Abstract
Background
The insulin-like growth factor 1 (IGF-1) pathway is involved in cell growth and proliferation and is associated with tumorigenesis and therapy resistance. This study aims to elucidate whether variation in the IGF-1 pathway is predictive for pathologic response in early HER2 negative breast cancer (BC) patients, taking part in the phase III NEOZOTAC trial, randomizing between 6 cycles of neoadjuvant TAC chemotherapy with or without zoledronic acid.
Methods
Formalin-fixed paraffin-embedded tissue samples of pre-chemotherapy biopsies and operation specimens were collected for analysis of IGF-1 receptor (IGF-1R) expression (n = 216) and for analysis of 8 candidate single nucleotide polymorphisms (SNPs) in genes of the IGF-1 pathway (n = 184) using OpenArray® RealTime PCR. Associations with patient and tumor characteristics and chemotherapy response according to Miller and Payne pathologic response were performed using chi-square and regression analysis.
Results
During chemotherapy, a significant number of tumors (47.2 %) showed a decrease in IGF-1R expression, while in a small number of tumors an upregulation was seen (15.1 %). IGF-1R expression before treatment was not associated with pathological response, however, absence of IGF-1R expression after treatment was associated with a better response in multivariate analysis (P = 0.006) and patients with a decrease in expression during treatment showed a better response to chemotherapy as well (P = 0.020). Moreover, the variant T allele of 3129G > T in IGF1R (rs2016347) was associated with a better pathological response in multivariate analysis (P = 0.032).
Conclusions
Absent or diminished expression of IGF-1R after neoadjuvant chemotherapy was associated with a better pathological response. Additionally, we found a SNP (rs2016347) in IGF1R as a potential predictive marker for chemotherapy efficacy in BC patients treated with TAC.
Trial registration
ClinicalTrials.gov NCT01099436. Registered April 6, 2010.
Similar content being viewed by others
Background
Insulin-like growth factor (IGF)-1 and other members of the IGF-1 pathway have been associated with development, progression, and metastasis of several cancers [1, 2]. Additionally, epidemiologic studies have shown a relation between high circulating IGF-1 levels, breast density [3], and risk of breast cancer (BC) [4]. Increased IGF-1 levels are associated with an elevated BC mortality [5] and with inherent resistance to anti-tumor treatments in preclinical models [6–9]. Furthermore, the IGF-1 receptor (IGF-1R), a transmembrane tyrosine kinase, is frequently upregulated in BC [10, 11]. The biological activity of IGF-1 and IGF-2 depends on binding with the insulin-like growth factor binding proteins (IGF-BPs), mainly IGF-BP3 [12, 13]. Both IGFs bind the IGF-1R and activate the Ras/mitogen-activated protein kinase (MAPK) and phosphatidylinositol-3-kinase (PI3K)/AKT pathways, through which cell proliferation is stimulated and apoptosis is inhibited, respectively [14, 15]. Additionally, the IGF-1R and the estrogen receptor (ER) have been shown to work synergistically, whereby activated ER binds to the promoter regions of IGF1R to promote transcription and IGF-1 is able to activate unliganded ER [16, 17].
Previous research has shown that low IGF-1R expression in the tumor is predictive for pathological complete response (pCR) in ER-positive tumors [10] and that upregulation of IGF-1R during chemotherapy predicts a poor outcome in a relative small, heterogeneous group of BC patients [18]. Moreover, genes encoding members of the IGF-1 pathway are known to harbor several single nucleotide polymorphisms (SNPs) that influence the activity of the pathway. SNPs associated with IGF-1 and IGF-BP3 plasma levels and breast density are described [19, 20] as well as SNPs associated with therapy resistance and outcome [21, 22].
Neoadjuvant chemotherapy has been demonstrated to be equivalent to adjuvant chemotherapy for BC survival. This treatment has the advantage of more frequent breast-conserving therapy [23] and offers the opportunity for translational research of molecular predictors of tumor response. Additionally, the Miller and Payne (MP) histological grading system can be used to assess response to neoadjuvant chemotherapy because it is associated with patients’ disease-free and overall survival [24, 25]. This study evaluates the expression of the IGF-1R of the tumor before and after neoadjuvant chemotherapy and whether it predicts pathological response according to MP classification after neoadjuvant chemotherapy in human epidermal growth factor receptor 2 (HER2)-negative early BC patients treated in the NEOZOTAC trial [26]. Moreover, we aim to identify SNPs, which have been described to influence the activity of the IGF-1 pathway, to predict chemotherapy efficacy in this cohort. In addition, these SNPs are tested for association with the occurrence of side effects.
