Abstract
Background
Black flies (Diptera: Simuliidae) are haematophagous insects that can cause allergic reactions and act as vectors of pathogens. Although their saliva has been thought to contain a diverse array of physiologically active molecules, little information is available on antimicrobial factors in black fly salivary glands, especially no defensins have been reported so far.
Methods
A novel cationic defensin designated SibaDef was purified using reverse phase high-performance liquid chromatography (RP-HPLC) from the salivary glands of the black fly Simulium bannaense. The amino acid sequence of SibaDef was determined by a combination method of automated Edman degradation and cDNA sequencing. The morphologic changes of Gram-positive bacteria Staphylococcus aureus or Bacillus subtilis treated with SibaDef were assessed by scanning electron microscopy (SEM). Quantitative PCR (qPCR) was performed to analyze the expression of SibaDef mRNA in whole bodies of insects after oral infection with the bacteria S. aureus or B. subtilis.
Results
Surprisingly, the phylogenetic analysis of defensin-related amino acid sequences demonstrated that SibaDef is most closely related to defensins from the human body louse Pediculus humanus corporis (Anoplura: Pediculidae), rather than to other dipteran defensins. SibaDef showed potent antimicrobial activities against Gram-positive bacteria with minimal inhibitory concentrations (MICs) ranging from 0.83 μM to 2.29 μM. SEM analysis indicated that SibaDef killed microorganisms through the disruption of cell membrane integrity. The transcript levels of SibaDef in the bacteria-immunized flies increased with the time course, reaching maximum at 36 h and then slowly decreased from that time point.
Conclusions
Our results indicate that SibaDef is involved in the innate humoral response of the black fly S. bannaense, and it might play a significant role in the defence against microorganisms in both sugar and blood meals.
Similar content being viewed by others
Background
Black flies (Diptera: Simuliidae) are closely related to some blood-sucking insects such as mosquitoes and biting midges [1,2]. They are not only a biting nuisance for humans and livestock but also transmit diseases including human onchocerciasis (river blindness) caused by the nematode Onchocerca volvulus and livestock disease caused by vesicular stomatitis virus [3-5]. To facilitate a blood meal, haematophagous Diptera have developed an extraordinary array of salivary proteins that can overcome the host’s hemostatic barriers, as well as suppressing inflammatory and immunologic reactions [6-9]. In addition, these Diptera also take sugar meals.
Several salivary anti-haemostatic factors have been identified from black fly, including inhibitors of coagulation factors (Factor Xa, V and thrombin), potent vasodilators (Simulium vittatum erythema proteins, SVEPs) and anti-platelet aggregation factors (apyrase) [10-16]. Hyaluronidase and immunomodulatory activities have also been described in S. vittatum salivary gland extract [5,17,18].
As an important hematophagous arthropod, there was not much information available about pharmacologically active compounds in black fly salivary glands, until salivary transcriptomes have been made and described from three black fly species (S. guianense, S. vittatum and S. nigrimanum) [19-21]. In these studies, many more substances with potential anti-haemostatic functions or immunity-related activities have been uncovered. Immunity-related gene products including six antimicrobial peptides of the cecropin family, nine lysozymes, and three members of the Gram-negative bacteria-binding protein, have been identified from these three species. However, no antimicrobial peptide (AMP) belonging to the defensin family has so far been biochemically characterized from black fly.
Insect defensins are a class of gene-encoded effector molecules of innate immunity. They have six strictly conserved cysteine residues linked in the 1–4, 2–5, 3–6 pattern, except for the antifungal peptide drosomycin from Drosophila melanogaster, which has eight cysteine residues forming four stabilizing disulfide bridges [22]. So far, more than 60 defensins have been identified from different species of insect orders (Diptera, Lepidoptera, Hymenoptera, Hemiptera, Isoptera, Coleoptera and Odonata) [22,23], The majority of insect defensins were isolated from the haemolymph, fat body or midgut of bacteria-immunized larvae [24-27], while such defensins were seldom reported from the saliva or salivary glands. In the current work, we firstly report the purification and characterization of the defensin from the black fly salivary glands.
Methods
Black fly salivary gland dissection
Adult S. bannaense (about 2,000 flies) were collected near streams in Xishuangbanna, Yunnan, China (21.556°N 101.162°E). The collections were made in five months (April-May, September-October 2013; May 2014). The black fly salivary glands (1,800 pairs) used for protein extraction (1,660 pairs) or total RNA extraction (140 pairs), were dissected in ice cold HEPES saline (10 mM HEPES pH 7.2, 150 mM NaCl) using fine entomological needles under a tereomicroscope, and stored in liquid nitrogen until use. The study was approved by the Animal Care and Use Ethics Committee of Kunming Medical University.
