C, modified from the protocol in Zhou et al. [65]. Amplification was confirmed by visualizing products on an agarose gel. Products were purified using USB Exo-sap-it® (Affymetrix Inc., Santa Clara, CA, USA) PCR cleanup kit. Each forward and reverse sequence reaction was prepared using 1 µM primers, 2 µl purified water, and 10 µl purified PCR product per reaction and sequenced on an Applied Biosystems 3730xl DNA Analyzer at the UC Berkeley DNA Sequencing Facility. Multilocus sequence typing (MLST) was also used to characterize supergroups (described below).
Strain characterization by multilocus sequence typing (MLST)
Species with samples which were successfully sequenced for supergroup A or B were also sequenced by multilocus sequence typing (MLST) using the standard primers for five ubiquitous bacterial housekeeping genes: coxA, gatB, ftsZ, fbpA and hcpA [57] (Table 1). The PCR mix for each gene used a mixture of 2 µl of DNA, 1 µl of each forward and reverse primer at 10 µM concentration, 1 µl of Taq polymerase, 5 µl of buffer, 1 µl of dNTPs (2.5 µM) (Takara-Clonetech Bio Inc., Mountain View, CA, USA) and 40 µl of sterile water to make the reaction volume of 51 µl. The PCR temperature profile for four of the genes (coxA, gatB, ftsZ and hcpA) was the following: initial denaturation for 2 min at 94 °C, followed by 37 cycles of 30 s at 94 °C, 45 s at 54 °C, and 1.5 min at 72 °C, a final elongation for 10 min at 72 °C and a final hold at 4 °C [57]; the PCR program for the fbpA gene was identical except the annealing was for 45 s at 59 °C. PCR amplification was visually confirmed on agarose gels, products purified by USB Exo-sap-it®, and sequencing reactions were similar to those previously described.
Sequence analysis
Wolbachia surface protein (wsp) and the MLST genes (coxA, gatB, ftsZ, hcpA and fbpA) sequence files were viewed, edited, and aligned in Geneious Prime 2020.05. Consensus sequences were generated and exported as FASTA files. Consensus sequences were queried using the BLASTn program to find sequences with the highest similarity.
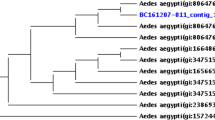
Wolbachia supergroup sequences from this study were combined with high similarity sequences from GenBank and others from a study by Carvajal et al. [38] to produce a Neighbor-Joining tree. Included in the tree were consensus sequences of 18 samples from this study [Cx. pipiens (n = 8) Culex stigmatosoma (n = 9) and Culiseta inornata (n = 1)] and an additional 15 wsp sequence files from GenBank which represented 11 genera previously confirmed with detections of Wolbachia. The species selected for comparison were Aedes albopictus (AF020058, AF020059), Brugia malayi (AJ252061), Culex pipiens (AF020061), Culex quinquefasciatus (AF020060), Dirofilaria imitis (AJ252062), Drosophila melanogaster (AF020072), Drosophila simulans (AF020070), Glossina austeni (AF020077), Glossina morsitans (AF020079), Muscidifurax uniraptor (AF020071) and Phlebotomus papatasi (AF020082) [38], and three additional sequences (Loxoblemmus sp. MG97910, Myrmecophilus sp. MK995471 and Cerapachys augustae KC137155) of high similarity. These 33 sequences were subjected to multiple sequence alignment using the ClustalW algorithm in MEGA 7.0. The Gamma distributed, Tamura 3-parameter substitution model was chosen based on the lowest Bayesian information criterion. A Neighbor-Joining tree was constructed using 1000 bootstraps in MEGA 7.0 [66].
Wolbachia strains were characterized by concatenating the coxA, gatB, ftsZ, hcpA and fbpA gene sequences from each sample in Geneious. Following concatenation, each sequence was exported in FASTA format and queried against the Wolbachia MLST database (https://pubmlst.org/Wolbachia/) to determine allelic profiles [57, 67]. An exact match with the queried database was necessary to distinguish profile composition. All sequences were submitted to Genbank.
Results
Mosquito collections, identification and abundance
In total, 12 mosquito species from 4 genera were collected from 85 sites within Merced county in 2017 and 2018 (Table 2, Additional file 1: Table S1). There was a total of 7150 mosquitoes identified to species. The species collected were the following: Aedes melanimon, Aedes vexans, Aedes nigromaculis, Aedes aegypti, Culex stigmatosoma, Culex pipiens, Culex tarsalis, Anopheles franciscanus, Anopheles freeborni, Anopheles punctipennis, Culiseta incidens, Culiseta inornata (Table 2). These species represent the diversity of nearly every mosquito from the region where trapping occurred [59]. The 85 trap sites were in the vicinity of 8 cities within Merced county: Atwater, Ballico, Hilmar, Le Grand, Los Banos, Merced, Snelling and Winton (Fig. 1, Table 2). Each mosquito species was trapped from two to five different regions of the county (Table 2, Additional file 1: Table S1), to provide geographic diversity in samples which were tested. Some mosquito species were more abundant than others. For example, Cx. pipiens and Cx. tarsalis were trapped in cities as well as in rural sites (Additional file 1: Table S1). Ae. melanimon and Ae. vexans adults were most abundant within rural wetland habitats. Aedes aegypti was found in several Merced neighborhoods and near the Merced Zoo. Anopheles franciscanus, An. freeborni and An. punctipennis were found at rural riparian sites. Culex stigmatosoma were numerous at a semi-natural rural site near dairy runoff. Aedes nigromaculis, Cs. incidens and Cs. inornata were collected from rural and residential properties.
