Abstract
Background
Several antimicrobial peptides (AMPs) belonging to the cecropin family have been identified from the salivary glands of different black fly species, however, the immunological functions for these molecules were poorly understood.
Methods
A novel cecropin-like antimicrobial peptide (SibaCec) was purified using reverse phase high-performance liquid chromatography (RP-HPLC) from the salivary glands of the black fly Simulium bannaense. The amino acid sequence of SibaCec was determined by a combination method of automated Edman degradation and cDNA sequencing. The morphologic changes of Gram-negative bacteria Escherichia coli treated with SibaCec were assessed by scanning electron microscopy (SEM). Quantitative PCR (qPCR) was performed to analyze the mRNA expression of the inducible NO synthase (iNOS) and pro-inflammatory cytokines. Nitric oxide (NO) generation was examined using a Griess assay and the secretion of pro-inflammatory cytokines was determined by an enzyme-linked immunosorbent assay (ELISA). The activation of extracellular signal-regulated kinase (ERK), p38, and the nuclear translocation of nuclear factor-kappaB (NF-κB) were assessed by Western blotting analysis. Circular dichroism (CD) spectroscopy was performed to evaluate the secondary structure of SibaCec in solvent environment. Interaction of SibaCec with lipopolysaccharide (LPS) was studied using fluorescein isothiocyanate (FITC)- conjugated LPS aggregates. Neutralization of LPS by SibaCec was assayed with the chromogenic limulus amebocyte lysate (LAL) test. qPCR was also used to analyze the expression of SibaCec mRNA in the salivary glands of insects after oral infection with the bacteria E.coli.
Results
SibaCec possessed potent antimicrobial activity against Gram-negative bacteria, and showed low cytotoxicity toward mammalian cells. SEM analysis indicated that SibaCec killed bacteria through the disruption of cell membrane integrity. Furthermore, SibaCec significantly inhibited lipopolysaccharide (LPS)-induced production of NO and pro-inflammatory cytokines such as tumor necrosis factor-α (TNF-α), interferon-1β (IL-1β) and interferon-6 (IL-6) by blocking the activation of MAPKs and NF-κB signaling pathways. It mainly adopted an α-helix conformation in membrane-mimetic environments. SibaCec could interact and neutralize LPS. Infection of black flies with bacteria caused an upregulation of the expression of SibaCec.
Conclusions
These results demonstrated that in addition to the bactericidal capacity, SibaCec can function as immune regulator, inhibiting host secretion of inflammatory factors.
Similar content being viewed by others
Background
Black flies (Diptera: Simuliidae) are annoying biting pests of humans and animals. They serve as obligate vectors for serious diseases such as human onchocerciasis (river blindness) and numerous arboviruses of livestock [1]. A number of salivary peptides/proteins have been characterized from hematophagous arthropods such as horseflies [2, 3], mosquitoes [4] and ticks [5]. There was relatively limited information available on pharmacologically active compounds in black fly salivary glands, until when the salivary transcriptomes were published for three black fly species (S. vittatum, S. nigrimanum and S. guianense) [6–8]. In these studies, some transcripts related to immunity, including six AMPs of cecropin family, have been revealed by sequence similarities with known peptides/proteins from other organisms. However, there have been very few subsequent efforts to establish the functions of these molecules in black flies. In addition, other immune-related molecules including lectins, prophenoloxidase and antimicrobial peptides/proteins have also been identified from black flies [9–11].
AMPs, including cecropins, defensins, and cathelicidins, are a unique and diverse group of effector molecules that play an important role in humoral immunity in all living organisms [12]. Cecropin was the first insect AMP isolated from the bacteria-challenged Hyalophora cecropia pupa [13], and then a number of cecropin-like peptides have been identified from different species of insect orders (Diptera, Lepidoptera, Hymenoptera, Coleoptera and Isoptera) [14, 15] and other organisms including mammals [16]. In insects, cecropin-like peptides have been shown to have effects on bacteria [17], fungi [18], parasites [19] and viruses [20].
The immune-stimulatory/modulatory functions and mechanisms of vertebrate AMPs, especially mammalian AMPs, such as defensins (i.e., hBD-2, hBD-3, and hBD-4) and cathelicidins (i.e., LL-37 and PR-39), have been well studied [21–23]. These AMPs are involved in modulating chemokine and cytokine production in immune cells, altering gene expression in host cells, limiting sepsis, improving wound healing and angiogenesis in vitro and in vivo [24, 25]. Some insect AMPs, such as the AMPs from blood-sucking triatomine bug and midges, have been shown to be involved in the immune responses [26–28]. In fact, there are relatively few studies that focus on the anti-inflammatory functions for these AMPs. To date, two AMPs of cecropin family (papiliocin and cecropin A) with anti-inflammatory activity have been characterized from the swallowtail butterfly Papilio xuthus [29] and cecropia moth Hyalophora cecropia [30]. Additionally, several hybrid peptides that are composed of cecropin A and other AMPs also showed the same activity [31–33].
We report herein the purification and characterization of a novel cecropin-like peptide with both antimicrobial and anti-inflammatory activities from the salivary glands of the hematophagous insect black fly S. bannaense.
Methods
Salivary gland dissection
Adult S. bannaense were collected near streams in Xishuangbanna, Yunnan, China. As our previous report [11], the black fly salivary glands were dissected in ice cold HEPES saline (10 mM HEPES pH 7.2, 150 mM NaCl) using fine entomological needles under a stereomicroscope, and stored in liquid nitrogen until use.