Peptide purification
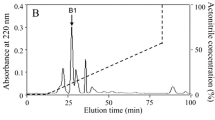
1,660 pairs of black fly salivary glands in HEPES saline were thawed and homogenized. After a centrifugation at 12,000 × g for 15 min at 4°C, the supernatant was prepurified through a 10-kDa cut-off Centriprep filter (Millipore, CA). The filtrate was then subjected to RP-HPLC on an Inertsil C4 column (25 × 0.46 cm) as illustrated in Figure 1A. The linear gradient elution was performed in a 0–70% acetonitrile containing 0.1% (v/v) trifluoroacetic acid for 80 min. The eluted peaks of A1 and A2 showed antimicrobial activities. The protein peak of A2 was pooled, lyophilized, and further purified by RP-HPLC on a Wondasil C18 column (25 × 0.46 cm) as indicated in Figure 1B. Elution was performed with a linear gradient of 0-60% acetonitrile in acidified water over 70 min at a flow rate of 0.7 ml/min. The antimicrobial activity of fractions was determined as indicated below. The interesting eluted peaks were subjected to automated Edman degradation analysis on an Applied Biosystems pulsed liquid-phase sequencer (model ABI 491, USA).
Isolation of Siba Def from the salivary gland of S. bannaense and MALDI–TOF MS. (A) The filtrate of the salivary gland homogenate of S. bannaense by 10 kDa cut-off was divided by an Inertsil C4 RP-HPLC column (25 × 0.46 cm) equilibrated with 0.1% (v/v) trifluoroacetic acid/water. The elution was performed with the indicated gradient of acetonitrile at a flow rate of 1 ml/min. (B) The eluted peak of A2 containing antimicrobial activity was further purified by C18 RP-HPLC column (25 × 0.46 cm) developed with a linear gradient of 0 to 70% acetonitrile in acidified water at a flow rate of 0.7 ml/min. The purified antimicrobial peptide is indicated by an arrow. (C) MALDI-TOF mass spectrometry analysis of the antimicrobial peptide.
MALDI-TOF MS analysis
1 μl of the eluted peak with antimicrobial activity was spotted onto a matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) plate with 1 μl of α-cyano-4-hydroxycinnamic acid matrix (10 mg/ml in 60% acetonitrile) and analyzed by an UltraFlex I mass spectrometer (Bruker Daltonics, Germany) in a positive ion mode.
cDNA library construction and screening of cDNA encoding defensin
Total RNA was extracted using TRIzol reagent (Invitrogen, USA) from salivary glands of S. bannaense. mRNA was purified from the total RNA by affinity chromatography in oligo(dT) cellulose columns (Promega, USA) and then used for cDNA library construction by the In-Fusion SMARTer™ Directional cDNA Library Construction Kit (Takara, Japan) according to the instructions of the manufacturer.
The synthesized second-strand cDNAs was used as a template for PCR to screen the cDNAs encoding defensin. Primers used in this research are shown in Table 1. SibaDef-F1 and 3′ PCR primer were used in PCR reaction to screen the 5′ fragments of cDNAs encoding defensin. SibaDef-F1 is designed from the amino acid sequence of SibaDef determined by Edman degradation, and 3′ PCR primer is based on the adaptor sequence of 3′ In-Fusion SMARTer CDS Primer provided in the kit. The PCR conditions were: 95°C for 5 min and 30 cycles of 95°C (30 s), 58°C (40 s), 72°C (1 min) followed by an extension step at 72°C for 10 min. The PCR product was purified by gel electrophoresis, cloned into pMD19-T vector (Takara, Japan) for sequencing. After the 3′ fragments of cDNA had been obtained, an antisense primer (SibaDef-R1) was designed based on the 3′-coding region of defensin cDNA and coupled with 5′ PCR primer provided in the kit to screen the full length cDNA encoding defensin. The PCR conditions were: 95°C for 5 min and 30 cycles of 95°C (30 s), 56°C (30 s), 72°C (50 s) followed by an extension step at 72°C for 8 min. DNA sequencing was performed on an Applied Biosystems DNA sequencer (model ABI PRISM 377, USA).
