Abstract
Background
The present study, conducted in Zambia’s Luangwa valley where both animal African trypanosomiasis (AAT) and human African trypanosomiasis (HAT) are endemic, combined the use of microscopy and molecular techniques to determine the presence of trypanosome species in cattle, goats and tsetse flies.
Methods
This study was conducted between 2008 and 2010 in Petauke, Chama and Isoka districts, north-eastern Zambia. A total of 243 cattle, 36 goats and 546 tsetse flies, were examined for presence of trypanosome species using microscopy, PCR and loop-mediated isothermal amplification (LAMP).
Results
There was poor agreement among the test methods used for detection of trypanosomes species in animal blood and tsetse flies. Trypanosomes were observed in 6.1 % (95 % CI: 3.3-8.9 %) of the animals sampled by microscopy, 7.5 % (95 % CI: 4.4–10.6 %) by PCR and 18.6 % (95 % CI: 13.6–23.6 %) by PFR-LAMP. PFR-LAMP was more sensitive for detecting Trypanozoon than KIN-PCR. The highest occurrence of AAT was recorded in cattle from Petauke (58.7 %, 95 % CI: 44.7–72.7 %) while the lowest was from Isoka (5.4 %, 95 % CI: 0.8–10.0 %). Infection of both cattle and goats with Trypanosoma congolense and T. vivax was associated with clinical AAT.
Conclusion
When selecting molecular techniques for AAT surveillance in endemic regions, the KIN-PCR and species-specific PCR may be recommended for screening animal or tsetse fly samples for T. congolense and T. vivax, respectively. On the other hand, species-specific PCR and/or LAMP might be of greater value in the screening of animal and human body fluids as well as tsetse fly samples for Trypanozoon.
Similar content being viewed by others
Background
Animal African trypanosomiasis (AAT) is one of the most important arthropod-borne diseases of cattle and other domestic animals in sub-Saharan Africa (SSA). It has a major impact on livestock productivity in SSA, with 50 million cattle and 70 million small ruminants at risk, costing up to US$5 billion annually [1]. Trypanosoma congolense and T. vivax, mainly transmitted by tsetse flies (Glossina spp.), are the major causes of AAT in domestic ruminants [2–4]. The disease is usually chronic and debilitating and characterized by intermittent fever, progressive anaemia, emaciation, lymphadenopathy and may lead to death if untreated [4]. Anaemia is the most pathogenic consequence of AAT [5]. Trypanosoma brucei subspecies (T. b. brucei s.l) are considered to have low pathogenicity to domestic ruminants. Cattle have been documented to serve as reservoirs of T. b. rhodesiense, the aetiological agent of human African trypanosomiasis (HAT), in Uganda [6, 7], Kenya [8] and Tanzania [9–11]. HAT in Zambia is thought to be endemic in the old foci (Luangwa, Zambezi and Kafue river valleys), with sporadic cases being reported [12–15]. In Zambia, HAT is transmitted by tsetse flies species of Glossina morsitans morsitans, Glossina pallidipes, Glossina morsitans centralis and Glossina brevipalpis [16], with several domestic and wild animals acting as reservoir hosts [17–19] .
Field diagnosis of AAT is complex since the clinical signs are not pathognomonic. Visualization of parasites in body fluids by microscopy is currently the gold standard of AAT and/or HAT diagnosis in endemic regions [20, 21]. Animals often exhibit a low but persistent parasitamia which fluctuates below the levels of microscopic detection [5, 21]. Over the past two decades, polymerase chain reaction (PCR) and Loop-mediated isothermal amplification (LAMP) techniques have been applied to improve the sensitivity and accuracy of AAT and HAT diagnosis [22–25] that are often applied in large-scale epidemiological surveys. These methods differentiate between trypanosome species and subspecies using specific primers and have been adapted to be able to simultaneously detect multiple species [25]. The internal transcribed spacer (ITS) region of ribosomal DNA offers a good target for universal trypanosome tests with highly conserved flanking regions, size variability among trypanosome species and subspecies and high copy number of around 200 [25]. Kinetoplastid (KIN) primers amplify the ITS1 [23, 25]. Sensitivity and specificity of the KIN-PCR is high for most of the pathogenic trypanosomes, but exhibit low sensitivity for detection of east African T. vivax [25] which was subsequently improved [26]. In order to increase the sensitivity and specificity, Cox et al. [27] used nested ITS-PCR. Adams et al. [28] have employed ITS-PCR techniques for trypanosome species differentiation in tsetse flies using fluorescent labelling. By means of ITS-PCR and multiple samples taken from single FTA cards, Cox et al. [29] demonstrated that a single punch from an FTA card is not sufficient to confirm the infectivity status of an individual animal as parasite DNA is unevenly distributed across the card. Furthermore, Ahmed et al. [30] have recently demonstrated that the use of an elution step simultaneously from 10 punches using Chelex 100® shows higher sensitivity compared to PCR of multiple punches separately. Ahmed et al. [31] further modified the ITS-PCR designed by Cox et al. [27] to include a Chelex 100® elution stage for detection of trypanosome species in cattle samples. Thus the KIN and the ITS1 primers, both targeting the ITS1 of rDNA, offer promise in the routine diagnosis of pathogenic trypanosomes in clinical specimens from infected animals.