Methods
Study population
From July 2010 until April 2012, 250 women participated in the multicenter phase III NEOZOTAC trial, randomizing between TAC chemotherapy (75 mg/m2 docetaxel, 50 mg/m2 doxorubicin, and 500 mg/m2 cyclophosphamide) with or without zoledronic acid (4 mg within 24 hours after chemotherapy). Eligible patients had a histologically confirmed diagnosis of HER2-negative stage II or III BC. Other inclusion and exclusion criteria have been described elsewhere [26]. Tumor regression was scored according to the MP classification [24]. pCR was defined as the absence of residual invasive cancer within the breast and lymph nodes [24]. Side effects and hematological toxicity were graded according to the Common Terminology Criteria for Adverse Events version 4.0 (CTCAE v.4.0) [27]. All patients gave written informed consent. The study was conducted in accordance with the Declaration of Helsinki (2008) and approved by the Ethics Committee of the Leiden University Medical Center in agreement with the Dutch law for medical research involving humans.
Immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) tumor tissue samples of prechemotherapy biopsies and operation specimens were collected for analysis of IGF-1R expression using immunohistochemistry (IHC). From each FFPE tumor tissue sample, one section of 4 μm was cut and deparaffinized with xylene, rehydrated through graded alcohol, and rinsed in distilled water. After blocking of endogenous peroxidase activity with 0.3 % H2O2 for 20 minutes, heat-induced antigen retrieval was performed in the EnVision™ Flex Target Retrieval Solution in PT Link (Dako, Glostrup, Denmark) at low pH. After blocking with 5 % normal goat serum to reduce aspecific binding by the primary antibody, the sections were incubated overnight at room temperature in a humidified chamber with the IGF-1R antibody (IGF-1 receptor β, D4O6W, rabbit monoclonal; Cell Signaling Technology, Danvers, MA, USA) diluted in phosphate-buffered saline (PBS)/bovine serum albumin (BSA) 1 % at a dilution of 1:200. After the primary antibody incubation, the sections were washed with PBS and incubated with a secondary anti-rabbit antibody EnVision™ (Dako, Glostrup, Denmark) for 30 minutes and visualized using liquid DAB+ (Dako, Glostrup, Denmark). Eventually, sections were counterstained with Mayer’s hematoxylin, dehydrated, and subsequently permanently mounted with Pertex (Histolab, Gothenburg, Sweden). Breast and placenta sections that had previously been identified to express the IGF-1R were used as positive controls, and sections that underwent the IHC staining procedure without application of primary antibodies served as negative controls. Membranous IGF-1R expression was scored on a scale of 0–3+ (see Fig. 1). Samples were considered negative if 0 or 1+ was scored, and positive if 2+ and 3+ was given. The staining was scored by two independent researchers (SdG and ALM).
Examples of the membranous IGF-1R staining in breast tumor tissue sections. Score 0: staining is observed in <10 % of the tumor cells. Score 1+: incomplete staining is observed in >10 % of the tumor cells, Score 2+: weak or moderate complete staining is observed in >10 % of the tumor cells, Score 3+: strong complete staining is observed >10 % of tumor cells. Samples were considered negative if 0 or 1+ was scored, and positive if 2+ and 3+ was given
SNP selection
To select relevant SNPs in the IGF-1 pathway, a PubMed search with the keywords “IGF-1”, “IGF-2”, “IGF-BP3”, “IGF-1R”, “single nucleotide polymorphism”, “breast cancer”, and/or “clinical outcome” was conducted in July 2013. SNPs that were associated with IGF-1 or IGF-BP3 plasma levels, BC risk, or clinical outcome in cancer patients treated with chemotherapy, were selected. SNPs with a minor allele frequency (MAF) >0.01 in a Caucasian population according to the HapMap project database and with a potential functionality according to the literature review or using national institutes of health functionality database were selected [28]. To minimize the number of tested associations, tagging SNPs were selected for SNPs that were in linkage disequilibrium (r 2 > 0.7). The selected SNPs in the IGF1, IGF2, IGFBP3, and IGF1R genes are summarized in Table 1.