Sequence analysis
Deduced defensin sequence was performed with ExPASy Translate Tool (http://web.expasy.org/translate/). Database searches were performed with Blastx (http://www.ncbi.nlm.nih.gov/), and the amino acid sequence identity between defensin sequences was aligned using ClustalW (http://embnet.vital-it.ch/software/ClustalW.html) [28]. The theoretical isoelectric point (pI) and molecular weight (Mw) were carried out using ExPASy Compute pI/Mw tool (http://web.expasy.org/compute_pi/) [29]. The dendrogram was drawn using the neighbor-joining (NJ) method in the Mega 5 package. A total of 1,000 bootstrap replicates were used to test the reliability of each branch.
Antimicrobial assay
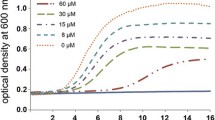
The microbicidal activity of SibaDef was evaluated as described in our previous papers [6,30]. Briefly, bacteria were cultured in Mueller-Hinton broth (MH broth) at 37°C to exponential phase and diluted with fresh MH broth to 5 × 105 colony-forming units (CFUs)/ml. Aliquots (50 μl) of serial dilutions of sample were dispensed into a 96-well microtiter plate and mixed with 50 μl of bacteria inoculums in MH broth. The microtiter plate was incubated at 37°C for 18 h, and the absorbance at 600 nm was measured using an automatic microplate spectrophotometer. The minimal concentrations at which no growth of microorganisms occurred were recorded as minimal inhibitory concentration (MIC).
Hemolytic assay
Hemolytic assay was conducted as previously reported [31]. Serial dilutions of SibaDef were incubated with washed human erythrocytes at 37°C for 30 min and then the cells were centrifuged at 1,000 × g for 5 min. The absorbance of supernatant was measured at 540 nm. 1% (v/v) Triton X-100 was used to determine the maximal hemolysis and 0.9% saline was used as negative control.
SEM
The morphologic changes of the bacteria treated with SibaDef were assessed by SEM as previously reported [31]. Gram-positive bacteria S. aureus ATCC 6538 and B. subtilis ATCC 6633 were cultured in MH broth to exponential phase respectively, and then incubated with SibaDef (1 × MIC) at 37°C for 45 min. After a centrifugation at 1,000 × g for 10 min, bacteria pellets were fixed with 2.5% glutaraldehyde solution for 2 h at 4°C. The bacteria were postfixed in 1% osmium tetroxide for 2 h at 4°C, and dehydrated in a graded series of alcohols. After being mounted onto aluminium stubs and vacuum sputter-coated with gold, the samples were observed with a Hitachi S-4800 SEM under standard operating conditions.
Bacterial feeding
Bacterial feed experiment was carried out as previously described [32]. The collected S. bannaense (200 flies) were fed with 70% sucrose solution ad libitum. After starving for 12 hour, black flies were fed through cotton wool with 20% sucrose solution (OD600 = 0.2) containing Gram-positive bacteria S. aureus ATCC 6538 or B. subtilis ATCC 6633. All the black flies, including the naïve (sugar fed controls), were kept under controlled conditions of temperature (26 ± 2°C), humidity (85-90%), and photoperiod (12 h/12 h). Total RNA was extracted from whole bodies of immune stimulated or naive insects at 12, 24, 36, 48 and 72 h after feeding and processed immediately as described below.
qPCR
qPCR was performed to analyze the expression of SibaDef mRNA in whole bodies of immune stimulated or naive insects, with the housekeeping gene β-actin as an endogenous control. As listed in Table 1, primers for SibaDef amplification were designed on the SibaDef cDNA sequence, and β-actin was amplified using primers based on the sequence from black fly S. vittatum (GenBank accession number AY083375.1). PrimeScript® Reverse Transcriptase (Takara, Japan) and SYBR green master mix (Takara, Japan) were used following the manufacturer’s instruction.
q-PCR was performed on a Realplex Mastercycler real-time PCR system (Eppendorf, Germany) with the following parameters: 95°C for 2 min, and 40 cycles of 95°C for 30 s,60°C for 30 s. SibaDef mRNA expression level was calculated following normalization to β-actin by ΔΔCt method. The accuracy of qPCR was verified by melt curve analysis.
Homology modeling
Defensin homology modeling was performed by Easymodeller version 2.0 [33]. The solution NMR structure of Sapecin (PDB entry code 1L4V) from Sarcophaga peregrine (Diptera: Sarcophagidae) was used as the template because this defensin antimicrobial peptide shared the highest identity of 44% with SibaDef. The comparative three-dimensional structure model of SibaDef was optimized using PYMOL software (http://www.pymol.org).