Ethical approval
The study was approved by the Animal Care and Use Ethics Committee of Kunming Medical University.
Peptide purification
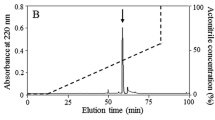
According to the methods in our previous report [11], the eluted peak of A1 (Fig. 1a) containing antimicrobial activity was pooled, lyophilized, and further purified by RP-HPLC on a Wondasil C18 column (25 × 0.46 cm). The elution was performed using a linear gradient of 0–60 % acetonitrile containing 0.1 % (v/v) trifluoroacetic acid in 0.1 % (v/v) trifluoroacetic acid/water over 70 min. N-terminal sequence of the purified peptide was done by Edman degradation on an Applied Biosystems pulsed liquid-phase sequencer (model ABI 491).
Purification of SibaCec from the salivary gland of S. bannaense and MALDI–TOF MS. a The filtrate of the salivary gland homogenate of S. bannaense was divided by an Inertsil C4 RP-HPLC column. b The eluted peak of A1 containing antimicrobial activity was further purified by C18 RP-HPLC column. The purified SibaCec is indicated by an arrow. c MALDI-TOF mass spectrometry analysis of the purified SibaCec
MALDI-TOF MS analysis
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) was used to analyze the purified peptide. AXIMA CFR mass spectrometer (Kratos Analytical) was operated in linear and positive ion mode. Polypeptide mass standard (Kratos Analytical) is used as an external calibration.
cDNA library construction and cDNA cloning
As our previous report [11], the In-Fusion SMARTerTM Directional cDNA Library Construction Kit (Takara, Japan) was used to prepare cDNAs. A sense degenerate primer (SibaCec-F1, according to the sequence determined by Edman degradation) was coupled with a 3’ PCR primer (the adaptor sequence of 3’ In-Fusion SMARTer CDS Primer provided in the kit) to screen the 3’ fragment of cDNA encoding SibaCec. The full length cDNA was finally obtained from primers of SibaCec-R1(antisense primer, according to the 3’-coding region of cDNA) and 5’ PCR primer from the library kit. The PCR conditions were: 95 °C for 5 min and 30 cycles of 95 °C (30 s), 56 °C (30 s), 72 °C (60 s) followed by an extension step at 72 °C for 8 min. Primers used in this research are shown in Additional file 1: Table S1.
Sequence analysis
The theoretical isoelectric point (pI) and molecular weight of SibaCec were analyzed through Bioinformatics Resource Portal (http://www.expasy.org/tools/) [34]. Multi-sequence alignment was performed using ClustalW (http://embnet.vital-it.ch/software/ClustalW.html) [35]. The phylogenetic tree was constructed by the neighbor-joining method in Mega 5 package.
Antimicrobial assay
The microbicidal activity of SibaCec was tested as described in our previous paper [36]. Standard and clinically isolated drug resistance bacteria were obtained from the First Affiliated Hospital of Kunming Medical University. Minimal inhibitory concentration (MIC) was defined as the lowest concentration at which no visible growth of microorganisms occurred.
SEM
Bacteria E. coli ATCC 25922 were incubated with SibaCec (1 × MIC) diluted in phosphate buffered saline (PBS) at 37 °C for 45 min. Bacteria incubated with PBS was used as negative control. After centrifugation, bacteria pellets were fixed with 2.5 % glutaraldehyde solution for 2 h at 4 °C and then postfixed in 1 % osmium tetroxide for 2 h. Dehydration was carried out with a graded series of alcohols. Bacteria were mounted onto aluminium stubs and sputtered with gold. Images were visualized in a Hitachi S-4800 electron microscope.
Cytotoxic and hemolytic assay
The cytotoxic activity was carried out as described in our previous work [37]. Briefly, 2 ml of 3 % (w/v) Brewer thioglycollate medium was injected into the peritoneal cavity of C57BL/6 mice. Three days later, peritoneal macrophages were harvested and cultured in 96-well plates (2 × 104 cells/well) in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 2 % fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. SibaCec dissolved in serum-free RPMI 1640 medium were added to wells, and the serum-free RPMI 1640 medium without SibaCec was used as control. After incubation at 37 °C for 24 h, 20 μl of MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) solution (5 mg/ml) was added to each well, and the cells were further incubated for 4 h at 37 °C. The cells were dissolved in 200 μl of Me2SO, and the absorbance at 570 nm was measured on a microplate reader (Epoch Etock, BioTek, USA).
Hemolytic assay was conducted as previously reported [36]. Serial dilutions of SibaCec were incubated with the washed human erythrocytes at 37 °C for 30 min. After centrifugation, the absorbance of supernatant was measured at 540 nm. 1 % (v/v) Triton X-100 was used to determine the maximal hemolysis and PBS was used as negative control.