LAMP is a novel strategy which amplifies DNA with high sensitivity and rapidity under isothermal conditions (60-65 °C), producing large quantities of DNA within an hour [24]. LAMP has the advantage over PCR of being cheaper and user-friendly and the LAMP products can be visualized by naked eyes or through measurement of turbidity or fluorescence [22, 32].
The present study, conducted in Zambia’s Northern, Muchinga and Eastern provinces where both AAT and HAT are endemic, combined microscopy, PCR and LAMP to examine the presence of different trypanosome species in cattle, goats and tsetse flies.
Methods
Study area and study design
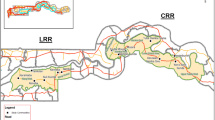
Studies were conducted between May and October in 2008 and 2010 in Petauke (Eastern Province), Chama (Muchinga Province) and Isoka (Northern Province) districts, north-eastern Zambia (Fig. 1). A total of 243 indigenous and mixed breeds of cattle from Isoka (92), Petauke (46) and Chama (105) and 36 indigenous breeds of goats (all from Chama), were examined for presence of trypanosome species using microscopy, PCR and LAMP assays. In addition, PCR and LAMP assays were used to examine the presence of trypanosome species in 546 tsetse samples (caught from Mbambanda Zaro sanctuary, Chama) comprising 492 Glossina m. morsitans (27 males and 465 females) and 54 G. pallidipes (all males). Mbambanda Zaro sanctuary, lying on the border with Malawi, is part of the Zambia wildlife authority (ZAWA). It has abundant wildlife and tsetse flies and is surrounded by human settlements that maintain limited livestock, mainly goats.
Sample collection and DNA preparation
Blood was collected from the jugular vein of cattle and goats whose owners consented to participate in the survey. Using vaccutainer tubes with EDTA-2Na (Terumo, Japan), 5 ml of blood was drawn from each animal, loaded into capillary tubes, packed cell volume (PCV) values determined and the buffy coat immediately examined for the presence of trypanosomes under light microscope [33]. In addition, total DNA of each blood sample (1 ml) was isolated using DNA Isolation Kit for Mammalian Blood (Roche Diagnostics K.K., Japan), following the manufacturer’s suggested protocol. The resultant DNA was stored at minus 30 °C until use.
Tsetse flies were trapped from Mbambanda Zaro sanctuary, Chama district, using baited epsilon trap which were set in tsetse fly favorable biotopes (mainly in shrubs adjacent to roads and footpaths within the sanctuary, and adjacent to homesteads in the immediate vicinity of the sanctuary). The traps, typically deployed at a distance of 100 m apart in a shade of a tree to reduce fly mortality due to heat, were emptied twice a day during 3 days of capture at Mbambanda Zaro sanctuary. The caught tsetse flies were separated according to sex and species and stored in 1.5 ml tubes containing silica gel. After grinding the dried tsetse flies using a disposable homogenizer (BioMasher I, Polysciences, Inc. PA. USA), total DNA of each tsetse fly was isolated using specific DNA isolation kit for Cells and Tissues (Roche Diagnostics, Germany) and stored at minus 30 °C until use.
Detection of trypanosome species by PCR and LAMP
This study applied 3 different PCR techniques to detect trypanosome DNA in cattle, goats and tsetse flies: (i) KIN-PCR that amplifies the internal transcribed spacer 1 (ITS-1) of African trypanosomes [25], (ii) TviCatL-PCR that amplifies the highly conserved Cathepsin L-like gene among T. vivax isolates [34] and (iii) SRA-PCR that specifically amplifies the human serum resistance-associated (SRA) gene uniquely expressed by T. b. rhodesiense [6]. In addition, we also used PFR-LAMP that amplifies the paraflagella rod A gene of Trypanozoon subgenus (T. b. brucei, T. b. gambiense, T. b. rhodesiense, T. evansi, T. equiperdum) [35].