Wolbachia screening with qPCR
For each species, 30–50 mosquitoes were typically screened for the presence or absence of Wolbachia, except for a few species which had smaller numbers of individuals collected (Table 2). A total of 406 mosquitoes were screened for Wolbachia prevalence using qPCR, and all mosquitoes screened were females. Wolbachia was detected within 73 of the 406 samples tested, and sites with mosquitoes positive for Wolbachia were found throughout the county (Table 2, Additional file 1: Table S1). Eight species within four genera tested positive for Wolbachia (Table 2). The frequency and percent of samples positive for each species from highest to lowest was the following: Cx. stigmatosoma (30/34; 88.2%), Cx. pipiens (31/37; 83.8%), Cs. inornata (1/7; 14.3%), Ae. melanimon (6/55: 10.9%), An. punctipennis (1/19: 5.3%), Cx. tarsalis (1/26: 3.9%), Ae. vexans (2/52; 3.9%) and Cs. incidens (1/42; 2.4%) (Table 2). Species where no Wolbachia was detected were An. freeborni, An. franciscanus, Ae. nigromaculis and Ae. aegypti (Table 2).
Each species was screened by qPCR for Wolbachia with two primers. For Cx. pipiens and Cx. stigmatosoma, all individuals were positive for Wolbachia when tested with both genes (16S rRNA and wsp) (Table 2). In a few cases, one primer would detect Wolbachia, while another would not (Table 2). For Ae. melanimon, Cx. tarsalis, Cs, incidens, Cs. inornata, An. punctipennis and Ae. vexans, Wolbachia was detected in very few individuals (Table 2). For Cs. incidens and Cs. inornata, both primers detected only one positive individual (Table 2). Six individuals were positive detections with the 16S rRNA primer set but were negative with wsp (one An. punctipennis, one Cx. tarsalis, two Ae. vexans and two Ae. melanimon). Only one sample was negative with 16S rRNA but positive for wsp (Ae. melanimon) (Table 2, Additional file 1: Table S1).
To evaluate the relative Wolbachia density of the two Culex spp., we conducted a relative comparison using qPCR for 30 individuals each of Culex stigmatosoma and Cx. pipiens, the later which was used as a control. The relative Wolbachia density comparison indicated no significant difference between the two species (16S, t-test, t = 0.80, df = 48, P = 0.43; wsp, t-test, t = − 1.34, df = 48, P = 0.18).
Wolbachia supergroup identification
Wolbachia supergroup identification was carried out by PCR of samples using general wsp supergroup A and supergroup B primers. A total of 18 Wolbachia surface protein sequences were generated from three species, Cx. pipiens (n = 8), Cx. stigmatosoma (n = 9) and Cs. inornata (n = 1). Wsp sequences were not successfully obtained from the other species with low frequency Wolbachia detections (Table 2). The sequences produced in this study were combined with an additional 15 wsp sequences from GenBank for supergroup comparison (described above). The Cx. pipiens and Cx. stigmatosoma individuals grouped with the reference supergroup B samples, and Cs. inornata grouped with supergroup A reference samples (Fig. 2).
Neighbor-joining tree for Cx. pipiens, Cx. stigmatosoma and Cs. inornata (this study) combined with 15 Wolbachia-surface-protein (wsp) supergroup A and B sequences from GenBank and Carvajal et al. [38]. Sequences from this study are shown in red. The tree was based on the Tamara 3-parameter, Gamma distributed model with 1000 bootstrap replications
Wolbachia strain characterization
There were five individual Cx. pipiens which had 5 MLST genes (coxA, gatB, ftsZ, hcpA and fbpA) successfully sequenced (Cx. pipiens nos. 29, 31, 32, 34 and 35) and they were matches with strain type 9, wPip supergroup B Wolbachia in the MLST database (Table 3). Four additional Cx. pipiens were similar at 3 or 4 of the five gene sequences to strain type 9 wPip; however, these had a low quality hcpA sequences and exact match of that allele could not be confirmed.
For Cx. stigmatosoma, two samples had complete gene sequences for the five MLST genes (coxA, gatB, ftsZ, hcpA and fbpA); the allelic profile for Cx. stigmatosoma samples 10 and 15 from Ballico were a match for the five sequences retrieved from several Cx. pipiens samples (nos. 29, 31, 32, and 35), and these were characterized as Wolbachia wPip supergroup B strain-type 9 (Table 3). The two Cx. stigmatosoma individuals had four of the five MLST genes sequenced and also matched strain type 9, but only partial sequences were obtained for the hcpA gene. The hcpA locus has been observed with variable sequence lengths, ranging from 435 to 477 bp (pubmlst.org/Wolbachia). Five additional samples (Cx. stigmatosoma nos. 16,17, 20, 25, 38) were sequenced at 3 or 4 of the 5 genes, which also had matching profiles to Wolbachia housekeeping genes (coxA, ftsZ and fbpA) from this study.