NO detection
Mouse peritoneal macrophages were incubated either with LPS (100 ng/ml, from Escherichia coli 0111:B4, Sigma-Aldrich, USA) and SibaCec (0, 5, 10, and 20 μg/ml) dissolved in serum-free RPMI 1640 medium or incubated with SibaCec (10 μg/ml) alone in 24-well plates (2.5 × 105 cells/well) for 24 h. The cells incubated with serum-free RPMI 1640 and 100 ng/ml LPS were used as control. The culture medium was harvested to detect NO production. NO production was determined by detecting the nitrite level using Griess reagent (Beyotime, China) according to the manufacturer’s instructions.
qPCR
Mouse peritoneal macrophages were cultured in 6-well plates (2 × 106 cells/well) with RPMI 1640 (2 % FBS). The cells were incubated either with LPS (100 ng/ml) and SibaCec (0, 5, 10, and 20 μg/ml) dissolved in serum-free RPMI 1640 medium or incubated with SibaCec (10 μg/ml) alone. The cells incubated with serum-free RPMI 1640 and 100 ng/ml LPS were used as control. After treatment for 6 h, the cells were collected and total RNA was isolated. PrimeScript® Reverse Transcriptase (Takara, Japan) and SYBR green master mix (Takara, Japan) were used following the manufacturer’s instructions. qPCR was performed on a Realplex Mastercycler real-time PCR system (Eppendorf, Germany). The cycle counts of the target genes were normalized to the GAPDH gene, and accordingly the fold changes of the target genes were calculated. The primers used for qPCR are listed in Additional file 1: Table S1.
Pro-inflammatory cytokine determination
Mouse peritoneal macrophages were incubated either with LPS (100 ng/ml) and SibaCec (0, 5, 10, and 20 μg/ml) dissolved in serum-free RPMI 1640 medium or incubated with SibaCec (10 μg/ml) alone in 24-well plates (2.5 × 105 cells/well). The cells incubated with serum-free RPMI 1640 and 100 ng/ml LPS were used as control. After treatment for 6 h, the cell culture supernatants were collected and assessed for TNF-α, IL-1β, and IL-6 by using ELISA kits (Dakewei, China).
Western blot analysis
Mouse peritoneal macrophages were incubated either with LPS (100 ng/ml) and SibaCec (0, 5, 10, and 20 μg/ml) dissolved in serum-free RPMI 1640 medium or incubated with SibaCec (10 μg/ml) alone in 6-well plates (2 × 106 cells/well). The cells incubated with serum-free RPMI 1640 and 100 ng/ml LPS were used as control. After incubation for 30 min, the cells were washed twice with ice-cold PBS and lysed with RIPA lysis buffer (Beyotime, China). Then the cytoplasmic or nuclear proteins were extracted for Western blot analysis according to our previously described method [36]. Primary antibodies of phospho-ERK/ERK, phospho-p38/p38, NF-κB p65 (1:2000, Cell Signaling Technology, USA), GAPDH/Lamin B (1:5000, Santa Cruz Biotechnology, USA), and secondary antibody (1:5000, Cell Signaling Technology, USA) were used in Western blot analysis.
Circular dichroism spectroscopy
CD spectroscopy was performed using a Jasco J-715 spectrophotometer (Jasco, Japan) to evaluate the secondary structure of SibaCec in solvent environment. SibaCec was dissolved in sodium dodecyl sulfate (SDS)/H2O solutions (0, 5, 10, 20, 40 mM) or LPS/H2O solution (0, 50, 100, 200, 400 ng/ml) to an ultimate concentration of 0.2 mg/ml. The spectra were measured at 298 K between 190 and 250 nm using 0.1 cm path-length cell with 1 nm bandwidth, 1 s response time, and a scan speed of 100 nm/min. Three consecutive scans were performed and averaged, followed by subtraction of the solvent signal.
Dissociation of FITC-LPS aggregates
The ability of SibaCec to dissociate LPS micelles was studied using FITC-conjugated LPS (1 μg/ml, Sigma-Aldrich, USA). FITC-LPS was excited at 480 nm and the change in the emission of FITC at 515 nm was monitored with different concentrations of SibaCec (0, 50, 100, 200 μg/ml) dissolved in PBS. PBS was used as control. The fluorescence experiment was performed using a Cary Eclipse fluorescence spectrophotometer (Varian, Inc., USA).
LPS neutralization assay
The ability of SibaCec to neutralize LPS was assayed with the chromogenic LAL test which was performed according to the manufacturer’s instruction (GenScript, Nanjing, China). Briefly, different concentrations of SibaCec (0, 25, 50, 100 μg/ml) dissolved in PBS were incubated with LPS at 37 °C for 30 min. PBS was used as control. After incubation, 100 μl of LAL solution was added to LPS-peptide mixtures (100 μl) in a pyrogen-free tube and incubated for 10 min followed by addition of pre-warmed substrate. After incubation of 6 min for the reaction, the absorbance was recorded at 545 nm on a microplate reader. The percentage of LPS neutralization was calculated as (A blank − A sample)/A blank × 100, where A blank represents the absorbance of blank control (50 μl of LAL water + 50 μl of LPS solution).
Bacterial feeding
Bacterial feed experiment was carried out as previously described [38]. In brief, the collected S. bannaense (~250 flies) were fed with 70 % sucrose solution ad libitum. After starving for 12 h, black flies were fed through cotton wool with 20 % sucrose solution (OD600 = 0.2) containing Gram-negative bacteria E.coli ATCC 25922. Total RNA was extracted from the salivary glands of immune stimulated or naive insects (sugar fed controls) at 12, 24, 36, 48 and 72 h after feeding. qPCR was performed to analyze the expression of SibaCec mRNA as described above, with the housekeeping gene β-actin as an endogenous control.
Statistics
Statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software Inc., San Diego, CA, USA) and Stata 10.0 software (StataCorporation, College Station, TX, USA). Data were presented as mean ± standard errors of mean, and compared using two-tailed equal variance Student’s t-test. *P < 0.05 and **P < 0.01 were considered as statistical significance.