The reaction mixtures of LAMP (25 μl) and PCR (50 μl) and their respective amplification conditions were previously described [23, 25, 36]. Genomic DNA for each trypanosome species was used as positive control in the corresponding species-specific PCR or LAMP while T. b. rhodesiense (IL1501) and T. congolense Savannah (IL3000) DNA were used as positive controls in the KIN-PCR. In the case of KIN-PCR, the results were positive when the specific size product was observed [25], while in the case of LAMP, positive samples exhibited a bright fluorescent green colour when visualized under the transilluminator [12]. Double distilled water (DDW) was used as negative control. All samples detected as positive for Trypanozoon by either KIN-PCR or PFR-LAMP assays were screened for T. b. rhodesiense using SRA-PCR.
Statistical analysis
Mean PCV values in trypanosome infected and non-infected livestock were compared using the student t-test. P values <0.05 were considered statistically significant. The kappa coefficient (K) was used to determine the agreement between two diagnostic tests. Calculations and interpretations of K values followed the methods of Viera and Garrett [37], with a value of 1 indicating perfect agreement and 0 indicating agreement equivalent to chance.
Ethical clearance
This study received approval from the Provincial and District Veterinary Officers in the respective provinces and districts and from ZAWA. Informed consent was sought from livestock owners to participate in the survey and collect blood from their animals.
Results
Detection of trypanosome species in cattle and goat blood
Trypanosome infections detected in cattle and goats by microscopy, PCR and LAMP are summarized in Table 1. The overall proportion of animals (cattle and goats) infected with AAT was 6.1 % (95 % CI: 3.3-8.9 %) by microscopy, 7.5 % (95 % CI: 4.4–10.6 %) by PCR and 18.6 % (95 % CI: 13.6–23.6 %) by PFR-LAMP. PRF-LAMP was more sensitive for detecting Trypanozoon (~17 times) than KIN-PCR. Surprisingly, Trypanosoma theileri was parasitologically detected in one cow in Chama which tested negative for both KIN-PCR and PRF-LAMP. Although a number of parasitologically AAT positive animals were also positive for PCR and/or LAMP, there was poor agreement between Microscopy and both PCR (Kappa = 0.26) and LAMP (Kappa = 0.04).
Petauke district recorded the highest proportion of animals infected with AAT, mainly caused by Trypanozoon (32.6 %, 95 % CI: 18.6–46.6 %, by PFR-LAMP), T. congolense (savannah type mainly) (17.4 %, 95 % CI: 6.4–28.4 %, by KIN-PCR) and T. vivax (8.7 %, 95 % CI: 4.7–12.5 %, by TviCatL-PCR) while the lowest proportion was recorded in Isoka (5.4 %, 95 % CI: 0.8–10.0 %, by PFR-LAMP) exclusively caused by Trypanozoon (Table 1). Of those, 8.7 % (95 % CI: 5.6–11.8 %) and 0.7 % (95 % CI: 0.4-1.0 %) were mixed infections of Trypanozoon (PFR-LAMP) and T. congolense (KIN-PCR) in Petauke and Chama, respectively, while 2.2 % (95 % CI: 1.1–3.3 %) were mixed infections of Trypanozoon (PFR-LAMP), T. congolense (KIN-PCR) and T. vivax (TviCatL-PCR) in Petauke.
Data obtained from Chama district suggest that the overall occurrence of AAT was similar in cattle (28.6 %, 95 % CI: 20.0–37.4 %) and goats (27.8 %, 95 % CI: 12.8–42.4 %) although cattle recorded more Trypanozoon infection (26.7 %, 95 % CI: 18.4–35.2 %, PFR-LAMP) than goats which recorded greater infection with T. congolense (8.3 %, 95 % CI: 3.0–13.6 %, by KIN-PCR) and T. vivax (5.5 %, 95 % CI: 1.3–9.5 %, by TviCatL-PCR). Zoonotic T. b. rhodesiense was only detected in cattle (0.95 %, 95 % CI: -1.1–2.8 %, SRA-LAMP) from Chama district (Table 1).