DNA isolation and preamplification
DNA was extracted from FFPE tissue samples. Preferentially, tissue from tumor-negative breast tissue and tumor-negative lymph nodes was used (N = 95); however, when this was unavailable or unclear from the pathology report tissue from tumor-containing blocks was used. Three sections of 4 μm were incubated overnight at 50 °C in 500 μl lysis buffer (NH4Cl 8.4 g/l, KHCO3 1.0 g/l, proteinase K 0.25 mg/ml). Next, 300 μl was taken to extract DNA using the Maxwell forensic DNA isolation kit (Promega, Leiden, the Netherlands) according to the manufacturer’s protocol. DNA isolated from FFPE tissue is cross-linked and fragmented into pieces with a length of a few hundred base pairs. To make DNA isolated from FFPE tissue more suitable for genotyping, preamplification was accomplished for enrichment of the target DNA [29]. The preamplification step consisted of a PCR reaction with eight diluted TaqMan assays (LifeTechnologies, Nieuwerkerk aan den IJssel, the Netherlands) and was performed using the following protocol; to 2.5 μl DNA, 1 μl of a dilution of eight TaqMan assays (pooled at a final concentration of 0.2×) and 3.5 μl HotStarTaq DNA polymerase was added and amplified on a conventional PCR machine. The following PCR conditions were used; 10 minutes at 95 °C followed by 18 cycles each consisting of 15 seconds at 95 °C and 4 minutes at 60 °C. The mixture was diluted 15 times and 2 μl was used for real-time PCR analysis. The selected SNPs were analyzed using TaqMan OpenArray® technology (Life Technologies); however, in case of low call rate, missing samples were reanalyzed separately using the Viia7 RealTime PCR system (Life Technologies).
Statistical analysis
Possible associations between parameters were analyzed using Pearson’s chi-square test and logistic regression. Univariate and multivariate odds ratios (ORs), 95 % confidence intervals (CIs), and P values were derived from logistic regressions. IGF-1R expression and clinical variables, which have been reported to be associated with pCR, were tested in univariate analysis (e.g., hormone receptor (HR) status and clinical T status). The association between IGF-1R expression and MP classification were tested using a logistic ordinal regression where MP classification groups were treated as ordered. In multivariate analyses, parameters were adjusted for covariates with P <0.1. We also reanalyzed the latter model using linear regression to check for linearity of relationship between IGF-1R expression and MP classification.
Genotype distributions were tested for adherence to Hardy–Weinberg equilibrium and SNPs significant at the 0.05 level after Bonferroni correction were excluded from the analysis. Genotypes found to be (borderline) significant in the univariate logistic regression models were carried forward to the multivariate model, adjusting for covariates with P < 0.1. To correct for multiple testing, a global score test including all SNPs was performed [30]. The score test assumes that the regression coefficients of the SNPs are normally distributed and tests whether the variance of this distribution is bigger than zero. In that case at least one regression coefficient has to be unequal to zero. To investigate the individual, relative contribution of SNPs, a classification and regression tree (CART) was computed (Statistical Package for Social Sciences (SPSS): classify, tree; (IBM Corp., Armonk, NY, USA)). A receiver operating characteristic curve and the area under the curve (AUC) were computed for the predicted probabilities of the CART. The global P value was computed using the package globaltest in R version 3.1.3 (The R Foundation for Statistical Computing, Institute for Statistics and Mathematics, Vienna, Austria). All other analyses were computed using SPSS software™ 20.0 (IBM Corp.). A significance level of 0.05 was used for all tests.
Results
Patient characteristics
Patients of both study arms, chemotherapy with or without zoledronic acid, were included in this study, as no differences were found between both arms regarding pathological response [26]. FFPE tissue was available from 216 (86.4 %) of 250 patients. Clinical characteristics of the 216 patients are presented in Table 2, which are comparable with the characteristics of the entire cohort of the NEOZOTAC trial [26]. Almost 12 % of the patients had a pCR.
