Abstract
Background
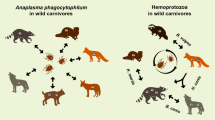
Anaplasma phagocytophilum is an obligate parasitic intracellular bacterium. It is the causative agent of granulocytic anaplasmosis, with effects on human and animal health. In Europe, the pathogen is mainly transmitted among a wide range of vertebrate hosts by blood-sucking arthropods. The aim of this study was to determine the presence of A. phagocytophilum in wild carnivores, viz raccoon dogs (Nyctereutes procyonoides), badgers (Meles meles), foxes (Vulpes vulpes), martens (Martes sp.) and European polecats (Mustela putorius), using molecular methods.
Methods
In the present study, 174 spleen samples were collected from adult, wild carnivores hunted in the years 2013–2016. A short fragment (383 bp) of the 16S ribosomal RNA gene partial sequence was used as a marker to identify A. phagocytophilum in spleen samples collected from carnivores using nested PCR.
Results
The prevalence of A. phagocytophilum in wild carnivores was 31.61% (55/174). Seven sequences of A. phagocytophilum were generated from two raccoon dogs, two badgers, one marten, one red fox and one European polecat. Six identical nucleotide sequences were obtained from one raccoon dog, two badgers, one marten, one red fox and one European polecat (A. phagocytophilum sequences 1: MH328205–MH328209, MH328211), and these were identical to many A. phagocytophilum sequences in the GenBank database (100% similarity). The second sequence (A. phagocytophilum sequence 2: MH328210) obtained from the raccoon dog shared 99.74% identity with A. phagocytophilum sequence 1.
Conclusions
To our knowledge, this is the first study to use molecular methods to determine the presence of A. phagocytophilum in wild carnivores, viz raccoon dog, badger, marten and European polecat, in Poland. The detected A. phagocytophilum sequences (1 and 2) were closely related with those of A. phagocytophilum occurring in a wide range of wild and domestic animals and vectors.
Similar content being viewed by others
Background
Anaplasma phagocytophilum is a Gram-negative bacterium belonging to the family Anaplasmataceae [1]. It is the causative agent of tick-borne fever in ruminants, also known as bovine or ovine granulocytic anaplasmosis, and of human granulocytic anaplasmosis. Anaplasma phagocytophilum is typically detected in wild animals (such as rodents, carnivores and ungulates) and domestic animals (such as cattle, goats and sheep) in the natural environment [2,3,4,5,6]. In the northern hemisphere, A. phagocytophilum is transmitted mainly by hard ticks of the genus Ixodes [7, 8]. Species of other tick genera, such as Dermacentor spp. and Rhipicephalus spp., can also serve as biological vectors; however, their significance currently remains unknown. In addition, while A. phagocytophilum is known to demonstrate transstadial transmission in tick vectors, transovarial transmission has not been observed [7].
Anaplasma phagocytophilum has also been detected in other arthropods such as blood-sucking flies, specifically deer flies (Lipoptena cervi) and horse flies (Diptera: Tabanidae) [9]. In addition, wild carnivores can also play a key role in pathogen transmission between wildlife, domestic animals and, possibly, humans. The growing degree of contact between free-living animals, domestic animals and human populations due to climate change and population growth has resulted in an elevated risk of human and bovine anaplasmosis outbreaks [10].
Anaplasma phagocytophilum has been detected by molecular methods in many carnivore species. In red foxes, it has been detected in Italy [11, 12], Germany [13], the Netherlands [14], Romania [15], Hungary [16], Switzerland [17], the Czech Republic [18, 19] and Austria [20]. In raccoon dogs, A. phagocytophilum has been detected in Germany [13] and in South Korea [21].
Knowledge about A. phagocytophilum infection in badgers, polecats and martens is still limited. It has been detected in European polecats (Mustela putorius) in Germany [22] and in the Netherlands [23], as well as in badgers (Meles meles) in the Netherlands and Spain [23, 24]. It has also been identified among pine martens (Martes martes) in Belgium. [23].
With the exception of foxes, data regarding the levels of A. phagocytophilum infection in many wild carnivores, such as raccoon dogs, European polecats, martens and badgers, is scarce in Poland [25]. While the majority of these carnivores are native to central Europe, the raccoon dog (Nyctereutes procyonoides) is an introduced species from Asia. In Poland, they were first identified in the Białowieża Primeval Forest in 1955 [26]; they are currently widespread throughout central Europe, and their range is extending into western Europe [27].