Results
Characterization of SibaCec
As illustrated in Fig. 1a, the fractions with antimicrobial activity (marked by A1) were collected, lyophilized, and further purified by C18 RP-HPLC. One peptide was purified from this step (Fig. 1b), and it was designated SibaCec. After Edman degradation, the initial 22 N-terminal amino acid residues of SibaCec was identified with the following sequence: GKLTKDKLKRGAKKALNVASKV. Due to the majority of insect cecropin-like peptides containing a C-terminally amidated residue, we presumed that the same structure feature exists in the SibaCec. After carboxypeptidase Y treatment, no free amino acid can be detected by HPLC (data not shown). The result indicated that the C-terminal end of SibaCec was amidated, which was further confirmed by mass spectrometry analysis. MALDI-TOF MS analysis (Fig. 1c) indicated that SibaCec with the C-terminal amidation had a measured molecular mass of 3431.21 Da, matching well with the calculated molecular mass 3432.16 Da.
The cDNA clone encoding the precursor of SibaCec was screened and sequenced from the salivary glands cDNA library of S. bannaense (GenBank accession number: KP642081). As shown in Fig. 2a, the deduced amino acid sequence of SibaCec precursor is completely consistent with the result of Edman degradation sequencing. It is composed of 57 amino acid residues, including a predicted 22 amino acid signal peptide, a 34 amino acid mature peptide, and a C-terminal glycine for enzymic amidation. Analysis using the ExPASy MW/pI tool showed that SibaCec has a predicted pI of 11.30. In addition, SibaCec has the net charge of +8. BLAST search indicated that the SibaCec precursor shared the highest identity of 70 % (40/57) with the salivary expressed cecropin precursor (GenBank accession number: ACH56893) from the black flies S. vittatum.
The cDNA sequence of SibaCec precursor and the alignment of the amino acid sequence of SibaCec with six known cecropin-like peptide precursors from three black fly species. a Deduced amino acid sequence is shown below the cDNA sequence. The putative signal peptide is italicized and the amino acid sequence of mature peptide is underlined and bold. The stop codon is indicated by an asterisk. Amino acid numbers or nucleotide numbers are shown after the sequences. b Six known cecropin-like peptide precursors in three black fly species are from Refs. [6–8]. The symbols under the alignment indicate: (*) identical sites; (:) conserved sites; (.) less conserved sites. Dashes are inserted to optimize the alignment
Multi-sequence alignment of the known cecropin-like peptide precursors from four black fly species (Fig. 2b) indicated that the putative signal peptide of these sequences are divergent, and nine amino acids residues are highly conserved within the mature peptide (Lys28, Asp29, Lys30, Leu31, Gla34, Ala35, Lys37, Ala38 and Leu39).
Phylogenetic analysis of SibaCec
The phylogenetic tree was generated from the amino acid sequences of 46 insect cecropin precursors (28 insect species including 20 Diptera, 7 Lepidoptera, 1 Coleoptera). As showed in Fig. 3, the cecropin sequences are divided between two distinct groups: one cluster comprising 41 sequences derived from different insect orders (Diptera, Lepidoptera and Coleoptera) and the second cluster comprising the five sequences derived from Simuliidae, including SibaCec.
Antimicrobial activity of SibaCec
The MICs of SibaCec against Gram-positive and Gram-negative bacteria were determined. As listed in Table 1, SibaCec exhibited broad spectrum antimicrobial activities against all ten bacterial strains tested, especially against Gram-negative bacteria E.coli (either standard strains or clinically isolated drug-resistance strains). SibaCec showed much higher antimicrobial activities against Gram-negative bacteria (MICs ranging from 0.87 to 2.33 μM) than against Gram-positive bacteria (MICs ranging from 14.56 to 43.70 μM).
SibaCec alters the morphology of E. coli
SEM was performed to study the possible mechanisms of action of SibaCec on Gram-negative bacteria E. coli ATCC 25922. The cells treated with SibaCec (1 × MIC) showed obvious morphological alterations by SEM analysis (Fig. 4). The membrane integrity of cells seemed to be disrupted, and cell shrinkage was obvious.
SibaCec shows low cytotoxicity and hemolytic activity
SibaCec exhibited very low cytotoxicity toward the mouse peritoneal macrophages. At concentrations up to 200 μg/ml (58.27 μM), which is almost 67-fold higher than the MIC value of SibaCec against E.coli ATCC 25922, SibaCec induced cell death percentages as low as 4.26 %. As to hemolytic activity, SibaCec yielded a hemolysis of 3.25 % at the same concentration of 200 μg/ml.