One Cs. inornata sample had a detection of Wolbachia with 16S rRNA gene, and this individual was used to generate sequence data for the five MLST genes. The one Cs. inornata had four sequences (fbpA, gatB, coxA, ftsz) which had mlst allele matches; these sequences matched fbpA allele 277, gatB 312, coxA 236, and ftsz 154, while hcpA had no match [67]. For Cs. inornata, the wsp sequence grouped with others in supergroup A.
Discussion
This study screened 12 field-collected mosquito species in the Central Valley of California for the presence or absence of Wolbachia, and for species with Wolbachia detections, attempted to characterize the supergroup and strain type. The 12 mosquito species identified and screened were the following: Ae. melanimon, Ae. nigromaculis, Ae. vexans, Ae. aegypti, Cx. pipiens, Cx. stigmatosoma, Cx. tarsalis, An. franciscanus, An. freeborni, An. punctipennis, Cs. incidens and Cs. inornata. Wolbachia was detected in eight of the mosquito species. To our knowledge, this study is the first to report Wolbachia detection in five of these species (Ae. melanimon, Cx. stigmatosoma, Cx. tarsalis, Cs. incidens and Cs. inornata), while three species which were positive in this study have been previously reported in the literature (Ae. vexans, Cx. pipiens and An. punctipennis). The Wolbachia supergroup was determined for two of these new records (Cx. stigmatosoma and Cs. inornata), and the strain was characterized for Cx. stigmatosoma using MLST. The other species with detections of Wolbachia had a very low prevalence (frequency) and could not be sequenced.
The two mosquito species which were positive for Wolbachia at high frequencies (prevalence) were Cx. pipiens and Cx. stigmatosoma. The other six species showed detections of Wolbachia at low prevalence (< 13%). Furthermore, when the relative Wolbachia density was compared between Cx. pipiens and Cx. stigmatosoma, there was no statistical difference indicating that these two species potentially hold similar Wolbachia densities. Further assessment via absolute quantification of Wolbachia would further confirm this finding. In addition, future work with Cx. stigmatosoma could investigate maternal transmission to provide supporting evidence for Wolbachia infection. The inability to sequence Wolbachia in the species with low Wolbachia prevalence could be due to a low Wolbachia density. One species, Cs. inornata, had a low Wolbachia prevalence (13%), yet the wspA sequence was obtained which allowed it to be tentatively classified into supergroup A. Four of five MLST genes were sequenced for Cs. inornata in this study. This Wolbachia isolate may potentially represent a new Wolbachia strain, but further research would be needed with additional samples collected to replicate detection of Wolbachia.
Wolbachia infections were previously reported in Cx. pipiens [58, 68], An. punctipennis [69], and Ae vexans [70]. Although Wolbachia has been previously detected in An. punctipennis and Ae. vexans, currently there is no description of a strain type for these mosquitoes. Our study did not detect Wolbachia in several mosquito species including An. freeborni, An. franciscanus, Ae. nigromaculis and Ae. aegypti. Although a few studies have indicated Wolbachia detection in Ae. aegypti [36,37,38,39], others found absence of infection in this species [40, 71] and suggest that the variability of strains found in previous studies on Ae. aegypti may indicate environmental contamination rather than a true Wolbachia infection. Ross et al. [40] recommend that to confirm Wolbachia infection, experiments should be run to demonstrate maternal transmission or to visualize Wolbachia in the mosquito using a method such as florescent in situ hybridization (FISH), in addition to determining sequences. Culex pipiens is well known for its infection with Wolbachia, as Wolbachia pipientis was first described from this mosquito species [1, 2]. Previous research identified wPip supergroup B infections in the Cx. pipiens species complex in five California populations [58]. Since then more than 60 wPip haplotypes have been identified [31, 72]. Our study screened Cx. pipiens from four sites and found individuals from all sites carrying Wolbachia. In the present study, the MLST results for Cx. pipiens found strain type 9 supergroup B among samples with complete allelic profile data. These were all acquired from the Ballico collection site. Isolates of strain type 9 have been documented in Cx. pipiens and Cx. quinquefasciatus [67]. Other studies have found Cx. pipiens with strain type 9 in Placer County, California and Tompkins County, New York; while Wolbachia-infected Cx. quinquefasciatus were found in Hawaii, Midway and Kenya [57, 73].
Interestingly, our study also found a new Wolbachia detection record for Cx. stigmatosoma. This species is highly ornithophilic [74] and often found in urban residential areas and near farms. It prefers foul water sources like street drains and dairy lagoons for oviposition [75]. These types of habitat are similar to those where Cx. pipiens can also be found. This species is known to occur throughout the western USA to Mexico, Central America and northern South America [75, 76]. Culex stigmatosoma is a competent vector of West Nile virus, and is capable of transmitting St Louis encephalitis and avian malaria [76, 77].