These wild carnivores could potentially act as vectors of A. phagocytophilum in the natural environment [8]. The aim of the present study was therefore to determine the prevalence of A. phagocytophilum in five species of predatory wild animals in Poland.
Methods
Spleen samples were collected from 174 wild carnivores [68 raccoon dogs: 28 females (F)/40 males (M); 49 badgers: 21F/28M; 29 foxes: 13F/16M; 24 martens: 7F/17M; 4 polecats: 2F/2M] hunted in Poland around the Głęboki Bród Forest District (53°98′N, 23°29′E) during the years 2013–2016. These were stored at −20 °C until further processing. DNA was isolated from the middle part of the spleen using a Genomic Mini AX Tissue kit (A&A Biotechnology, Gdynia, Poland) and according to the manufacturer’s instructions.
Molecular detection of A. phagocytophilum was based on semi-nested PCR amplification of the partial 16S rRNA gene according to Werszko et al. [9]. Three primers were used to molecular detection of A. phagocytophilum: A480F, A520F and A900R. The first PCR reaction was performed with primers A480F and A900R. For the second reaction (semi nested PCR), 1 μl of the first reaction product and primers A520F and A900R were used. PCR reactions were conducted in 50 µl of reaction mixture containing the following: 40 μl of deionized water in first reaction (41 μl in second), 1 μl of a 25 mM solution of MgCl2, 0.5 μl of Allegro Taq DNA Polymerase (5 U/μl; Novazym, Poznań, Poland), 0.5 μl of dNTPmix (10 mM), 5 μl of 10× Taq DNA Polymerase Buffer (with 25 mM MgCl2), 0.5 μl of each primer (20 pmol/μl), 2 μl of template DNA in the first reaction and 1 μl of first reaction PCR product in the second reaction. DNA isolated from red deer (Cervus elaphus) infected with A. phagocytophilum (GenBank: GQ450278) was used as the positive control. Nuclease-free water was added to the PCR mix as a negative control. The PCR reaction was performed in an automated DNA Engine PTC-200 Peltier Thermal Cycler (BioRad, Hercules, CA, USA). The thermocycling profile was as follows: denaturation at 92 °C for 2 min, followed by 35 cycles with 30 s denaturation at 94 °C, 10 s primer annealing at 60 °C, and 1 min at 72 °C for primer extension, with a final extension step of 5 min at 72 °C.
The PCR products were subjected to electrophoresis (Bio-Rad Power Pac Basic 85 V, 45 min) on a 1.0% agarose gel stained with ethidium bromide and visualized using ChemiDoc, MP Lab software (Imagine, BioRad); a Nova 100 bp DNA Ladder (Novazym, Poznań, Poland) was used for comparison. The PCR amplicons were purified using a QIAEX II Gel Extraction Kit (Qiagen, Hilden, Germany), sequenced by Genomed (Warszawa, Poland) and assembled into contigs using ContigExpress, Vector NTI Advance v.11.0 (Invitrogen Life Technologies, New York, NY, USA). The obtained sequences were compared with sequences available on GenBank using the Basic Local Alignment Search Tool (BLAST). Statistical analyses were performed using the Chi-square test in GraphPad online (https://www.graphpad.com).
Results
The overall prevalence of A. phagocytophilum infection was 31.60% (55/174); however, among all tested groups, the prevalence of infection varied according to the host species. The highest prevalence was observed in martens (41.70%; 10/24), followed by raccoon dogs (35.30%; 24/68) and foxes (34.48%; 10/29), with the lowest prevalence observed in badgers (18.70%; 9/49). Anaplasma phagocytophilum was detected in two of four investigated European polecats. The prevalence of A. phagocytophilum was significantly higher in martens than in badgers (χ2 = 4.5422, df = 1, P = 0.033; 95% CI: 0.16–0.36). The prevalence in raccoon dogs was significantly higher than in badgers (χ2 = 4.0295, df = 1, P = 0.045; 95% CI: 0.20–0.36). During the tests, a large difference was observed between the first and the second reaction in nested PCR. After the first reaction only 4 samples were positive in all tested animals (2.30%). After the second amplification step, 55 samples out of 174 were positive (31.60%).