Data and statistical analysis
Statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software Inc., San Diego, CA, USA) and Stata 10.0 software (StataCorporation, College Station, TX, USA). Data were presented as mean ± standard errors of mean, and compared using two-tailed equal variance Student’s t-test. P < 0.05 was considered as statistical significance.
Results
Characterization of SibaDef
The fractions with antimicrobial activity (marked by A2) were collected, lyophilized, and further purified by C18 RP-HPLC as illustrated in Figure 1B. After Edman degradation, a primary structure of 18 amino acid residues was identified with the following sequence: ATCDLLSISTPWGSVNSA. MALDI-TOF MS analysis (Figure 1C) indicated that the peptide (SibaDef) had a measured molecular mass of 4795.23 Da, matching well with the calculated molecular mass 4795.55 Da. The complete nucleotide sequence of cDNA (GenBank accession KJ842485) and deduced amino acid sequence of SibaDef precursor are shown in Figure 2. The N-terminal deduced sequence of SibaDef precursor is completely consistent with the result of Edman degradation sequencing. The cDNA encoding protein precursor is composed of 105 amino acid residues, including a predicted 22 amino acid signal peptide, a 37 amino acid propeptide region and a 46 amino acid mature SibaDef peptide. There is a characteristic dipeptide cleavage site (−R58R59-) for trypsin-like proteases between propeptide and mature peptide. Analysis using the ExPASy MW/pI tool showed that it has a predicted pI of 8.94. The eluted peak of A1 containing antimicrobial activity was also purified, sequenced and aligned well with other insect cecropins (data not shown).
The cDNA sequence of Siba Def precursor and the deduced amino acid sequence. Deduced amino acid sequence is shown below the cDNA sequence. The amino acid sequence of mature peptide is underlined and the stop codon is indicated by an asterisk. The dibasic cleavage site and the putative polyadenylation consensus signal are italicized and gray shaded. Amino acid numbers or nucleotide numbers are shown after the sequences.
BLAST search indicated that the amino acid sequence identity between SibaDef and their homologues from different insect species varied widely, ranging from 52% to 67%. Surprisingly, SibaDef shared the highest identity of 67% (31/46) with the defensin-2 from the human body louse P. humanus corporis (Anoplura: Pediculidae).
Multi-sequence alignment of insect defensin precursors (Figure 3) indicated that the signal peptide and propeptide region of these sequences are divergent. However, sixteen amino acids residues within the mature peptides are highly conserved, including a signature motif of six conserved cysteines and an additional ten residues (Ala60, Thr61, Asp63, Ser66, His76, Ala80, His82, Gly92, Gly93 and Arg104). A characteristic feature of all the mature peptides is the presence of an alanine residue and a threonine residue (−AT-) at the N-terminus. In addition, there are two basic residues (−RR- or -RK-) at the C-terminus of the mature peptide, except for the defensin A from Nilaparvata lugens, which possesses an arginine residue and an asparagine residue (−RN-) at the C-terminus.
Alignment of the amino acid sequence of Siba Def with different insect defensins. These sequences were based on BLAST search results. The symbols under the alignment indicate: (*) identical sites; (:) conserved sites; (.) less conserved sites. The six conserved cysteine residues involved in disulfide bridges are grey shaded and activation peptide cleavage sites are marked with a triangle. GenBank accession numbers for the analyzed sequences are shown in Figure 4.
Phylogenetic analysis
The phylogenetic tree was generated from 53 defensin-related amino acid sequences (25 insect species including 11 Diptera, 6 Hymenoptera, 4 Hemiptera, 2 Coleoptera, 1 Anoplura and 1 Homoptera). As showed in Figure 4, all defensin sequences are divided into two distinct clusters including 47 sequences derived from different orders of insects (Diptera, Hemiptera, Coleoptera, Anoplura and Homoptera) and 6 sequences derived from hymenopteran insects, respectively. SibaDef was grouped together with the anopluran defensins (defensin-2 and defensin) from the human body louse P. humanus corporis.
Antimicrobial activity
The MICs of SibaDef against Gram-positive and Gram-negative bacteria were determined. As listed in Table 2, SibaDef showed strong antimicrobial activities against four tested Gram-positive bacteria, with MICs ranging from 0.83 μM to 2.29 μM. However, no effect was observed against Gram-negative bacteria E. coli and P. aeruginosa.