Culex stigmatosoma had MLST sequences produced from two different collection sites. One site was a rural semi-natural habitat near a dairy (Ballico), and another was a rural farm in Atwater. At the first site, Cx. stigmatosoma had sequences from the five MLST genes that were an identical match for those from Wolbachia strain type 9 (ST-9) wPip infection in the MLST database, sequences which were identical to those characterized from Cx. pipiens tested in this study (Table 3). Although the five MLST housekeeping genes sequenced from Cx. stigmatosoma matched those of Cx. pipiens for strain type 9 wPip, it would be worthwhile to examine differences in Wolbachia from these two species using a more comprehensive method such as comparative genomics before concluding the two species harbor the same strain [78]. Bleidorn & Gerth (2018) discussed the limits of the MLST for Wolbachia strain characterization; one of these is that several of the MLST genes used to characterize Wolbachia strains evolve slowly, and may not sufficiently differentiate among strains where significant biological differences may exist. In this study, Cx. stigmatosoma was not likely to be misidentified as adult Cx. pipiens. Adult Cx. stigmatosoma more closely resemble Cx. tarsalis, but the two species are distinguished by distinct markings on ventral abdominal segments [59]. Culex stigmatosoma had a high prevalence (frequency) of individuals with detections of Wolbachia. The second collection site (Atwater) where Cx. stigmatosoma was positive for Wolbachia in this study also had an individual with MLST alleles match those of Cx. pipiens Wolbachia strain type as well (strain 9). This species could represent a new Wolbachia infection, not just a detection of Wolbachia. However, further studies would be needed to provide evidence of infection which are complementary to sequencing, such as FISH or loop mediated isothermal amplification (LAMP) [40].
The Wolbachia similarity observed between the two species above (Cx. pipiens and Cx. stigmatosoma) is not unusual. In fact, several studies have documented high similarity among some Wolbachia-infections in hosts within the same genus [57, 79, 80]. In Italy, evidence of natural wPip Wolbachia infections have been identified within Culex modestus and Culex torrentium mosquito species, and there was no observable divergence in wsp sequences when compared to field collected Cx. pipiens [79]. Another example was documented in Portugal, where low prevalence Wolbachia-infections were found in Culex theileri and indistinguishable from Cx. pipiens by 16S rRNA, ank2 and pk1 genes [80]. Furthermore, the results of restricted fragment length polymorphisms (RFLP) suggested a shared wPip haplotype I infection among both Cx. theileri and Cx. pipiens. Thus, it is not surprising that two closely related Culex species in the present study could harbor very similar or closely related Wolbachia strains.
Culiseta inornata had several MLST genes match those in the MLST database. When grouped in the supergroup phylogeny with other vector species, Cs. inornata was closely related to supergroup A infections previously reported in a dipteran, Phlebotomos papatasi (sand fly) and an orthopteran (Loxoblemmus spp.) (Fig. 2). Culiseta inornata in this study was collected in a semi-natural riparian habitat along the Merced River. This species is predominant in rural areas, and is capable of vectoring West Nile virus, western equine encephalitis, St Louis encephalitis, Japanese encephalitis, California encephalitis and avian malaria [76, 81, 82]. This species occurs throughout the United States, with a known presence in 46 states from California to New York and the range also expands north into Canada [75, 83]. Culiseta inornata persists through the winter months, which could have implications for the seasonality of arbovirus transmission. Given that Cs. inornata transmits a number of vector-borne diseases, further study to investigate Wolbachia within this species could be worthwhile, since Wolbachia can influence vector competence. Moreover, future research could investigate whether Wolbachia in this species persist within other populations in California or other regions.
Several other mosquito species had Wolbachia at a low frequency or density. Some of these species have been previously tested through traditional PCR, but perhaps escaped detection due to the lower sensitivity of traditional PCR compared to qPCR [58]. Our study detected Ae. melanimon with Wolbachia at a low frequency. This study is the first record of Ae. melanimon with detection of Wolbachia, but additional tests as previously described would be needed to confirm infection [40]. Aedes melanimon are widely distributed throughout western and southwestern USA and Canada [75, 84, 85]. This species prefers to oviposit in or around irrigated pastures, ponds and fields. Aedes melanimon is the primary vector of California encephalitis and is capable of transmitting western equine encephalitis and West Nile virus [76, 86, 87]. Past literature has identified Ae. melanimon to have a secondary role in maintenance of western equine encephalitis virus within the Central Valley of California, and has identified this mosquito as preferentially feeding on humans and other mammals [88, 89]. Along with Ae. melanimon, several other species (An. punctipennis, Cx. tarsalis and Cs. incidens) had very low prevalence (all less than 10%), perhaps due to horizontal transmission [90]. Recently, Shaikevich et al. [90] suggested that Wolbachia diversity is likely attributed to horizontal transfer and strain recombination. By utilizing one-allele-criterion (OAC) phylogenetic networks, the authors suggest a link between the Ae. albopictus (wAlbB) Wolbachia strain and Wolbachia from ants; furthermore, that supergroup B strains from mosquitoes are linked with Wolbachia from Lepidoptera [90]. Routes of horizontal transmission have been shown to occur through parasitism, shared habitats, and predation [11, 91,92,93].