Anaemia is one of the pathogenic consequences of infection with animal infective trypanosomes [5]. Because low PCV is one of the indicators of anaemia, we examined the association between infection with various trypanosome species and the level of PCV in such infected livestock. PCV values in cattle and goats ranged from 13.0 % to 50.0 % and 17.0 % to 46.0 %, respectively (Fig. 2). A significant reduction of PCV in animals with detectable parasitaemia of trypanosomes was observed in cattle (p < 0.05, Fig. 2a). Cattle and goats detected with Trypanozoon parasites by PFR-LAMP generally had similar PCV values to those of uninfected animals (Trypanozoon positive cattle: PCV = 35.0 ± 5.4 %; Trypanozoon negative cattle: PCV = 34.3 ± 4.9 %; Trypanozoon positive goat: PCV = 33.3 ± 5.7 %; Trypanozoon negative goat: PCV = 35.2 ± 3.3 %). In contrast, infection of cattle and goats (positive by either microscopy or PCR) with T. congolense (p < 0.001) or T. vivax (p < 0.05) was associated with significant reduction in PCV values (Fig. 2).
Comparison of PCV values between trypanosome infected and non-infected animals. a Cattle blood samples from all the 3 districts (n = 243). b Goat blood samples from Chama (n = 36). Values are mean ± SD. * Significant difference by Student’s t-test (*p < 0.05, **p < 0.001). Note: only 1 goat was detected to be trypanosome positive by KIN-PCR
Detection of trypanosome species in tsetse flies
The 546 tsetse flies from Mbambanda Zaro sanctuary, Chama district, were examined for the presence of trypanosome species by PCR and LAMP. Table 2 summarizes the obtained data. Trypanosome DNA was identified in 36.4 %, 95 % CI: 23.8–49.0 % (by PCR) and 40.1 %, 95 % CI: 25.0–55.2 % (by PFR-LAMP) of the tsetse flies, respectively. The main trypanosome DNA detected was Trypanozoon (30.4 %, 95 % CI: 19.8–41.0 %, by PFR-LAMP), T. vivax (25.1 %, 95 % CI: 15.6–34.6 %, by TviCatL-PCR) and T. congolense (mainly savannah type and to a lesser extent kilifi type) (11.2 %, 95 % CI: 4.9–17.5 %, by KIN-PCR). PFR-LAMP was more sensitive for detecting Trypanozoon (>21 times) than KIN-PCR. There was poor agreement between LAMP and KIN-PCR (Kappa = 0.38). Trypanozoon DNA was only detected in G. m. morsitans, with T. b. rhodesiense DNA being detected in 3.5 % (95 % CI: 1.9–5.1 %, by SRA-PCR) of the flies.
Discussion
Diagnosis of AAT and HAT in endemic regions remains a big challenge. In the present study, several tests were applied to determine AAT infection in cattle and goats and the presence of trypanosome species in tsetse vectors. As expected, microscopy exhibited relatively lower sensitivity than PCR and LAMP. Of note, there was poor agreement among the test methods used for detection of trypanosomes species in animal blood and tsetse flies (Kappa <0.40). For instance, the failure of PCR and LAMP to detect and identify 6 parasitologically positive cattle samples in this study was unexpected. These discrepancies may be resolved by the use of more sensitive trypanosome-species-specific PCR and/or LAMP. Furthermore, T. theileri was only detected in one cow in Chama district by microscopy. Although cattle frequently habour T. theileri [27, 29, 31], the KIN-PCR was unable to detect that parasite in the present study. This may be a result of lower sensitivity of the test [26] and/or possible occurrence of inhibitors in cattle blood [35]. In agreement with the former notion, detection of T. theileri has mainly been reported when more sensitive ITS-PCR such as nested ITS-PCR [27] or modified nested ITS-PCR [31] were used.
The major advantage of molecular tests over microscopy is for epidemiological studies to identify trypanosome species. ITS-PCR can identify several pathogenic trypanosome species in a single PCR reaction, reducing on the cost of PCR diagnosis and allowing for a greater number of field samples to be tested in epidemiological studies [25, 26]. However, in the present study, in agreement with previous reports [25, 26], there was little T. vivax detected by KIN-PCR. T. vivax is very diverse, comprising 3 main groups including (i) East African, (ii) West African and (iii) South American isolates [34], and makes diagnosis difficult. TviCatL-PCR was required to complement on the low sensitivity of KIN-PCR for T. vivax detection [34]. Furthermore, this study confirmed that the KIN-PCR has significantly lower sensitivity for Trypanozoon detection than the PFR-LAMP [36].
In this study, some cattle were given curative treatments for AAT several days before sampling. Since longevity of trypanosome DNA after treatment remains to be established, we cannot rule out the possibility of the LAMP assay being better at picking up residual DNA from dead trypanosomes in cattle blood than PCR. All KIN-PCR positive samples were also PFR-LAMP positive, suggesting that the KIN-PCR may be recommended for epidemiological surveillance of T. congolense in AAT endemic regions, but it would not be appropriate for T. vivax or Trypanozoon surveillance. Finally, whereas both the KIN-PCR and the PFR-LAMP are unable to distinguish between the closely related T. brucei subspecies, SRA-PCR is needed to specifically distinguishing the human-infective T. b. rhodesiense from the animal-infective T. b. brucei [6].