Seven sequences were obtained from A. phagocytophilum PCR-positive samples and submitted to the GenBank database (from two raccoon dogs, two badgers, one marten, one red fox and one European polecat). Six sequences (Anaplasma phagocytophilum sequence 1: MH328205-MH328209, MH328211) were found to be identical (100% similarity) with many A. phagocytophilum sequences obtained from Europe, Asia, Africa and North America. Anaplasma phagocytophilum sequences were obtained from a wide range of hosts: human, dog, raccoon dog, fox, sheep, black-striped field mouse, northern red-backed vole, tick, cow, goat and horse. More detailed data are shown in Table 1. Anaplasma phagocytophilum sequence 2 was highly similar to all examples of A. phagocytophilum sequence 1 obtained in this study (99.74% identity) and differed only by one nucleotide from sequence 1.
Discussion
Studies performed in Europe confirm that a wide range of mammals can serve as competent animal reservoirs for A. phagocytophilum, and that the composition of this reservoir varies according to geographical regions [8, 28]. The high prevalence of A. phagocytophilum infection in the tested animals provides compelling evidence for the involvement of wild carnivores in the enzootic cycle. So far in Europe, A. phagocytophilum has been detected in red foxes in Germany (8.20%), Italy (16.60%) and Hungary (12.50%) [11, 13, 16]. It has also been detected in Poland, but only with an infection rate of 2.70% [25]. The higher prevalence identified in the present study (34.48%) may be associated with infection rate in the vectors [29], or the geographical distribution: the previous study was performed in central Poland, whereas the present study was performed in the north-east. Additionally, the prevalence was found to be 2.7% in the earlier study in Poland [25] which is close to prevalence levels obtained after the first PCR reaction in presented study. Anaplasma phagocytophilum can be detected in many various organs such as spleen, lung and liver tissue [4, 12, 13]. Our present findings confirm that semi-nested PCR offers a high level of accuracy in detecting Anaplasma in spleen samples, and that it is more sensitive than standard PCR: while the detected prevalence was only 4/174 (2.30%) in all tested animals after the first reaction, the detection level was increased to 55/174 (31.60%) after the second stage.
Foxes are well adapted to the urban environment and are accustomed to the presence of humans. In addition, their frequent exposure to tick bites and their potential to act as reservoirs or maintenance hosts for pathogens infecting humans and domestic animals highlights their importance in public health [15]. The prevalence of A. phagocytophilum observed in raccoon dogs in Poland (35.30%) is higher than in Germany (23.00%) [13]. Interestingly, while Han et al. [21] and Kang et al. [30] reported the prevalence of A. phagocytophilum infection in this host to be 1.00% in Korea, Hildebrandt et al. [31] reported no infection in raccoon dogs in western Poland.
According to the present findings, 18.70% of the tested badgers in north-eastern Poland displayed A. phagocytophilum infection; however, Garcia-Pérez et al. [24] reported a prevalence of 1.50% among badgers in Spain, and Hofmeester et al. [23] found it to be 1.80% in the Netherlands. While Anaplasma infection was found in two of the four tested European polecats in the present study, previous studies have found the prevalence to be 4.30% in Germany and 4.90% in the Netherlands [22, 23]. Similarly, A. phagocytophilum was found to be more common in pine martens (Martes martes) in the present study (41.70%) than in Belgium (22.00%) [23].
Conclusions
All five tested species of carnivore may serve as suitable reservoir hosts in the ecology of Anaplasma phagocytophilum in the natural environment. Most isolates obtained in this study were identical to A. phagocytophilum sequences deposited in GenBank. Our findings represent the first detection of A. phagocytophilum in badgers, raccoon dogs, martens and polecats from Poland. The nested PCR used in the present study was more sensitive to detection of A. phagocytophilum than standard PCR for spleen tissue samples.
Availability of data and materials
The data supporting the conclusions of this article are included within the article. The sequences were submitted to the GenBank database under the accession numbers MH328205-MH328211 (Anaplasma phagocytophilum).
Abbreviations
- rDNA:
-
ribosomal DNA
- PCR:
-
polymerase chain reaction
- BLAST:
-
basic local alignment search tool
References
Dumler JS, Barbet AF, Bekker DPJ, Dasch GA, Palmer GH, Ray SC, et al. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‛HGE agentʼ as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001;51:2145–65.