Hemolysis
Human fresh erythrocytes were used to evaluate the hemolytic activity of SibaDef. The result showed SibaDef displayed negligible hemolytic activity on human erythrocyte even with peptide concentrations up to 41.71 μM, which is almost 40-fold higher than their corresponding MIC values.
SEM
SEM was performed to study the possible mechanisms of action of SibaDef on Gram-positive bacteria S. aureus and B. subtilis. In contrast to the untreated S. aureus cells (Figure 5A) and B. subtilis (Figure 5C), cells treated with SibaDef (1 × MIC) showed obvious morphological alterations (Figure 5B, D). The membrane integrity of cells seemed to be disrupted, and there were a large number of filaments on the surface of cells. In addition, exposure of S. aureus to SibaDef resulted in aggregation (Figure 5B).
Scanning electron microscopy analysis of Siba Def-treated bacteria. (A) Control, untreated S. aureus. (B) 1 × MIC (0.83 μM) SibaDef-treated S. aureus. (C) Control, untreated B. subtilis. (D) 1 × MIC (1.04 μM) SibaDef-treated B. subtilis. Arrow indicates severe leakage of cellular cytoplasmic contents.
Transcription of SibaDef in black flies fed on bacteria
After S. aureus or B. subtilis ingestion, the expression levels of SibaDef mRNA in whole bodies of bacteria-immunized or naive insects were compared at the different time course. As illustrated in Figure 6A, the levels of SibaDef mRNA were up-regulated by bacterial-challenge at 12, 24, 36, 48 and 72 h after S. aureus ingestion (9.8, 17.4, 31.1, 22.6 and 18.5 fold, respectively). After B. subtilis ingestion, the fold increase in defensin transcription at different time course (12.3, 20.9, 34.7, 26.8 and 21.7 fold, respectively) was shown in Figure 6B. The expression of defensin mRNA peaked at 36 h (31.1 and 37.4 fold, respectively) and relatively decreased with time.
Fold increase of Siba Def in whole bodies of insects after oral infection with bacteria at different time course. (A) Fold increase of sibaDef in insects after S. aureus ingestion. (B) Fold increase of SibaDef in insects after B. Subtilis ingestion. Expression levels in whole bodies of bacteria-immunized insects were calculated relative to the level of SibaDef in corresponding naive insects, which was arbitrarily defined as 1. Values for infection treatment are significantly different from control values. *P < 0.05, **P < 0.01 significantly different compared to the control (n = 9).
3D structure analysis of SibaDef
The homology modeled structures of SibaDef are shown in Figure 7. The common motifs are preserved in the sequence alignment of the template and SibaDef structures. It consists of one α-helix (residues Gly13-His23, in red), two antiparallel β-sheets (residues Ala26-Phe31 and Tyr35-Lys39, in green) and some random coils (in blue) locating at both terminal end of SibaDef and regions between α-helix and β-sheets (Figure 7A). It also shows the positive charges distribution of SibaDef (five basic residues) in the surface of the three-dimensional structure (Figure 7B, in red). Electrostatic surface analysis revealed that several regions of the solution structure surface are positively charged at a neutral pH (Figure 7C, in blue). Taken together, SibaDef shared common structural features and electrostatic characteristics with a variety of insect defensins.
Homology modeling of Siba Def. (A) Representation of the homology-derived solution structure of SibaDef. It consists of one α-helix (in red), two antiparallel β-sheets (in green) and some random coils (in blue). (B) Basic residues (Lys and Arg, in red) are displayed in the structures. (C) Electrostatic potential map of SibaDef, positively charged region and negatively charged region are shown in blue and red, respectively. Homology model was performed by Easymodeller version2.0 and optimized by using PYMOL software.
Discussion
Insects lack an acquired immune response, but they have an unspecific cellular response (phagocytosis and encapsulation of invading microorganisms by blood cells) and humoral immune reactions (activation of proteolytic pathways and the rapid synthesis of immune-related peptides) [34]. These peptides are synthesized either by the fat body and various epithelia in holometabolous insects, or by hemocytes in heterometabolous insects [23]. Extensive research in the past decades has established that insect AMPs are ubiquitous and ancient contributors to immune defense against bacterial, fungal and parasitic infections [23,26,35]. As an important blood-sucking insect, there have been comparatively few studies on antimicrobial substances in black fly, especially no defensins have been reported so far.