Our data show that Petauke district (58.7 %) reported the highest occurrence of AAT, followed by Chama (28.6 %), with Isoka (5.4 %) having the lowest. Of note, the animals in all the districts were mostly infected with Trypanozoon parasites such as T. b. brucei. However, such Trypanozoon-infected animals either exhibited only subclinical symptoms of AAT or were asymptomatic altogether, suggesting that those trypanosomes do not have significant effect on the general animal health [3, 6]. In sharp contrast, infection with T. congolense (mainly Savannah type) and T. vivax seems to be associated with clinical AAT as evidenced by significantly lower PCV values. In particular, T. congolense strains within the Savannah type are regarded to be the most pathogenic/virulent and widespread throughout the savannah ecosystem of SSA [38]. Accordingly, Marcotty et al. [39] suggested that low PCV could be used as an indicator of infection with T. congolense. On the other hand, however, it is important to note that there were several anaemic animals that were trypanosome negative, suggesting that infection or co-infection with other haemoparasites or worms could also induce anaemia [40]. Interestingly, there was hardly any case of clinical AAT in Isoka district (recorded no case of T. congolense or T. vivax infection) and Chama district (recorded very low prevalence of both parasite species [~1.0 %]). According to the District Veterinary Officers, most of the sampled cattle in the two districts were treated with isometamidium chloride a few weeks prior to the sampling exercise. Our data is in agreement with previous reports that T. congolense is the major cause of clinical AAT in both cattle and goats [3–5].
The detection of the human-infective T. brucei subspecies in cattle in Chama district is a significant finding albeit the low prevalence (~1.0 %). This is in view of the increased cases of HAT that were being reported from Chama around the time of sampling [12]. According to recent reports from Uganda [6, 7], cattle have been implicated to be the principal domestic reservoirs of HAT. Cattle have also been previously documented as reservoirs of T. b. rhodesiense in Kenya [8] and Tanzania [9–11]. Furthermore, we also recently documented the detection of T. b. rhodesiense in dogs from Zambia’s Luangwa and Zambezi valley foci where HAT is re-emerging [17, 18]. Although we did not investigate the presence of trypanosome species in wildlife, several wild animals have previously been documented to act as reservoirs of T.b. rhodesiense in Zambia’s Luangwa valley ecosystem, including greater kudus (Tragelaphus strepsiceros), warthogs (Phacochoerus africanus), bushbuck (Tragelaphus scriptus), duiker (Sylvicapra grimmia), giraffe (Girraffa camelopardalis), impala (Aepyceros melampus), lion (Panthera leo), waterbuck (Kobus ellipsiprymnus), zebra (Equus quagga boehmi) and African buffalo (Syncerus caffer) [19, 41–43]. The evidence of detection of T. b. rhodesiense in G. m. morsitans in the present study is intriguing in view of the latter’s ability to take blood meals from multiple hosts including wildlife, domestic animals and humans (Homo sapiens) (mainly wildlife staff and hunters), facilitating the circulation of the parasite within the ecosystem. Dennis et al. [44] also recently detected T. b. rhodesiense, mainly in G. m. morsitans (35.3 %), and to a lesser extent in G. pallidipes (0.5 %), in their studies in Zambia’s Luambe National Park, within the Luangwa valley ecosystem. Taken together, these data suggest that G. m. morsitans flies are the main vectors of T. b. rhodesiense in the Luangwa valley ecosystem. It is, however, noteworthy that the actual prevalence of T. b. rhodesiense in both cattle blood and tsetse flies may be higher in view of the fact that SRA is a single copy gene which may be difficult to detect in low parasitaemias where there is not enough DNA to amplify it [6, 45]. Moreover, we may not establish from our assays whether we were detecting active infection in the tsetse flies or residual DNA from dead trypanosomes that the flies may have picked following a blood meal from treated animals.
In agreement with Anderson et al. [19], results obtained in the present study indicate that various trypanosome species circulate within a wide and diverse host community in the bio-diverse Luangwa valley ecosystem. The epidemiological stability of the diverse Luangwa valley ecosystem is being eroded by an influx of people and their livestock, encroaching into the Luangwa valley. Overall, these data should be used to trigger a “One Health” approach towards HAT control through disease intervention in livestock, wildlife and tsetse vectors [46].