Conclusions
Our survey of Wolbachia infections in Merced county mosquitoes identified new Wolbachia detections, providing information to support current and future Wolbachia-mediated vector control applications. As noted, it will be important to confirm Wolbachia detections are true infections by providing evidence in addition to Wolbachia sequences. Wolbachia-based approaches have been implemented within vector control strategies by propagation of a desired strain within an uninfected population, or by inducing cytoplasmic incompatibility through mating incompatibility. Successful integration depends on the strain chosen for its effects on the novel host [94]. Characterizing new Wolbachia strains and determining their mosquito host species are critical to efforts to further develop Wolbachia-mediated vector control applications.
Availability of data and materials
The datasets generated during this study consist of sequences submitted to GenBank under the accession numbers MW125593- MW125610 (wsp), MW133153-MW133170 (ftsz), MW133171-MW133187 (coxA), MW133188-MW133204 (fbpA), MW133205-MW133220 (gatB), and MW133221-MW133228 (hcpA).
References
Hertig M, Wolbach SB. Studies on Rickettsia-like micro-organisms in insects. J Med Res. 1924;44:329–74.
Hertig M. The rickettsia, Wolbachia pipientis (gen. et sp. N.) and associated inclusions of the mosquito, Culex pipiens. Parasitology. 1936;28:453–86.
Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol Lett. 2008;281:215–20.
Zug R, Hammerstein P. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE. 2012;7:38544.
Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–51.
Axford JK, Ross PA, Yeap HL, Callahan AG, Hoffmann AA. Fitness of wAlbB Wolbachia infection in Aedes aegypti: parameter estimates in an outcrossed background and potential for population invasion. Am J Trop Med Hyg. 2016;94:507–16.
Ma Y, Chen W, Li Z, Zhang F, Gao Y, Luan Y. Revisiting the phylogeny of Wolbachia in Collembola. Ecol Evol. 2017;7:2009–17.
Werren JH, Zhang W, Guo LR. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc R Soc B-Biol Sci. 1995;261:55–63.
Werren JH. Biology of Wolbachia. Ann Rev Entomol. 1997;42:587–609.
Zug R, Hammerstein P. Bad guys turned nice? A critical assessment of Wolbachia mutualisms in arthropod hosts. Biol Rev. 2014;90:89–111.
Ahmed MZ, Breinholt JW, Kawahara AY. Evidence for common horizontal transmission of Wolbachia among butterflies and moths. BMC Evol Biol. 2016;16:118.
Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, Muzzi F, et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476:451–7.
O’Neill SL, Ryan PA, Turley AP, Wilson G, Retzki K, Iturbe-Ormaetxe I, et al. Scaled deployment of Wolbachia to protect the community from dengue and other Aedes transmitted arboviruses. Gates Open Res. 2019;2:36.
Frentiu FD, Zakir T, Walker T, Popovici J, Pyke AT, van den Hurk A, et al. Limited dengue virus replication in field-collected Aedes aegypti mosquitoes infected with Wolbachia. PLoS Negl Trop Dis. 2014;8:2688.
Saridaki A, Bourtzis K. Wolbachia-induced reproductive parasitism and applications. Entomol Hell. 2017;18:3.
Sinkins SP. Wolbachia and cytoplasmic incompatibility in mosquitoes. Insect Biochem Mol Biol. 2004;34:723–9.
Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6:e2.
Brownlie JC, Johnson KN. Symbiont-mediated protection in insect hosts. Trends Microbiol. 2009;17:348–54.
Bian G, Xu Y, Lu P, Xie Y, Xi Z. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 2010;6:e1000833.
Schultz MJ, Isern S, Michael SF, Corley RB, Connor JH, Frydman HM. Variable inhibition of Zika virus replication by different Wolbachia strains in mosquito cell cultures. J Virol. 2017;91:339.
Dutra HLC, Rocha MN, Dias FBS, Mansur SB, Caragata EP, Moreira LA. Wolbachia blocks currently circulating Zika virus isolates in Brazilian Aedes aegypti mosquitoes. Cell Host Microbe. 2016;19:771–4.
Yeap HL, Mee P, Walker T, Weeks AR, O’Neill SL, Johnson P, et al. Dynamics of the “popcorn” Wolbachia infection in outbred Aedes aegypti informs prospects for mosquito vector control. Genetics. 2011;187:583–95.
Ross PA, Endersby NM, Hoffmann AA. Costs of three Wolbachia infections on the survival of Aedes aegypti larvae under starving conditions. PLoS Negl Trop Dis. 2016;10:e0004320.
Turley AP, Moreira LA, O’Neill SL, McGraw EA. Wolbachia infection reduces blood-feeding success in the dengue fever mosquito, Aedes aegypti. PLoS Negl Trop Dis. 2009;9:e516.
Duan X, Sun J, Wang L, Shu X, Guo Y, Keiichiro M, et al. Recent infection by Wolbachia alters microbial communities in wild Laodelphax striatellus populations. Microbiome. 2020;8:104.
Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell. 2009;139:1268–78.
Dodson BL, Hughes GL, Paul O, Matacchiero AC, Kramer LD, Rasgon JL. Wolbachia enhances West Nile virus (WNV) infection in the mosquito Culex tarsalis. PLoS Negl Trop Dis. 2014;8:e2965.