Here, a novel cationic defensin designated SibaDef was purified from the salivary glands of the black fly S. bannaense, The structural organization of SibaDef precursor (Figure 2) is similar to other insect defensin precursors, comprising a signal peptide sequence, an N-terminal propeptide region containing several aspartic and glutamic acid residues, and the mature peptide at the C-terminus of the precursor. These sequences also share the conserved enzymatic processing sites (−KR- or -RR-) to release the mature peptides. The dibasic cleavage site (Figure 3) has been found in many insect defensins identified from the different orders (Diptera, Anoplura, Coleoptera, Homoptera and Hemiptera) [22,23,26]. The first two amino acid residues (AT) at the N-terminus of SibaDef is conserved in phylogenetically higher insects such as mosquitos and triatomines [36]. The consensus motifs of SibaDef are C-X16-C-X3-C-X11-C-X5-C-X1-C (where C is a cysteine, and X is any amino acid except cysteine), which is consistent with the spacing pattern of insect defensins (C-X5–16-C-X3-C-X9–11-C-X4–7-C-X1-C) [37].
Phylogenetic analysis (Figure 4) showed that SibaDef is most closely related to anopluran defensins from the human body louse P. humanus corporis, rather than to other dipteran defensins. The evolutionary trends of insect/mosquito defensins have revealed the similar outcomes, in which two dipteran defensins (Agd3 and Agd4) from Anopheles gambiae are grouped with lepidopterans more than with mosquitoes. In addition, a lepidopteran defensin (Mbd1) from cabbage moth Mamestra brassicae is clustered with the members of the mosquito specific cluster [38]. However, no meaningful explanation for these associations can be found. Previous research on evolution of invertebrate defensins has shown that the available data in hand is inadequate to provide an integrated view of the evolutionary history of AMPs [39]. We suggest that defensins in P. humanus corporis and SibaDef, possibly perform similar functions in vivo due to tremendous evolutionary pressure such as the immune pressure imposed by the vertebrate hosts.
Insect defensins are classified into antimicrobial defensins and antifungal defensins according to their in vitro activity against bacteria or filamentous fungi [23]. The antimicrobial defensins are known to be active mainly against Gram-positive bacteria at different concentrations (MICs ranging from 0.4 μM to 100 μM) [26,40]. They interact with negatively charged bacterial membranes and insert into membrane bilayers to form pores, leading to membrane permeabilization and disruption [41]. Homology modeling of SibaDef (Figure 7) shows that it has a cationic structure with one α-helix and two antiparallel β-strands. The structure contributes to the ability of antimicrobial defensins to kill bacteria [23]. As expected, SibaDef shows strong activities (Table 2) against Gram-positive bacteria (MICs ranging from 0.83 to 2.29 μM). SEM analysis indicated that such activities are carried out with a lytic effect on the bacterial membranes (Figure 5). These results confirm that the microbial membrane is a key target for cationic defensins. The potent antimicrobial effect of SibaDef facilitates the prevention of bacterial contamination in sugar or blood meal acquisition. However, the actual functions of defensin in the salivary gland of haematophagous insects remain to be elucidated.
The transcript levels of SibaDef in whole bodies of insects increase after oral infection with Gram-positive bacteria S. aureus or B. subtilis, and peak at 36 h post-feeding (Figure 6). In these experiments, black flies challenged with B. subtilis express relatively higher levels of defensin mRNA when compared to those insects challenged with S. aureus at different time course. Meanwhile, we also observe increased levels of transcription for cecropin in S. bannaense after infection (data not shown). These results suggest that SibaDef involves in S. bannaense innate humoral response and cooperates with other immune-related peptides such as cecropin to control bacterial infection.
Conclusions
In conclusion, the black fly defensin was first identified in the present work by peptide purification and molecular cloning procedures. This defensin exhibited potent antimicrobial activity against Gram-positive bacteria through the disruption of microbial membrane. Further work needs to be done to investigate what is the actual functions of this immune-related peptide during the meal and bacterial infection.
References
Nayduch D, Lee MB, Saski CA. Gene discovery and differential expression analysis of humoral immune response elements in female Culicoides sonorensis (Diptera: Ceratopogonidae). Parasit Vectors. 2014;7:388.
Nasar F, Haddow AD, Tesh RB, Weaver SC. Eilat virus displays a narrow mosquito vector range. Parasit Vectors. 2014;7:595.
Mead DG, Howerth EW, Murphy MD, Gray EW, Noblet R, Stallknecht DE. Black fly involvement in the epidemic transmission of vesicular stomatitis New Jersey virus (Rhabdoviridae: Vesiculovirus). Vector Borne Zoonotic Dis. 2004;4:351–9.