SibaCec reduces LPS-induced iNOS transcription and NO production
To determine the effect of SibaCec on the LPS-induced NO production in mouse peritoneal macrophages, we used qPCR to determine the mRNA level of iNOS. As shown in Fig. 5a, incubation with SibaCec for 6 h significantly reduced the mRNA level of iNOS induced by 100 ng/ml LPS in a dose-dependent manner. At a concentration of 20 μg/ml, SibaCec inhibited 51.9 % of the iNOS transcription. Furthermore, we determined the NO production by examining the nitrite concentration in the culture supernatants of mouse peritoneal macrophages. As illustrated in Fig. 5b, 100 ng/ml LPS induced 31.2 μM nitrate production. The addition of SibaCec significantly reduced LPS-induced nitrite production. At the concentration of 20 μg/ml, SibaCec inhibited 77.4 % of nitrite production
Effects of SibaCec on iNOS transcription and NO production induced by LPS. a iNOS mRNA. b Nitrite production. Data are mean ± S.E. values from three independent experiments. *p <0.05, **p <0.01, significantly different compared with the control that treated with serum-free RPMI 1640 and 100 ng/ml LPS
SibaCec inhibits LPS-induced pro-inflammatory cytokine production
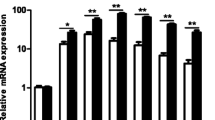
To evaluate the effect of SibaCec on LPS-induced pro-inflammatory cytokine production in mouse peritoneal macrophages, we used qPCR to determine pro-inflammatory cytokine gene expression. SibaCec significantly blocked LPS-induced expression of TNF-α, IL-1β, and IL-6 in a dose-dependent manner. 20 μg/ml SibaCec inhibited the expression of all three of the pro-inflammatory cytokine genes by 64.5, 51.4 and 68.8 %, respectively (Fig. 6a, b, and c). Furthermore, we used ELISA to confirm the effect of SibaCec on pro-inflammatory cytokine production induced by LPS in mouse peritoneal macrophages. 100 ng/ml LPS alone induced the production of TNF-α, IL-1β, and IL-6 for about 1500, 1270, and 464 pg/ml, respectively. Fig. 6d, e and f showed a dose-dependent effect, which revealed activities similar to those obtained in the qPCR experiments. 20 μg/ml SibaCec inhibited LPS-induced TNF-α, IL-1β, and IL-6 production by 65, 61.9, and 50.5 %, respectively.
Effects of SibaCec on pro-inflammatory cytokines transcription and secretion induced by LPS. a TNF-α mRNA. b TNF-α secretion. c IL-1β mRNA. d IL-1β secretion. e IL-6 mRNA. f IL-6 secretion. Data are mean ± S.E. values from three separate experiments. *p <0.05, **p <0.01, significantly different compared with the control that incubated with serum-free RPMI 1640 and 100 ng/ml LPS
SibaCec inhibits LPS-induced inflammatory response pathways
The above mentioned data indicate that SibaCec significantly inhibited the transcription and production of NO, TNF-α, IL-1β, and IL-6, which were induced by LPS in mouse peritoneal macrophages. MAPKs and NF-κB signal pathways play important roles in cytokine production. Therefore, we studied the effect of SibaCec on LPS-induced inflammatory signaling pathways. As shown in Fig. 7, 100 ng/ml LPS induced the phosphorylation of ERK, p38, and the translocation of the NF-κB p65 subunit from cytoplasm to nucleus. The incubation of SibaCec (10 μg/ml) markedly inhibited the LPS-induced phosphorylation of ERK and p38 and translocation of the NF-κB p65 subunit, which indicates that SibaCec exerts its anti-inflammatory effect through inhibition of MAPKs and NF-κB inflammatory signaling pathways.
Effects of SibaCec on LPS-induced inflammatory response pathways. Western blot of phosphorylation of ERK and p38, and the translocation of the NF-κB p65 subunit from cytoplasm to nucleus in peritoneal macrophages. The cells were incubated with LPS (100 ng/ml) and different concentrations of SibaCec (5, 10 and 20 μg/ml). After incubation for 30 min, the cells were collected, and the cytoplasmic or nuclear proteins were extracted for Western blot analysis
Secondary structure of SibaCec
To detect the secondary structures of SibaCec in membrane-mimetic or hydrophobic environments, we analyzed the CD spectra of the peptide dissolved under increasing concentrations of SDS or LPS. As shown in Fig. 8, the CD spectra of SibaCec dissolved in H2O showed a strong negative peak at 198 nm, which indicates that SibaCec adopts a random coil conformation. In different concentrations of SDS solutions (5, 10, 40 mM), the CD spectra of SibaCec exhibited a strong positive peak at 192 nm and two negative peaks at 208 and 222 nm (Fig. 8a), which indicates that SibaCec mainly adopts an α-helix conformation in membrane-like environments.
In various proportions of LPS solutions (50, 100, 200, 400 ng/ml) (Fig. 8b), the conformation of SibaCec gradually changed from a random-coil (percent decrease from 57.6 to 31.9) to an α-helix structure (percent increase from 0 to 35.1) in a relatively hydrophobic environment (Table 2).
SibaCec dissociates FITC-LPS aggregates
LPS forms micellar aggregates in water and FITC fluorescence is highly quenched in FITC-LPS micelles [29, 39]. Interactions of some AMPs with LPS may cause an enhancement of the FITC fluorescence because of de-quenching, which indicates the dissociation of large LPS aggregates into smaller sizes [39]. As shown in Fig. 9a, SibaCec had a strong effect on the FITC-LPS aggregates, and the addition of SibaCec caused a dose-dependent increase of FITC-LPS fluorescence. At the concentrations of 25, 50, 100 μg/ml, SibaCec increased 29.6, 48.1 and 68.4 % of fluorescence intensity, respectively.
Interaction and neutralization of LPS by SibaCec. a The changes in fluorescence intensity of FITC-labeled LPS as a function of various concentrations of SibaCec, (b) LPS neutralization by SibaCec. A chromogenic LAL assay was used to evaluate the neutralizing activity. Data are mean ± S.E. values from three separate experiments. *p <0.05, **p <0.01, significantly different compared with the control (PBS)
SibaCec neutralizes LPS
The chromogenic LAL assay was used to determine whether SibaCec is active in neutralization of endotoxin. In parallel with the dissociation of FITC-LPS aggregates assay, SibaCec exhibited LPS neutralizing activity in a concentration dependent manner (Fig. 9b). At the concentrations of 25, 50, 100 μg/ml, SibaCec inhibited 23.8, 33.2 and 49.4 % of LPS, respectively.