Conclusions
The present study employed several diagnostic tools to examine the presence of different trypanosome species in cattle, goats and tsetse flies. There was poor agreement among the test methods used for detection of trypanosomes species in animal blood and tsetse flies (Kappa <0.40). Microscopy generally exhibited relatively lower sensitivity than PCR and LAMP. Among the molecular tests, the KIN-PCR was found to be sensitive for the detection of T. congolense but not for T. vivax or Trypanozoon which were better detected by TviCatL-PCR and PFR-LAMP, respectively. Thus the KIN-PCR may be recommended in epidemiological surveillance of T. congolense in AAT endemic regions, while species-specific PCR and/or LAMP might be of greater value in the detection of T. vivax or Trypanozoon, including human-infective T. b. rhodesiense.
Abbreviations
- AAT:
-
Animal African trypanosomiasis
- DDW:
-
Double distilled water
- HAT:
-
Human African trypanosomiasis
- SRA:
-
Serum resistance-associated
- ITS:
-
internal transcribed spacers of the ribosomal DNA
- KIN:
-
primers that target the internal transcribed spacer region of kinetoplastid ribosomal DNA
- LAMP:
-
Loop-mediated isothermal amplification
- PCR:
-
Polymerase chain
- PCV:
-
Packed cell volume reaction
- PFR:
-
Paraflagella rod A gene
- SRA:
-
Human serum resistance-associated antigen
References
Kristjanson PM, Swallow BM, Rowlands GJ, Kruska RL, de Leeuw PN. Measuring the costs of African animal trypanosomosis, the potential benefits of control and returns to research. Agric Syst. 1999;59:79–98.
Snow WF, Wacher TJ, Rawlings P. Observations on the prevalence of trypanosomosis in small ruminants, equines and cattle, in relation to tsetse challenge, in The Gambia. Vet Parasitol. 1996;66:1–11.
Van den Bossche P, Chitanga S, Masumu J, Marcotty T, Delespaux V. Virulence in Trypanosoma congolense Savannah subgroup. A comparison between strains and transmission cycles. Parasite Immunol. 2011;33:456–60.
Simukoko H, Marcotty T, Phiri I, Geysen D, Vercruysse J, Van den Bossche P. The comparative role of cattle, goats and pigs in the epidemiology of livestock trypanosomiasis on the plateau of eastern Zambia. Vet Parasitol. 2007;147:231–8.
Van den Bossche P. Some general aspects of the distribution and epidemiology of bovine trypanosomosis in southern Africa. Int J Parasitol. 2001;31:592–8.
Welburn SC, Picozzi K, Fevre EM, Coleman PG, Odiit M, Carrington M, et al. Identification of human infective trypanosomes in animal reservoir of sleeping sickness in Uganda by means of serum-resistance-associated (SRA) gene. Lancet. 2001;358:2017–19.
von Wissmann B, Fyfe J, Picozzi K, Hamill L, Waiswa C, Welburn SC. Quantifying the association between bovine and human trypanosomiasis in newly affected sleeping sickness areas of Uganda. PLoS Negl Trop Dis. 2014;8:e2931.
Von Wissman B, Machila N, Picozzi K, Fe`vre EM, Bronsvoort BM, Handel IG, et al. Factors associated with acquisition of human infective and animal infective trypanosome infections in domestic livestock in western Kenya. PLoS Negl Trop Dis. 2011;5:e941.
Kaare MT, Picozzi K, Mlengeya T, Fèvre EM, Mtambo MM, Mellau LS, et al. Sleeping sickness - a re-emerging disease in the Serengeti? Travel Med Infect Dis. 2007;5:117–24.
Auty HK, Picozzi K, Malele I, Torr SJ, Cleaveland S, Welburn S. Using molecular data for epidemiological inference: assessing the prevalence of Trypanosoma brucei rhodesiense in tsetse in Serengeti Tanzania. PLoS Negl Trop Dis. 2012;6:e1501.
Welburn SC, Picozzi K, Coleman PG, Packer C. Patterns in Age-Seroprevalence Consistent with Acquired Immunity against Trypanosoma brucei in Serengeti Lions. PLoS Negl Infect Dis. 2008;2:e347.
Namangala B, Hachaambwa L, Kajino K, Mweene AS, Hayashida K, Simuunza M, et al. The use of Loop-mediated Isothermal Amplification (LAMP) to detect the re-emerging Human African Trypanosomiasis (HAT) in the Luangwa and Zambezi valleys. Parasit Vectors. 2012;5:282.