Mead DG, Ramberg FB, Besselsen DG, Mare CJ. Transmission of vesicular stomatitis virus from infected to noninfected black flies co-feeding on nonviremic deer mice. Science. 2000;287:485–7.
Tsujimoto H, Gray EW, Champagne DE. Black fly salivary gland extract inhibits proliferation and induces apoptosis in murine splenocytes. Parasite Immunol. 2012;32:275–84.
Xu XQ, Yang HL, Ma DY, Wu J, Wang YP, Song YZ, et al. Toward an understanding of the molecular mechanism for successful blood feeding by coupling proteomics analysis with pharmacological testing of horsefly salivary glands. Mol Cell Proteomics. 2008;7:582–90.
Ma DY, Wang YP, Yang HL, Wu J, An S, Gao L, et al. Anti-thrombosis repertoire of blood-feeding horsefly salivary glands. Mol Cell Proteomics. 2009;8:2071–9.
Billingsley PF, Baird J, Mitchell JA, Drakeley C. Immune interactions between mosquitoes and their hosts. Parasite Immunol. 2006;28:143–53.
Schneider BS, McGee CE, Jordan JM, Stevenson HL, Soong L, Higgs S. Prior exposure to uninfected mosquitoes enhances mortality in naturally- transmitted west nile virus infection. PLoS One. 2007;2:e1171.
Abebe M, Cupp MS, Champagne D, Cupp EW. Simulidin: a black fly (Simulium vittatum) salivary gland protein with antithrombin activity. J Insect Physiol. 1995;41:1001–6.
Abebe M, Ribeiro JM, Cupp MS, Cupp EW. Novel anticoagulant from salivary glands of Simulium vittatum (Diptera: Simuliidae) inhibits activity of coagulation factor V. J Med Entomol. 1996;33:173–6.
Jacobs JW, Cupp EW, Sardana M, Friedman PA. Isolation and characterization of a coagulation factor Xa inhibitor from black fly salivary glands. Thromb Haemost. 1990;64:235–8.
Tsujimoto H, Kotsyfakis M, Francischetti IM, Eum JH, Strand MR, Champagne DE. Simukunin from the salivary glands of the black fly Simulium vittatum inhibits enzymes that regulate clotting and inflammatory responses. PLoS One. 2012;7:e29964.
Cupp MS, Ribeiro JM, Champagne DE, Cupp EW. Analyses of cDNA and recombinant protein for a potent vasoactive protein in saliva of a blood-feeding black fly Simulium vittatum. J Exp Biol. 1998;201:1553–61.
Cupp MS, Ribeiro JM, Cupp EW. Vasodilative activity in black fly salivary glands. Am J Trop Med Hyg. 1994;50:241–6.
Cupp MS, Cupp EW, Ochoa AJ, Moulton JK. Salivary apyrase in New World blackflies (Diptera: Simuliidae) and its relationship to onchocerciasis vector status. Med Vet Entomol. 1995;9:325–30.
Ribeiro JM, Charlab R, Rowton ED, Cupp EW. Simulium vittatum (Diptera: Simuliidae) and Lutzomyia longipalpis (Diptera: Psychodidae) salivary gland hyaluronidase activity. J Med Entomol. 2000;37:743–7.
Cross ML, Cupp MS, Cupp EW, Galloway AL, Enriquez FJ. Modulation of murine immunological responses by salivary gland extract of Simulium vittatum (Diptera: Simuliidae). J Med Entomol. 1993;30:928–35.
Chagas AC, Calvo E, Pimenta PF, Ribeiro JM. An insight into the sialome of Simulium guianense (Diptera: Simuliidae), the main vector of river blindness disease in Brazil. BMC Genomics. 2011;12: 612.
Andersen JF, Pham VM, Meng Z, Champagne DE, Ribeiro JM. Insight into the Sialome of the Black Fly, Simulium vittatum. J Proteome Res. 2009;8:1474–88.
Ribeiro JM, Valenzuela JG, Pham VM, Kleeman L, Barbian KD, Favreau AJ. An insight into the Sialotranscriptome of Simulium nigrimanum, a black fly associated with Fogo Selvagem in South America. Am J Trop Med Hyg. 2010;82:1060–75.
Yi HY, Chowdhury M, Huang YD, Yu XQ. Insect antimicrobial peptides and their applications. Appl Microbiol Biotechnol. 2014;98:5807–22.
Bulet P, Stöcklin R. Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept Lett. 2005;12:3–11.