Transcript levels of SibaCec increase after infection with E.coli
After E. coli ingestion, the expression levels of SibaCec mRNA in the salivary glands of bacteria-immunized or naive insects were compared at the different time course, respectively. As illustrated in Fig. 10, the levels of SibaCec mRNA were up-regulated by bacterial-challenge at 12, 24, 36, 48 and 72 h after E. coli ingestion (38.2, 41.8, 33.5, 29.6 and 15.6 fold, respectively). The expression of SibaCec peaked at 24 h post-feeding and relatively decreased with time.
Fold increase of SibaCec in the salivary glands of insects after oral infection with E.coli at different time course. Expression levels in the salivary glands of bacteria-immunized insects were calculated relative to the level of SibaCec in corresponding naive insects, which was arbitrarily defined as 1. *p < 0.05, **p < 0.01, significantly different compared to the control that received the sucrose solution without E.coli
Discussion
Black flies are blood-sucking insects that can secrete various immunomodulatory molecules to suppress the host’s inflammatory and immunologic reactions, and to contribute to efficient transmission of fly-borne pathogens [7]. The salivary gland extract of black fly S. vittatum, has been shown to contain immunomodulatory activities that reduces expression of I-A (mouse MHC class II), IL-5 and IL-10 in splenocytes [40, 41], and inhibits mitogen-stimulated mouse splenocyte proliferation [42]. However, the component(s) responsible for immunomodulation has not been characterized from black flies so far.
The current work identified a novel cationic cecropin-like peptide (SibaCec) from the salivary glands of the black fly S. bannaense. The structural organization of SibaCec precursor (Fig. 2a) is similar to other insect cecropin precursors, comprising a putative signal peptide and the mature peptide. The alanine residue (Ala22) in the precursor is cleaved to release the mature peptide, which is consistent with most cecropins isolated from Diptera [43]. SibaCec presents a canonical feature of most characterized insect cecropins (an amidated C-terminus), but it is devoid of another feature (a tryptophan residue at position 1 or 2) [14]. The first two amino acid residues at the N-terminus of SibaCec are glycine and lysine. The absence of tryptophan residue within the N-terminal domain was also reported in some mosquito cecropins [17, 43].
The phylogenetic tree of cecropin precursors (Fig. 3) showed that the black fly cecropins appear in one branch and other insect cecropins are grouped in another branch. This suggests the cecropins occurred in the insects before the divergence of the Diptera, Lepidoptera and Coleoptera. The result also supports that the cecropin molecules have evolved independently between these insect taxa [44].
SibaCec, like other cationic AMPs, is a highly basic peptide with the net charge of +8, which implies that it would be readily attracted and adhered to the negative-charged bacterial surface. As expected, SibaCec has a broad spectrum of antibacterial activity (Table 1), and it is more effective against Gram-negative bacteria (MICs ranging from 0.87 to 2.33 μM) than against Gram-positive bacteria (MICs ranging from 14.56 to 43.7 μM). SEM analysis indicated that such activities are carried out with a lytic effect on the bacterial membranes (Fig. 4). Additionally, CD spectroscopy indicated that SibaCec mainly adopt an α-helical conformation in membrane-mimetic solutions (Fig. 8), which contributes to the ability of cationic peptide to kill bacteria. These results confirm that the microbial membrane is a key target for SibaCec.
Extensive research has established that MAPK signal transduction pathways are involved in regulating the transcriptions of cytokine genes [29, 37]. In this study, SibaCec significantly inhibited the transcription and production of pro-inflammatory factors induced by LPS in mouse peritoneal macrophages, including NO, TNF-α, IL-1β, and IL-6 (Figs. 5 and 6). Furthermore, Western blot analysis showed that SibaCec significantly suppressed the LPS induced activation of ERK, p38, and the translocation of NF-κB p65 subunit (Fig. 7). These data indicated that SibaCec executes the anti-inflammatory effect in LPS-stimulated murine macrophages by blocking the activation of MAPKs and NF-κB signaling pathways.
The analysis of interaction between SibaCec and LPS indicated that SibaCec can dissociate the aggregated form of LPS in a dose-dependent manner (Fig. 9a), and the LAL assay indicated that SibaCec also can neutralize LPS in a dose-dependent fashion (Fig. 9b). Since SibaCec is a membrane-targeting AMP with polycationic (basic-amino-acid-rich) and amphipathic α-helix structure, we rationally conclude SibaCec binds to negatively charged LPS mostly through electrostatic interaction and the amphiphilic helical structure [45]. All these results implied that the neutralization of LPS by SibaCec prevents LPS from binding to the LPS-binding protein, and hence suppresses the production of cytokines induced by LPS. Gram-negative bacteria and/or their endotoxins (LPS) may trigger a systemic inflammatory response, leading to some life-threatening systemic diseases, such as sepsis and septic shock [46]. The above results indicate that SibaCec is not only involved in suppressing Gram-negative bacteria growth but also that it attenuates inflammatory responses induced by LPS. These functions of SibaCec may favor resolution of infection and reverse potentially harmful inflammation.
The transcript levels of SibaCec in the salivary glands of insects increased after oral infection with Gram-negative bacteria E.coli, reaching maximum at 24 h and then slowly decreased from that time point (Fig. 10). These data suggested that SibaCec is involved in the innate humoral response of the black fly S. bannaense.