Bakken JS, Krueth J, Tilden RL, Dumler JS, Kristiansen BE. Serological evidence of human granulocytic ehrlichiosis in Norway. Eur J Clin Microbiol Infect Dis. 1996;15:829–32.
Foley JE, Nieto NC, Barbet A, Foley P. Antigen diversity in the parasitic bacterium Anaplasma phagocytophilum arises from selectively-represented, spatially clustered functional pseudogenes. PLoS One. 2009;4:e8265.
Dumler JS, Choi KS, Garcia-Garcia JC, Barat NS, Scorpio DG, Garyu JW, et al. Human granulocytic anaplasmosis and Anaplasma phagocytophilum. Emerg Infect Dis. 2005;11:1828–34.
Smetanová K, Schwarzová K, Kocianová E. Detection of Anaplasma phagocytophilum, Coxiella burnetii, Rickettsia spp., and Borrelia burgdorferi s.l. in ticks, and wild-living animals in western and middle Slovakia. Ann N Y Acad Sci. 2007;1078:312–5.
Nicholson WL, Allen KE, McQuiston JH, Breitschwerdt EB, Little SE. The increasing recognition of rickettsial pathogens in dogs and people. Trends Parasitol. 2010;26:205–12.
Woldehiwet Z. The natural history of Anaplasma phagocytophilum. Vet Parasitol. 2010;167:108–22.
Karbowiak G, Biernat B, Stańczak J, Werszko J, Szewczyk T, Sytykiewicz H. The role of particular ticks developmental stages in the circulation of tick-borne pathogens in central Europe. 4. Anaplasmataceae. Ann Parasitol. 2016;62:267–84.
Werszko J, Szewczyk T, Steiner-Bogdaszewska Ż, Laskowski Z, Karbowiak G. Molecular detection of Anaplasma phagocytophilum in blood-sucking flies (Diptera: Tabanidae) in Poland. J Med Entomol. 2019. https://doi.org/10.1093/jme/tjy217.
Duscher GG, Fuehrer HP, Kübber-Heiss A. Fox on the run–molecular surveillance of fox blood and tissue for the occurrence of tick-borne pathogens in Austria. Parasit Vectors. 2014;7:521.
Ebani VV, Verin R, Fratini F, Poli A, Cerri D. Molecular survey of Anaplasma phagocytophilum and Ehrlichia canis in red foxes (Vulpes vulpes) from central Italy. J Wildl Dis. 2011;43:699–703.
Ebani VV, Rocchigiani G, Nardoni S, Bertelloni F, Vasta V, Papini RA, et al. Molecular detection of tick-borne pathogens in wild red foxes (Vulpes vulpes) from central Italy. Acta Trop. 2017;172:197–200.
Härtwig V, von Loewenich FD, Schulze C, Straubiner RK, Daugschies A, Dyachenko V. Detection of Anaplasma phagocytophilum in red foxes (Vulpes vulpes) and raccoon dog (Nyctereutes procyonoides) from Brandenburg, Germany. Ticks Tick Borne Dis. 2014;5:277–80.
Jahfari S, Coipan EC, Fonville M, van Leeuwen AD, Hengeveld P, Heylen D, et al. Circulation of four Anaplasma phagocytophilum ecotypes in Europe. Parasit Vectors. 2014;7:365.
Dumitrache MO, Matei IA, Ionică MA, Kalmár Z, D’Amico G, Sikó-Barabási S, et al. Molecular detection of Anaplasma phagocytophilum and Borrelia burgdorferi sensu lato genospecies in red foxes (Vulpes vulpes) from Romania. Parasit Vectors. 2015;8:514.
Tolnai Z, Sréter-Lancz Z, Sréter T. Spartial distribution of Anaplasma phagocytophilum and Hepatozoon canis in red foxes (Vulpes vulpes) in Hungary. Ticks Tick Borne Dis. 2015;6:645–8.
Hofmann-Lehmann R, Wagmann N, Meli ML, Riond B, Novacco M, Joekel D, et al. Detection of “Candidatus Neoehrlichia mikurensis” and other Anaplasmataceae and Rickettsiaceae in Canidae in Switzerland and Mediterranean countries. Schweiz Arch Tierheilkd. 2016;224:44–51.
Hodžić A, Mitková B, Modrý D, Juránková J, Frgelecová L, Forejtek P, et al. A new case of the enigmatic Candidatus Neoehrlichia sp. (FU98) in a fox from the Czech Republic. Mol Cell Probes. 2017;31:59–60.