Laven H. Eradication of Culex pipiens fatigans through cytoplasmic incompatibility. Nature. 1967;216:383–4.
Zabalou S, Riegler M, Theodorakopoulou M, Stauffer C, Savakis C, Bourtzis K. Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc Natl Acad Sci USA. 2004;101:15042–5.
Alam U, Medlock J, Brelsfoard C, Pais R, Lohs C, Balmand S, et al. Wolbachia symbiont infections induce strong cytoplasmic incompatibility in the tsetse fly Glossina morsitans. PLoS Pathog. 2011;7:e1002415.
Atyame CM, Delsuc F, Pasteur N, Weill M, Duron O. Diversification of Wolbachia endosymbiont in the Culex pipiens mosquito. Mol Biol Evol. 2011;28:2761–72.
Bourtzis K, Dobson SL, Xi Z, Rasgon JL, Calvitti M, Moreira LA, et al. Harnessing mosquito–Wolbachia symbiosis for vector and disease control. Acta Trop. 2014;132:S150–63.
Zhang D, Lees RS, Xi Z, Gilles RL, Bourtzis K. Combining the sterile insect technique with Wolbachia-based approaches: II—a safer approach to Aedes albopictus population suppression programmes, designed to minimize the consequences of inadvertent female releases. PLoS ONE. 2015;10:e0135194.
Crawford JE, Clarke DW, Criswell V, Desnoyer M, Cornel D, Deegan B, et al. Efficient production of male Wolbachia-infected Aedes aegypti mosquitoes enables large-scale suppression of wild populations. Nat Biotechnol. 2020;38:484–92.
Coon KL, Brown MR, Strand MR. Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol Ecol. 2016;25:5806–26.
Thongsripong P, Chandler JA, Green AB, Kittayapong P, Wilcox BA, Kapan DD, Bennett SN. Mosquito vector-associated microbiota: metabarcoding bacteria and eukaryotic symbionts across habitat types in Thailand endemic for dengue and other arthropod-borne diseases. Ecol Evol. 2018;8:1352–68.
Balaji S, Jayachandran S, Prabagaran SR. Evidence for the natural occurrence of Wolbachia in Aedes aegypti mosquitoes. FEMS Microbiol Lett. 2019;366:fnz055.
Carvajal TM, Hashimoto K, Harnandika RK, Amalin DM, Watanabe K. Detection of Wolbachia in field-collected Aedes aegypti mosquitoes in metropolitan Manila, Philippines. Parasit Vectors. 2019;12:361.
Kulkarni A, Yu W, Jiang J, Sanchez C, Karna AK, Martinez KJ, et al. Wolbachia pipientis occurs in Aedes aegypti populations in New Mexico and Florida, USA. Ecol Evol. 2019;9:6148–56.
Ross PA, Callahan AG, Yang Q, Jasper M, Arif MAK, Afizah AN, et al. An elusive endosymbiont: does Wolbachia occur naturally in Aedes aegypti? Ecol Evol. 2020;10:1581–91.
Ruang-Areerate T, Kittayapong P. Wolbachia transinfection in Aedes aegypti: a potential gene driver of dengue vectors. Proc Natl Acad Sci USA. 2006;103:12534–9.
Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476:450–3.
Xi Z, Khoo CC, Dobson SL. Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science. 2005;310:326–8.
Fraser JE, De Bruyne JT, Iturbe-Ormaetxe I, Stephnell J, Burns RL, Flores HA, O’Neill SL. Novel Wolbachia-transinfected Aedes aegypti mosquitoes possess diverse fitness and vector competence phenotypes. PLoS Pathog. 2017;13:e1006751.
Ant TH, Herd CS, Geoghegan V, Hoffmann AA, Sinkins SP. The Wolbachia strain wAu provides highly efficient virus transmission blocking in Aedes aegypti. PLoS Pathog. 2018;14:e1006815.
Flores HA, O’Neill SL. Controlling vector-borne diseases by releasing modified mosquitoes. Nat Rev Microbiol. 2018;16:508–18.
Gloria-Soria A, Ayala D, Bheecarry A, Calderon-Argudas O, Chadee DD, Chiappero M, et al. Global genetic diversity of Aedes aegypti. Mol Ecol. 2016;25:5377–95.
Joyce AL, Torres MM, Torres R, Moreno M. Genetic variability of the Aedes aegypti (Diptera: Culicidae) mosquito in El Salvador, vector of dengue, yellow fever, chikungunya, and Zika. Parasit Vectors. 2018;11:637.
Porse CC, Kramer V, Yoshimizu MH, Metzger M, Hu R, Padgett K, Vugia DJ. Public health response to Aedes aegypti and Ae. albopictus mosquitoes invading California, USA. Emerg Infect Dis. 2015;21:1827–9.
CDPH (California Department of Public Health). Map and city list of Aedes aegypti and Aedes albopictus mosquitoes in CA, 2011–2020. 2020. https://www.cdph.ca.gov/Programs/CID/DCDC/CDPH%20Document%20Library/AedesDistributionMap.pdf.
Hoffmann AA, Iturbe-Ormaetxe I, Callahan AG, Phillips BL, Billington K, Axford JK, et al. Stability of the wMel Wolbachia infection following invasion into Aedes aegypti populations. PLoS Negl Trop Dis. 2014;8:e3115.