Mwanakasale V, Songolo P. Disappearance of some human African trypanosomiasis transmission foci in Zambia in the absence of a tsetse fly and trypanosomiasis control program over a period of forty years. Trans R Soc Trop Med Hyg. 2011;105:167–72.
Mwanakasale V, Songolo P, Daka V. Challenges in the control of Human African Trypanosomiasis in the Mpika district of Zambia. BMC Research Notes. 2013;6:180.
Hayashida K, Kajino K, Hachaambwa L, Namangala B, Sugimoto C. Direct blood dry LAMP: a rapid, stable, and easy diagnostic tool for Human African Trypanosomiasis. PLoS Negl Trop Dis. 2015;9:e0003578.
Robinson T, Rogers D, Williams B. Mapping tsetse habitat suitability in the common fly belt of southern Africa using multivariate analysis of climate and remotely sensed vegetation data. Med Vet Entomol. 1997;11:235–45.
Namangala B, Oparaocha E, Kajino K, Hayashida K, Moonga L, Inoue N, et al. Preliminary Investigation of Trypanosomosis in Exotic Dog Breeds from Zambia’s Luangwa and Zambezi Valleys Using LAMP. Am J Trop Med Hyg. 2013;89:116–8.
Lisulo M, Sugimoto C, Kajino K, Hayashida K, Mudenda M, Moonga L, et al. Determination of the prevalence of African trypanosome species in indigenous dogs of Mambwe district, eastern Zambia, by loop-mediated isothermal amplification. Parasit Vectors. 2014;7:19.
Anderson NE, Mubanga J, Fevre EM, Picozzi K, Eisler MC, Thomas R, et al. Characterisation of the wildlife reservoir community for human and animal trypanosomiasis in the Luangwa Valley Zambia. PLoS Negl Trop Dis. 2011;5:e1211.
Chappuis F, Loutan L, Simarro P, Lejon V, Büscher P. Options for field diagnosis of human African trypanosomiasis. Clin Microbiol Rev. 2005;18:133–46.
Bronsvoort BMC, Wissmann B, Fe`vre EM, Handel IG, Picozzi K, Welburn SC. No Gold Standard Estimation of the Sensitivity and Specificity of Two Molecular Diagnostic Protocols for Trypanosoma brucei species in Western Kenya. PLoS One. 2010;5:e8628.
Thekisoe OM, Kuboki N, Nambota A, Fujisaki K, Sugimoto C, Igarashi I, et al. Species-specific loop-mediated isothermal amplification (LAMP) for diagnosis of trypanosomosis. Acta Trop. 2007;102:182–9.
Desquesnes M, Dávila AM. Applications of PCR-based tools for detection and identification of animal trypanosomes: a review and perspectives. Vet Parasitol. 2002;109:213–31.
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63.
Desquesnes M, McLaughlin G, Zoungrana A, Dávila AMR. Detection and identification of Trypanosoma of African livestock through a single PCR based on internal transcribed spacer 1 of rDNA. Int J Parasitol. 2001;31:609–13.
Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RC, et al. The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res. 2005;95:186–92.
Cox A, Tilley A, McOdimba F, Fyfe J, Eisler MC, Hide G, et al. A PCR based assay for detection and differentiation of African trypanosome species in blood. Exp Parasitol. 2005;111:24–9.
Adams ER, Hamilton PB, Malele II, Gibson WC. The identification, diversity and prevalence of trypanosomes in field caught tsetse in Tanzania using ITS-1 primers and fluorescent fragment length barcoding. Infect Genet Evol. 2008;8:439–44.
Cox AP, Tosas O, Tilley A, Picozzi K, Coleman P, Hide G, et al. Constraints to estimating the prevalence of trypanosome infections in East African zebu cattle. Parasit Vectors. 2010;3:82.
Ahmed HA, MacLeod ET, Hide G, Welburn SC, Picozzi K. The best practice for preparation of samples from FTA®cards for diagnosis of blood borne infections using African trypanosomes as a model system. Parasit Vectors. 2011;4:68.
Ahmed HA, Picozzi K, Welburn SC, MacLeod ET. A comparative evaluation of PCR- based methods for species- specific determination of African animal trypanosomes in Ugandan cattle. Parasit Vectors. 2013;6:316.
Wastling SL, Picpzzi K, Kakembo AS, Welburn SC. LAMP for human African trypanosomiasis: a comparative study of detection formats. PLoS Negl Trop Dis. 2010;4:e865.
McDermott J, Woitag T, Sidibe I, Bauer B, Diarra B, Ouédraogo D, et al. Field studies of drug-resistant cattle trypanosomes in Kénédougou Province, Burkina Faso. Acta Trop. 2003;86:93–103.