Conclusions
In conclusion, the black fly cecropin-like peptide (SibaCec) was identified in the present work by peptide purification and molecular cloning procedures. SibaCec possesses potent antimicrobial activity against Gram-negative bacteria, and shows low cytotoxicity toward mammalian cells. It can neutralize LPS and exhibit strong anti-inflammatory activity. All the amalgamated properties make SibaCec a potentially potent candidate for the treatment of inflammatory and infectious diseases.
References
Adler PH, McCreadie JW. Black flies (Simuliidae). In: Mullen GR, Durden LA, editors. Medical and Veterinary Entomology. 2nd ed. San Diego: Elsevier; 2009. p. 189–206.
Ma D, Wang Y, Yang H, Wu J, An S, Gao L, et al. Anti-thrombosis repertoire of blood-feeding horsefly salivary glands. Mol Cell Proteomics. 2009;8(9):2071–9.
Xu X, Yang H, Ma D, Wu J, Wang Y, Song Y, et al. Toward an understanding of the molecular mechanism for successful blood feeding by coupling proteomics analysis with pharmacological testing of horsefly salivary glands. Mol Cell Proteomics. 2008;7(3):582–90.
Calvo E, Mans BJ, Andersen JF, Ribeiro JM. Function and evolution of a mosquito salivary protein family. J Biol Chem. 2006;281(4):1935–42.
Francischetti IM, Sa-Nunes A, Mans BJ, Santos IM, Ribeiro JM. The role of saliva in tick feeding. Front Biosci (Landmark Ed). 2009;14:2051–88.
Chagas AC, Calvo E, Pimenta PF, Ribeiro JM. An insight into the sialome of Simulium guianense (Diptera: Simuliidae), the main vector of river blindness disease in Brazil. BMC Genomics. 2011;12:612.
Andersen JF, Pham VM, Meng Z, Champagne DE, Ribeiro JM. Insight into the Sialome of the Black Fly, Simulium vittatum. J Proteome Res. 2009;8:1474–88.
Ribeiro JM, Valenzuela JG, Pham VM, Kleeman L, Barbian KD, Favreau AJ. An insight into the Sialotranscriptome of Simulium nigrimanum, a black fly associated with Fogo Selvagem in South America. Am J Trop Med Hyg. 2010;82:1060–75.
Kläger SL, Watson A, Achukwi D, Hultmark D, Hagen HE. Humoral immune response of Simulium damnosum s.l. following filarial and bacterial infections. Parasitology. 2002;125(Pt 4):359–66.
Ham PJ, Albuquerque C, Baxter AJ, Chalk R, Hagen HE. Approaches to vector control: new and trusted. 1. Humoral immune responses in blackfly and mosquito vectors of filariae. Trans R Soc Trop Med Hyg. 1994;88(2):132–5.
Wei L, Mu L, Wang Y, Bian H, Li J, Lu Y, et al. Purification and characterization of a novel defensin from the salivary glands of the black fly, Simulium bannaense. Parasit Vectors. 2015;8(1):71.
Koczulla AR, Bals R. Antimicrobial peptides: current status and therapeutic potential. Drugs. 2003;63:389–406.
Steiner H, Hultmark D, Engstrom A, Bennich H, Boman HG. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature. 1981;292:246–8.
Yi HY, Chowdhury M, Huang YD, Yu XQ. Insect antimicrobial peptides and their applications. Appl Microbiol Biotechnol. 2014;98:5807–22.
Bulet P, Stöcklin R. Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept Lett. 2005;12:3–11.
Boman HG. Innate immunity and the normal microflora. Immunol Rev. 2000;173:5–16.
Vizioli J, Bulet P, Charlet M, Lowenberger C, Blass C, Muller HM, et al. Cloning and analysis of a cecropin gene from the malaria vector mosquito, Anopheles gambiae. Insect Mol Biol. 2000;9:75–84.
Andra J, Berninghausen O, Leippe M. Cecropins, antibacterial peptides from insects and mammals, are potently fungicidal against Candida albicans. Med Microbiol Immunol. 2001;189:169–73.
Kokoza V, Ahmed A, Woon Shin S, Okafor N, Zou Z, Raikhel AS. Blocking of plasmodium transmission by cooperative action of cecropin A and defensin A in transgenic Aedes aegypti mosquitoes. Proc Natl Acad Sci U S A. 2010;107(18):8111–6.
Luplertlop N, Surasombatpattana P, Patramool S, Dumas E, Wasinpiyamongkol L, Saune L, et al. Induction of a peptide with activity against a broad spectrum of pathogens in the Aedes aegypti salivary gland, following infection with Dengue Virus. PLoS Pathog. 2011;7(1):e1001252.
Niyonsaba F, Ushio H, Nakano N, Ng W, Sayama K, Hashimoto K, et al. Antimicrobial peptides human beta-defensins stimulate epidermal keratinocyte migration, proliferation and production of proinflammatory cytokines and chemokines. J Invest Dermatol. 2007;127(3):594–604.
Heilborn JD, Nilsson MF, Kratz G, Weber G, Sorensen O, Borregaard N, et al. The cathelicidin anti-microbial peptide LL-37 is involved in re-epithelialization of human skin wounds and is lacking in chronic ulcer epithelium. J Invest Dermatol. 2003;120(3):379–89.
Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, et al. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414(6862):454–7.
Lai Y, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30(3):131–41.
Bowdish DM, Davidson DJ, Scott MG, Hancock RE. Immunomodulatory activities of small host defense peptides. Antimicrob Agents Chemother. 2005;49(5):1727–32.