Cecilia V, Peter JW, Débora PM, Daniele PC, Cícero BM, Norman AR, et al. Humoral responses in Rhodnius prolixus: bacterial feeding induces differential patterns of antibacterial activity and enhances mRNA levels of antimicrobial peptides in the midgut. Parasit Vectors. 2014;7:232.
Lambert J, Keppi E, Dimarcq JL, Wicker C, Reichhart JM, Dunbar B, et al. Insect immunity: isolation from immune blood of the dipteran Phormia terranovae of two insect antibacterial peptides with sequence homology to rabbit lung macrophage bactericidal peptides. Proc Natl Acad Sci U S A. 1989;86:262–6.
Boulanger N, Lowenberger C, Volf P, Ursic R, Sigutova L, Sabatier L, et al. Characterization of a defensin from the sand fly Phlebotomus duboscqi induced by challenge with bacteria or the protozoan parasite Leishmania major. Infect Immun. 2004;72:7140–6.
Seufi AM, Hafez EE, Galal FH. Identification, phylogenetic analysis and expression profile of an anionic insect defensin gene, with antibacterial activity, from bacterial-challenged cotton leafworm, Spodoptera littoralis. BMC Mol Biol. 2011;12:47.
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–500.
Bjellqvist B, Hughes GJ, Pasquali C, Paquet N, Ravier F, Sancez JC, et al. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14:1023–31.
Yang HL, Wang X, Liu XH, Wu J, Liu CB, Gong W, et al. Antioxidant peptidomics reveals novel skin antioxidant system. Mol Cell Proteomics. 2009;8:571–83.
Wang YP, Zhang ZY, Chen L, Guang H, Li Z, Yang HL, et al. Cathelicidin-BF, a snake cathelicidin-derived antimicrobial peptide, could be an excellent therapeutic agent for acne vulgaris. PLoS One. 2012;6:e22120.
Telleria EL, Sant'Anna MR, Alkurbi MO, Pitaluga AN, Dillon RJ, Traub-Csekö YM. Bacterial feeding, Leishmania infection and distinct infection routes induce differential defensin expression in Lutzomyia longipalpis. Parasit Vectors. 2013;6:12.
Bhusan KK, Polamarasetty A, Pallu R. EasyModeller: a graphical interface to MODELLER. BMC Res Notes. 2010;3:226.
Hoffmann JA. Immune responsiveness in vector insects. Proc Natl Acad Sci U S A. 1997;94:11152–3.
Ferrandon D, Imler JL, Hetru C, Hoffmann JA. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7:862–74.
Waniek PJ, Castro HC, Sathler PC, Miceli L, Jansen AM, Araújo CA. Two novel defensin-encoding genes of the Chagas disease vector Triatoma brasiliensis (Reduviidae, Triatominae): gene expression and peptide-structure modeling. J Insect Physiol. 2009;55:840–8.
Ceřovský V, Bém R. Lucifensins, the insect defensins of biomedical importance: the story behind maggot therapy. Pharmaceuticals (Basel). 2014;7:251–64.
Dassanayake RS, Silva Gunawardene YI, Tobe SS. Evolutionary selective trends of insect/mosquito antimicrobial defensin peptides containing cysteine-stabilized alpha/beta motifs. Peptides. 2007;28:62–75.
Rodriguez de la Vega RC, Possani LD. On the evolution of invertebrate defensins. Trends Genet. 2005;21:330–2.
Bulet P, Hetru C, Dimarcq JL, Hoffmann D. Antimicrobial peptides in insects; structure and function. Dev Comp Immunol. 1999;23:329–44.
Hancock REW. Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect Dis. 2001;1:156–64.
Acknowledgments
This work was supported by Chinese National Natural Science Foundation (81373380, 81360253, 81260258, 81402830) and Jiangsu Province Foundation (BK20140362, 14KJD350003).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: LW, LM, YW, JW, HY. Performed the experiments: LW, LM, YW, HB, JL, YL, YH, TL, JL, CF. Analyzed the data: LW, LM, JW, HY. Contributed reagents/materials/analysis tools: JW, HY. Wrote the paper: JW, HY. All authors participated in the revision of the manuscript, read and approved the final manuscript.
Lin Wei and Lixian Mu contributed equally to this work.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Wei, L., Mu, L., Wang, Y. et al. Purification and characterization of a novel defensin from the salivary glands of the black fly, Simulium bannaense . Parasites Vectors 8, 71 (2015). https://doi.org/10.1186/s13071-015-0669-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-015-0669-9