Cortez AP, Rodrigues AC, Garcia HA, Neves L, Batista JS, Bengaly Z, et al. Cathepsin L-like genes of Trypanosoma vivax from Africa and South America - characterization, relationships and diagnostic implications. Mol Cell Probes. 2009;23:44–51.
Thekisoe OMM, Bazie RSB, Coronel-Servian AM, Sugimoto C, Kawazu S-I, Inoue N. Stability of Loop-Mediated Isothermal Amplification (LAMP) reagents and its amplification efficiency on crude trypanosome DNA templates. J Vet Med Sci. 2009;71:471–5.
Laohasinnarong D, Thekisoe OM, Malele I, Namangala B, Ishii A, Goto Y, et al. Prevalence of Trypanosoma sp. in cattle from Tanzania estimated by conventional PCR and loop - mediated isothermal amplification (LAMP). Parasitol Res. 2011;109:1735–9.
Viera AJ, Garrett JM. Understanding interobserver agreement: the kappa statistic. Fam Med. 2005;37:360–3.
Bengaly Z, Sidibe I, Ganaba R, Desquesnes M, Boly H, Sawadogo L. Comparative pathogenicity of three genetically distinct types of Trypanosoma congolense in cattle: clinical observations and haematological changes. Vet Parasitol. 2002;108:1–19.
Marcotty T, Simukoko H, Berkvens D, Vercruysse J, Praet N, Van den Bossche P. Evaluating the use of packed cell volume as an indicator of trypanosomal infections in cattle in eastern Zambia. Prev Vet Med. 2008;87:288–300.
Muleya W, Namangala B, Simuunza M, Nakao R, Inoue N, Kimura K, et al. Population genetic analysis and sub-structuring of Theileria parva in the northern and eastern parts of Zambia. Parasit Vectors. 2012;5:255.
Rickman LR, Ernest A, Kanyangala S, Kunda E. Human serum sensitivities of Trypanozoon isolates from naturally infected hosts in the Luangwa Valley, Zambia. East Afr Med J. 1991;68:880–92.
Dillmann JS, Townsend AJ. Trypanosomiasis survey of wild animals in the Luangwa Valley, Zambia. Acta Trop. 1979;36:349–56.
Awan MA. Identification by the blood incubation infectivity test of Trypanosoma brucei subspecies isolated from game animals in the Luangwa Valley, Zambia. Acta Trop. 1979;36:343–7.
Dennis JW, Durkin SM, Horsley-Downie JE, Hamill LC, Anderson NE, MacLeod ET. Sodalis glossinidius prevalence and trypanosome presence in tsetse from Luambe National Park Zambia. Parasit Vectors. 2014;7:378.
Picozzi K, Carrington M, Welburn SC. A multiplex PCR that discriminates between Trypanosoma brucei brucei and zoonotic T. b. rhodesiense. Exp Parasitol. 2008;118:41–6.
Hamill CL, Magai TK, Welburn SC, Picozzi K. Domestic pigs as potential reservoirs of human and animal trypanosomiasis in Northern Tanzania. Parasit Vectors. 2013;6:322.
Acknowledgement
This study was supported by a Grant-in-Aid for scientific Research from JSPS to SK, CS and NI. The authors acknowledge the farmers, the Zambia wildlife authority (ZAWA), the respective Provincial and District Veterinary Officers, the technical staff at the University of Zambia, School of Veterinary Medicine, for their assistance and support and the logistic support provided by Yuka Suzuki, HUCZCZ-Lusaka, Zambia during the study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DL helped to conceive the study, participated in its design, collected samples, purified the DNA from cattle blood, performed Microscopy, PCR, LAMP and blood meal analysis assays, analyzed data and drafted the manuscript. YG, MA, RN, KH and KK, helped to conceive the study, participated in its design, sample collection and laboratory work. SK, CS and NI helped to conceive the study, participated in its design, field and laboratory work, analyzed data, helped to edit the manuscript and obtained funding. BN helped to conceive the study, participated in its design, field and laboratory work, analyzed data, helped to draft and edit the manuscript. All the authors read and approved the final manuscript.
An erratum to this article is available at http://dx.doi.org/10.1186/s13071-015-1173-y.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Laohasinnarong, D., Goto, Y., Asada, M. et al. Studies of trypanosomiasis in the Luangwa valley, north-eastern Zambia. Parasites Vectors 8, 497 (2015). https://doi.org/10.1186/s13071-015-1112-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-015-1112-y