Schmidt TL, Barton NH, Rašić G, Turley AP, Montgomery BL, Iturbe-Ormaetxe I, et al. Local introduction and heterogeneous spatial spread of dengue-suppressing Wolbachia through an urban population of Aedes aegypti. PLoS Biol. 2017;15:e2001894.
Nazni WA, Hoffmann AA, NoorAfizah A, Cheong YL, Mancini MV, Golding N, et al. Establishment of Wolbachia strain wAlbB in Malaysian populations of Aedes aegypti for dengue control. Curr Biol. 2019;29:4241-4248.e5.
O’Neill SL, Ryan PA, Turley AP, Wilson G, Retzki K, Iturbe-Ormaetxe I, et al. Scaled deployment of Wolbachia to protect the community from Aedes transmitted arboviruses. Gates Open Res. 2018;2:36.
Baldo L, Werren JH. Revisiting Wolbachia supergroup typing based on WSP: spurious lineages and discordance with MLST. Curr Microbiol. 2007;55:81–7.
O’Neill SL, Giordano R, Colbert AM, Karr TL, Robertson HM. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci USA. 1992;89:2699–702.
Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72:7098–110.
Rasgon JL, Scott TW. An initial survey for Wolbachia (Rickettsiales: Rickettsiaceae) infections in selected California mosquitoes (Diptera: Culicidae). J Med Entomol. 2004;41:255–7.
Meyer RP, Durso SL. Identification of the mosquitoes of California. Sacramento: Mosquito and Vector Control Association; 1993.
QGIS. QGIS Geographic Information System. Open Source Geospatial Foundation Project. 2019; https://qgis.org/en/site/
Qiagen. DNeasy tissue handbook. Valencia: Qiagen; 2006.
Li L, Fallon AM. Mosquito ribosomal protein S3 lacks a critical glutamine residue associated with DNA repair activity in homologous Drosophila proteins. Arch Insect Biochem Physiol. 2006;4:188–96.
Park K, Kwak I. Gene expression of ribosomal protein mRNA in Chironomus riparius: effects of endocrine disruptor chemicals and antibiotics. Comp Biochem Physiol C Toxicol Pharmacol. 2012;2:113–20.
Livak KJ, Schmittgen TD. Analysis of real gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–8.
Zhou W, Rousset F, O’Neill S. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc R Soc Lond B-Biol Sci. 1998;265:509–15.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
Jolley KA, Maiden MCJ. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010;11:595.
Rasgon JL, Scott TW. Wolbachia and cytoplasmic incompatibility in the California Culex pipiens mosquito species complex: parameter estimates and infection dynamics in natural populations. Genetics. 2003;165:2029–38.
Muturi EJ, Ramirez JL, Rooney AP, Kim C. Comparative analysis of gut microbiota of mosquito communities in Central Illinois. PLoS Negl Trop Dis. 2017;11:e0005377.
Wiwatanaratanabutr I. Geographic distribution of Wolbachia-infections in mosquitoes from Thailand. J Invertebr Pathol. 2013;114:337–40.
Gloria-Soria A, Chiodo TG, Powell JR. Lack of evidence for natural Wolbachia infections in Aedes aegypti (Diptera: Culicidae). J Med Entomol. 2018;55:1354–6.
Sinkins SP, Walker T, Lynd AR, Steven AR, Makepeace BL, Godfray HC, Parkhill J. Wolbachia variability and host effects on crossing types in Culex mosquitoes. Nature. 2005;436:252–60.
Atkinson CT, Watcher-Weatherwax W, Lapointe D. Genetic diversity of Wolbachia endosymbionts in Culex quinquefasciatus from Hawai`i, Midway Atoll, and Samoa. Technical Report. Hilo, Hi. Hawaii Cooperative Studies Unit (HCSU); 2016. https://hdl.handle.net/10790/2671.
McPhatter LP, Su T, Williams G, Cheng ML, Dhillon M, Gerry AC. Host-feeding patterns of Culex stigmatosoma (Diptera: Culicidae) in southern California. J Med Entomol. 2017;54:1750–7.
Carpenter SJ, LaCasse WJ. Mosquitoes of North America north of Mexico. Berkeley: University of California Press; 1955.
Goddard LB, Roth AE, Reisen WK, Scott TW. Vector competence of California mosquitoes for West Nile virus. Emerg Infect Dis. 2002;8:1385–91.
Reisen WK, Milby MM, Presser SB, Hardy JL. Ecology of mosquitoes and St. Louis encephalitis virus in the Los Angeles basin of California, 1987–1990. J Med Entomol. 1992;29:582–98.
Bleidorn C, Gerth M. A critical re-evaluation of multilocus sequence typing (MLST) efforts in Wolbachia. FEMS Microbiol Ecol. 2018;94:1–11.
Ricci I, Cancrini G, Gabrielli S, D’amelio S, Favia G. Searching for Wolbachia (Rickettsiales: Rickettsiaceae) in mosquitoes (Diptera: Culicidae): large polymerase chain reaction survey and new identifications. J Med Entomol. 2002;39:562–7.