IGF-1R expression
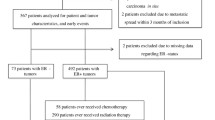
FFPE breast tumor tissue from 216 patients was available for analyzing at least one condition (biopsy and/or operation specimen), while both samples were available for 106 cases. Data of available tissue are summarized in the consort diagram (Fig. 2). Representative tissue examples with different scoring values can be found in Fig. 1. High IGF-1R expression in the prechemotherapy biopsy was associated with ER expression (P = 0.001) and the progesterone receptor (PR) expression (P = 0.035). ER and/or PR-positive tumors showed positive IGF-1R on the membrane in 78.0 % of the cases, while triple-negative tumors showed positivity for IGF-1R in only 50.0 % of the cases.
During chemotherapy, a significant subset (47.2 %), of tumors showed a decrease in IGF-1R expression while in a small subset of tumors the IGF-1R was upregulated (15.1 %). IGF-1R expression before treatment was not associated with pathological response (Fig. 3). However, the absence of IGF-1R expression (45 %) after treatment in the postchemotherapy operation specimens was associated with a better pathological response comparing ordinal MP classification response in univariate analysis (OR 2.60, 95 % CI 1.31–5.18, P = 0.006) (Fig. 3). This result remained significant in multivariate analysis when adjusting for HR status and clinical N stage (OR 2.64, 95 % CI 1.32–5.31, P = 0.006). With linear regression P = 0.008, indicating that the relationship between MP classification and IGF-1R expression is almost linear. Additionally, patients with a decrease in expression during treatment showed a better response to chemotherapy as well (OR 2.64, 95 % CI 1.17–5.98, P = 0.020 in multivariate analysis). Treatment with zoledronic acid had no influence on the IGF-1R expression in the operation specimen after treatment (P = 0.620) nor on diminished IGF-1R expression during treatment (P = 0.830) (data not shown).
IGF-1R pathway SNPs
FFPE tissue samples from 184 (74 %) of 250 patients were available for analysis of IGF-1 pathway polymorphisms (preferentially tumor-negative tissue, see Methods). Data of available tissue are summarized in the consort diagram (Fig. 2). Of the eight genotyped SNPs, two significantly deviated from the Hardy–Weinberg equilibrium (rs2946834 and rs1520220). After correction for multiple testing, rs2946834 still significantly deviated from the Hardy–Weinberg equilibrium and was therefore excluded from the analysis. The genotype frequencies of rs1520220 did not differ from those observed in a publicly available database of European subjects (e.g., from the HapMap project) [28]. All eight SNPs had a call rate above 85 %, which is shown in Table 3. Clinical T stage, clinical N stage, and HR status were associated with pCR, wherefore was adjusted in multivariate analyses (Table 4). The variant T allele of 3129G > T in IGF1R (rs2016347) was associated with pCR in multivariate analysis (4.4 % for GG vs. 16.7 % GT/TT, P = 0.032) and the variant C allele of rs2854746 in IGFBP3 tended to be associated with pCR in multivariate analysis (7.3 % for GG vs. 18.1 % GC/CC, P = 0.058). The global P value used for multiple testing correction for all eight SNPs together was P = 0.0095 for the dominant model (global score test). The CART derived from these SNPs is shown in Fig. 4. The corresponding AUC was 0.613 (95 % CI 0.518–0.707, P = 0.040).
CART analyses of pCR in BC patients treated with TAC chemotherapy. The tree is divided by the SNPs to predict pCR, which has a significant prediction score (AUC 0.613 95 % CI 0.518–0.707, P = 0.040). IGFBP3 insulin-like growth factor binding protein 3, IGF1R insulin-like growth factor 1 receptor, pCR pathological complete response
Moreover, the variant T allele of C > T in IGFBP3 (rs2270628) was associated with a higher occurrence of grade III/IV side effects in univariate analysis (OR 2.20, 95 % CI 1.04–4.67, P = 0.039) and multivariate analysis (18.1 % for CC vs. 32.7 % CT/TT, OR 2.30, 95 % CI 1.06–4.98, P = 0.034) (data not shown). The multivariate analysis was adjusted for body mass index, as it was significantly associated with grade III/IV side effects.
Genotype–phenotype associations
rs2016347 in IGF1R was not associated with IGF-1R expression before chemotherapy (78.3 % for GG vs. 65.9 % GT/TT, P = 0.115) or after chemotherapy (50.0 % for GG vs. 67.7 % GT/TT, P = 0.099).