Hulínská D, Langrová K, Pejcoch M, Paviásek I. Detection of Anaplasma phagocytophilum in animals by real-time polymerase chain reaction. APMIS. 2004;112:239–47.
Hodžić A, Mrowietz N, Cézanne R, Bruckschwaiger P, Punz S, Habler VE, et al. Occurrence and diversity of arthropod-transmitted pathogens in red fox (Vulpes vulpes) in western Austria, and possible vertical (transparental) transmission of Hepatozoon canis. Parasitology. 2018;145:335–44.
Kang JG, Chae JB, Cho YK, Jo YS, Shin NS, Lee H, et al. Molecular detection of Anaplasma, Bartonella, and Borrelia theileri in raccoon dogs (Nyctereutes procyonoides) in Korea. Am J Trop Med Hyg. 2018;98:1061–8.
Król N, Obiegala A, Kretschmar FM, Hamel D, Pfeffer M. Tick-borne pathogens in the European polecat, Mustela putorius and in attached Ixodes hexagonus ticks from Germany. Ticks Tick Borne Dis. 2019;10:594–7.
Hofmeester TR, Hofmeester TR, Krawczyk AI, van Leeuwen AD, Fonville M, Montizaan MGE, van den Berge, et al. Role of mustelids in the life-cycle of ixodid ticks and transmission cycles of four tick-borne pathogens. Parasit Vectors. 2018;11:600.
Garcia-Pérez AL, Oporto B, Espi A, del Cerro A, Barral M, Povedano I, et al. Anaplasmataceae in wild ungulates and carnivores in northern Spain. Ticks Tick Borne Dis. 2016;7:264–9.
Karbowiak G, Víchová B, Majlathová V, Hapunik J, Peťko B. Anaplasma phagocytophilum infection of red foxes (Vulpes vulpes). Ann Agric Environ Med. 2009;16:299–300.
Knaflewska J, Siemionowicz M. Ilustrated book of Polish nature. Poznań: Publicat SA; 2014.
Kamieniarz R, Panek M. Game animals in Poland at the turn of the 20th and 21st century. Czempiń: Ośrodek Hodowli Zwierzyny Polski Związek Łowiceki; 2008. p. 132.
Huhn C, Winter C, Wolfsperger T, Wüppenhorst N, Strašek Smrdel K, Skuballa J, et al. Analysis of the population structure of Anaplasma phagocytophilum using multilocus sequence typing. PLoS One. 2014;9:e93725.
Pusterla N, Deplazes P, Braun U, Lutz H. Serological evidence of infestation with Ehrlichia spp. in red foxes (Vulpes vulpes) in Switzerland. J Clin Microbiol. 1999;37:1168–9.
Han YJ, Park JH, Lee YS, Chae JS, Yu DH, Park BK, et al. Molecular identification of selected tick-borne pathogens in wild deer and raccoon dogs from Republic of Korea. Vet Parasitol Reg Stud Reports. 2017;7:25–31.
Hildebrand J, Buńkowska-Gawlik K, Adamczyk M, Gajda E, Merta D, Popiołek M, et al. The occurrence of Anaplasmataceae in European populations of invasive carnivores. Ticks Tick Borne Dis. 2018;9:934–7.
Acknowledgements
The authors would like to thank the local hunters and the staff from the Głęboki Bród Forest District for their help with collecting samples.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
TS planned and organized the study. TS, AM and ZL collected the samples. JW extracted the DNA. ZL and TS performed PCR, sequencing and analyzed sequence data. TS and JW drafted the manuscript, and wrote the final version together with GK. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All animals examined in this study were hunted within Project Life+”Active protection of lowland populations of capercaillie in the Bory Dolnoslaskie Forest and Augustowska Primeval Forest” (Life 11, NAT/PL/428); therefore, no separate agreement with Second Warsaw Local Ethics Committee for Animal Experimentation for Witold Stefański Institute of Parasitology was needed to conduct research on animal tissue (due to the Resolution no. 22/2006 of the National Ethics Committee for Animal Experiments, 7 November 2006).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Szewczyk, T., Werszko, J., Myczka, A.W. et al. Molecular detection of Anaplasma phagocytophilum in wild carnivores in north-eastern Poland. Parasites Vectors 12, 465 (2019). https://doi.org/10.1186/s13071-019-3734-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-019-3734-y