De Pinho MV, Mendes AM, Maurício IL, Calado MM, Novo MT, Belo S, Almeida APG. Molecular detection of Wolbachia pipientis in natural populations of mosquito vectors of Dirofilaria immitis from continental Portugal: first detection in Culex theileri. Med Vet Entomol. 2016;30:301–9.
Hammon WM, Reeves WC. Laboratory transmission of St. Louis encephalitis virus by three genera of mosquitoes. J Exp Med. 1943;78:241–53.
Work TM, Washino RK, Van Riper C. Comparative susceptibility of Culex tarsalis, Anopheles franciscanus and Culiseta inornata (Diptera: Culicidae) to Plasmodium relictum (Haemosporidia: Plasmodiiae). J Med Entomol. 1990;27:68–71.
Ward RD. Identification and geographical distribution of the mosquitoes of North America, North of Mexico. 2nd ed. Gainesville: University Press of Florida; 2005.
Richards CS. Aedes melanimon Dyar and related species. Can Entomol. 1956;88:261–9.
Burgess L. Note on Aedes melanimon Dyar, a mosquito new to Canada (Diptera: Culicidae). Can Entomol. 1957;89:532.
Hammon WM, Reeves WC, Sather G. California encephalitis virus, a newly described agent. II. Isolations and attempts to identify and characterize the agent. J Immunol. 1952;69:493–510.
Jensen T, Washino RK. An assessment of the biological capacity of a Sacramento Valley population of Aedes melanimon to vector arboviruses. Am J Trop Med Hyg. 1991;44:355–63.
Hardy JL. The ecology of western equine encephalomyelitis virus in the Central Valley of California, 1945–1985. Am J Trop Med Hyg. 1987;37:18S-32S.
Tempelis CH, Washino RK. Host-feeding patterns of Culex tarsalis, with notes on other species. J Med Entomol. 1967;4:315–8.
Shaikevich E, Bogacheva A, Rakova V, Ganushkkina L, Illinsky Y. Wolbachia symbionts in mosquitoes: intra- and intersupergroup recombinations, horizontal transmission and evolution. Mol Phylogenet Evol. 2019;134:24–34.
Huigens ME, de Almeida RP, Boons PA, Luck RF, Stouthamer R. Natural interspecific and intraspecific horizontal transfer of parthenogenesis-inducing Wolbachia in Trichogramma wasps. Proc R Soc Lond B. 2004;271:509–15.
Heath BD, Butcher RDJ, Whitfield WGF, Hubbard SF. Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr Biol. 1999;6:313–6.
Li S, Ahmed MZ, Lv N, Shi P, Wang X, Huang J, et al. Plant-mediated horizontal transmission of Wolbachia between whiteflies. ISME J. 2017;11:1019–28.
Hoffmann AA, Ross PA, Rašić G. Wolbachia strains for disease control: ecological and evolutionary considerations. Evol Appl. 2015;8:751–68.
Werren JH, Windsor DM. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc R Soc Lond B. 2000;267:1277–85.
Gomes FM, Hixson BL, Tyner MDW, Ramirez JL, Canepa GE, Alves e Silva TL, et al. Effect of naturally occurring Wolbachia in Anopheles gambiae s.l. mosquitoes from Mali on Plasmodium falciparum malaria transmission. Proc Natl Acad Sci USA. 2017;47:12566–71.
Osborne SE, Leong YS, O’Neill SL, Johnson KN. Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans. PLoS Pathog. 2009;5:e100656.
Lee SF, White VL, Weeks AR, Hoffmann AA, Endersby NM. High-throughput PCR assays to monitor Wolbachia infection in the dengue mosquito (Aedes aegypti) and Drosophila simulans. Appl Environ Microbiol. 2012;78:4740–3.
Acknowledgements
We thank the Merced County Merced Mosquito Abatement District (MCMAD) for supporting this research, and the following people from MCMAD for assistance with this project: Rhiannon Jones, Jason Bakken, and Allan Inman. The following individuals from the University of California Merced assisted with the project: Jocelyn Acosta, Hannah Parolini, Ricardo Hernandez, Mackenzie Hoyt and Michaela Hoyt. The Pacific Southwest Regional Center of Excellence for Vector-Borne Diseases assisted with funding, and was funded by the U.S. Centers for Disease Control and Prevention (Cooperative Agreement 1U01CK000516). The USDA is an equal opportunity employer.
Funding
This study was supported by funding from Merced County Mosquito Abatement District. In addition, RT was supported by a trainee grant from the Pacific Southwest Regional Center of Excellence for Vector-Borne Diseases, which is funded by the U.S. Centers for Disease Control and Prevention (Cooperative Agreement 1U01CK000516).
Author information
Authors and Affiliations
Contributions
Study design: ALJ and JLR. Data collection: RT, EH, VF, JLR and ALJ. Data analysis: RT, VF, EH, JLR and ALJ. Manuscript preparation: RT, ALJ and JLR. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Mosquito collections in Merced county.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Torres, R., Hernandez, E., Flores, V. et al. Wolbachia in mosquitoes from the Central Valley of California, USA. Parasites Vectors 13, 558 (2020). https://doi.org/10.1186/s13071-020-04429-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04429-z