Discussion
This translational study showed that IGF-1R expression changed in most of the tumors during treatment in stage II/III HER2-negative BC patients treated with neoadjuvant TAC chemotherapy and that absent or diminished expression after treatment was associated with a better pathological response according to MP classification. Additionally, we found that the variant T allele of 3129G > T in IGF1R (rs2016347) was significantly associated with a better pathological response according to MP classification after neoadjuvant chemotherapy.
Changes of IGF-1R expression of the tumor during chemotherapy have been described previously [18, 31]. Our study confirms these results in a larger and more homogeneous patient cohort. Moreover, in the current trial a greater part of the tumors showed a decline in IGF-1R expression (47.2 %) compared with the prior described 14.0 %. This might be explained by the difference in chemotherapy regimens used as well as the absence of HER2 expression in our cohort, as HER2-positive tumors show less IGF-1R expression [10, 11]. The decline of IGF-1R expression in the tumor during TAC treatment observed in our study might reflect chemotherapy efficacy, as patients with a decline in IGF-1R expression showed a significantly better pathological response than tumors with no change or an increase in expression. In keeping with this inference, downregulation of IGF-1R during chemotherapy treatment is associated with prolonged survival [18]. Bhargava et al. [10] showed that low IGF-1R expression before treatment was associated with a better response to neoadjuvant chemotherapy in ER-positive tumors, but not in triple-negative tumors. We could not reproduce this association, but this could be explained by the difference in cohort (e.g., differences in HER2 status and chemotherapy regimen).
In our exploratory analysis of IGF-1 pathway polymorphisms, the variant T allele of 3129G > T in IGF1R (rs2016347) was associated with a better pathological response according to MP classification after neoadjuvant chemotherapy. This is in accordance with studies that associated 3129G > T in IGF1R (rs2016347) with cancer prognosis and treatment outcome [22, 32, 33]. Winder et al. [22] found that the T allele was associated with a better overall survival in colorectal cancer patients treated with cetuximab [33] and a better overall survival in ER-positive BC patients treated with tamoxifen. rs2016347 is localized in the 3′-untranslated region of the IGF1R gene, functioning as a microRNA binding site [28]. Because microRNA binding sites are important for mRNA translation and degradation, the variant T allele of rs2016347 might disturb binding to this microRNA site [34]. Although the precise functional effect of IGF1R rs2016347 is unknown, it would be a plausible explanation that the T allele of rs2016347 may reduce IGF-1R expression. However, in our study rs2016347 in IGF1R was not associated with IGF-1R expression.
The variant T allele of C > T in IGFBP3 (rs2270628) was associated with the occurrence of grade III/IV side effects. Although the mechanism is unclear, several studies have shown that the variant T allele of rs2270628 is associated with decreased serum IGF-BP3 levels [35, 36]. IGF-1 activity depends on binding with IGF-BP3 [12, 13], so it may be that higher activity of IGF-1 due to lower levels of IGF-BP3 causes a higher incidence of toxicity of chemotherapy in our study [6].
Our study has some limitations. Using our approach, we could not investigate the best responders (MP5) after chemotherapy because inherently no tumor tissue was left to measure IGF-1R in the operation specimen. Moreover, the response of the lymph nodes is not evaluated in the MP grading system because it focuses only on the primary tumor. Although, the survival of patients with a partial response is affected by residual lymph node status [37]. Additionally, the number of evaluable triple-negative tumors was too small to evaluate for differences in response associated with IGF-1R between HR-positive tumors and triple-negative tumors. Our sample size for the explorative genotype–phenotype optional side study was small and this was probably the reason why we could not reproduce the associations between the serum IGF-1 and IGF-BP3 levels and SNPs. However, the results of our study provide further evidence for the importance of patient selection for (co)treatment with an IGF-1 inhibitor. Until now no convincing benefit of IGF-I pathway inhibitors was found in clinical studies in BC [38–40]. These studies lacked patient selection based on IGF-1 pathway activity. It may be hypothesized that patients with a diminished IGF-1R after chemotherapy will not benefit from an IGF-1R inhibitor, while a patient with upregulated IGF-1R might benefit.