Vieira CS, Waniek PJ, Mattos DP, Castro DP, Mello CB, Ratcliffe NA, et al. Humoral responses in Rhodnius prolixus: bacterial feeding induces differential patterns of antibacterial activity and enhances mRNA levels of antimicrobial peptides in the midgut. Parasit Vectors. 2014;7:232.
Vieira CS, Mattos DP, Waniek PJ, Santangelo JM, Figueiredo MB, Gumiel M, et al. Rhodnius prolixus interaction with Trypanosoma rangeli: modulation of the immune system and microbiota population. Parasit Vectors. 2015;8:135.
Nayduch D, Lee MB, Saski CA. Gene discovery and differential expression analysis of humoral immune response elements in female Culicoides sonorensis (Diptera: Ceratopogonidae). Parasit Vectors. 2014;7:388.
Kim JK, Lee E, Shin S, Jeong KW, Lee JY, Bae SY, et al. Structure and function of papiliocin with antimicrobial and anti-inflammatory activities isolated from the swallowtail butterfly, Papilio xuthus. J Biol Chem. 2011;286(48):41296–311.
Lee E, Shin A, Kim Y. Anti-inflammatory activities of cecropin A and its mechanism of action. Arch Insect Biochem Physiol. 2015;88(1):31–44.
Scott MG, Rosenberger CM, Gold MR, Finlay BB, Hancock RE. An alpha- helical cationic antimicrobial peptide selectively modulates macrophage responses to lipopolysaccharide and directly alters macrophage gene expression. J Immunol. 2000;165(6):3358–65.
Ryu S, Choi SY, Acharya S, Chun YJ, Gurley C, Park Y, et al. Antimicrobial and anti-inflammatory effects of Cecropin A(1–8)- Magainin2(1–12) hybrid peptide analog p5 against Malassezia furfur infection in human keratinocytes. J Invest Dermatol. 2011;131(8):1677–83.
Liu Y, Xia X, Xu L, Wang Y. Design of hybrid β-hairpin peptides with enhanced cell specificity and potent anti-inflammatory activity. Biomaterials. 2013;34(1):237–50.
Bjellqvist B, Hughes GJ, Pasquali C, Paquet N, Ravier F, Sancez JC, et al. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14:1023–31.
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–500.
Wei L, Che H, Han Y, Lv J, Mu L, Lv L, et al. The first anionic defensin from amphibians. Amino Acids. 2015;47(7):1301–8.
Wu J, Wang Y, Liu H, Yang H, Ma D, Li J, et al. Two immune-regulatory peptides with antioxidant activity from tick salivary glands. J Biol Chem. 2010;285(22):16606–13.
Telleria EL, Sant’Anna MR, Alkurbi MO, Pitaluga AN, Dillon RJ, Traub-Csekö YM. Bacterial feeding, Leishmania infection and distinct infection routes induce differential defensin expression in Lutzomyia longipalpis. Parasit Vectors. 2013;6:12.
Rosenfeld Y, Papo N, Shai Y. Endotoxin (lipopolysaccharide) neutralization by innate immunity host-defense peptides. Peptide properties and plausible modes of action. J Biol Chem. 2006;281(3):1636–43.
Cross ML, Cupp MS, Cupp EW, Galloway AL, Enriquez FJ. Modulation of murine immunological responses by salivary gland extract of Simulium vittatum (Diptera: Simuliidae). J Med Entomol. 1993;30:928–35.
Cross ML, Cupp EW, Enriquez FJ. Modulation of murine cellular immune responses and cytokines by salivary gland extract of the black fly Simulium vittatum. Trop Med Parasitol. 1994;45:119–24.
Tsujimoto H, Gray EW, Champagne DE. Black fly salivary gland extract inhibits proliferation and induces apoptosis in murine splenocytes. Parasite Immunol. 2012;32:275–84.
Lowenberger C, Charlet M, Vizioli J, Kamal S, Richman A, Christensen BM, et al. Antimicrobial activity spectrum, cDNA cloning, and mRNA expression of a newly isolated member of the cecropin family from the mosquito vector Aedes aegypti. J Biol Chem. 1999;274(29):20092–7.
Tassanakajon A, Somboonwiwat K, Amparyup P. Sequence diversity and evolution of antimicrobial peptides in invertebrates. Dev Comp Immunol. 2015;48(2):324–41.
Stewart I, Schluter PJ, Shaw GR. Cyanobacterial lipopolysaccharides and human health - a review. Environ Health. 2006;5:7.
Schulte W, Bernhagen J, Bucala R. Cytokines in sepsis: potent immunoregulators and potential therapeutic targets-an updated view. Mediators Inflamm. 2013;2013:165974.
Acknowledgments
This work was supported by Chinese National Natural Science Foundation (81373380, 81360253, 81260258, 81402830) and Jiangsu Province Foundation (BK20140362, 14KJD350003).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: JW, LM, LW, HY. Performed the experiments: JW, LM, YH, TL, JL, CF. Analyzed the data: JW, HY, LW. Contributed reagents/materials/analysis tools: JW, HY. Wrote the paper: JW, HY. All authors read and approved the final version of the manuscript.
Jing Wu and Lixian Mu contributed equally to this work.
Additional file
Additional file 1: Table S1.
Primer sequences used for cloning and qPCR in this study. (DOC 39 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wu, J., Mu, L., Zhuang, L. et al. A cecropin-like antimicrobial peptide with anti-inflammatory activity from the black fly salivary glands. Parasites Vectors 8, 561 (2015). https://doi.org/10.1186/s13071-015-1176-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-015-1176-8