Conclusions
IGF-1R expression in the tumor changed during chemotherapy and absent or diminished expression of IGF-1R after treatment was associated with a better pathological response. rs2016347 in IGF1R was associated with pCR after TAC chemotherapy. These observations may help to predict the efficacy of TAC chemotherapy and to select patients who might benefit from (co)treatment with an IGF-1 pathway inhibitor.
Abbreviations
- AUC:
-
Area under the curve
- BC:
-
Breast cancer
- BSA:
-
Bovine serum albumin
- CART:
-
Classification and regression tree
- CI:
-
Confidence interval
- CTCAE:
-
Common Terminology Criteria for Adverse Events
- ER:
-
Estrogen receptor
- FFPE:
-
Formalin-fixed paraffin-embedded
- HR:
-
Hormone receptor
- IGF:
-
Insulin-like growth factor
- IGF-1R:
-
Insulin-like growth factor 1 receptor
- IGF-BP:
-
Insulin-like growth factor binding protein
- IHC:
-
Immunohistochemistry
- MAF:
-
Minor allele frequency
- MP:
-
Miller and Payne
- OR:
-
Odds ratio
- PBS:
-
Phosphate-buffered saline
- pCR:
-
Pathological complete response
- PR:
-
Progesterone receptor
- SNP:
-
Single nucleotide polymorphism
- TAC:
-
Docetaxel, doxorubicin, and cyclophosphamide
References
Samani AA, Yakar S, LeRoith D, Brodt P. The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocr Rev. 2007;28:20–47.
Renehan AG, Zwahlen M, Minder C, O’dwyer ST, Shalet SM, Egger M. Insulin-like growth factor (IGF)-I, IGF binding protein-3, and cancer risk: systematic review and meta-regression analysis. Lancet. 2004;363:1346–53.
Diorio C, Pollak M, Byrne C, Masse B, Hebert-Croteau N, Yaffe M, et al. Insulin-like growth factor-I, IGF-binding protein-3, and mammographic breast density. Cancer Epidemiol Biomarkers Prev. 2005;14:1065–73.
Key TJ, Appleby PN, Reeves GK, Roddam AW. Insulin-like growth factor 1 (IGF1), IGF binding protein 3 (IGFBP3), and breast cancer risk: pooled individual data analysis of 17 prospective studies. Lancet Oncol. 2010;11:530–42.
Duggan C, Wang CY, Neuhouser ML, Xiao L, Smith AW, Reding KW, et al. Associations of insulin-like growth factor and insulin-like growth factor binding protein-3 with mortality in women with breast cancer. Int J Cancer. 2013;132:1191–200.
Lee C, Safdie FM, Raffaghello L, Wei M, Madia F, Parrella E, et al. Reduced levels of IGF-I mediate differential protection of normal and cancer cells in response to fasting and improve chemotherapeutic index. Cancer Res. 2010;70:1564–72.
Lin MZ, Marzec KA, Martin JL, Baxter RC. The role of insulin-like growth factor binding protein-3 in the breast cancer cell response to DNA-damaging agents. Oncogene 2012;33:88-96.
Zhang Y, Moerkens M, Ramaiahgari S, de Bont H, Price L, Meerman J, et al. Elevated insulin-like growth factor 1 receptor signaling induces antiestrogen resistance through the MAPK/ERK and PI3K/Akt signaling routes. Breast Cancer Res. 2011;13:R52.
Nahta R, Yuan LX, Zhang B, Kobayashi R, Esteva FJ. Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer Res. 2005;65:11118–28.
Bhargava R, Beriwal S, McManus K, Dabbs DJ. Insulin-like growth factor receptor-1 (IGF-1R) expression in normal breast, proliferative breast lesions, and breast carcinoma. Appl Immunohistochem Mol Morphol. 2011;19:218–25.
Yerushalmi R, Gelmon KA, Leung S, Gao D, Cheang M, Pollak M, et al. Insulin-like growth factor receptor (IGF-1R) in breast cancer subtypes. Breast Cancer Res Treat. 2012;132:131–42.
Jones JI, Clemmons DR. Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev. 1995;16:3–34.
Stewart CE, Rotwein P. Growth, differentiation, and survival: multiple physiological functions for insulin-like growth factors. Physiol Rev. 1996;76:1005–26.
Zhang X, Lin M, van Golen KL, Yoshioka K, Itoh K, Yee D. Multiple signaling pathways are activated during insulin-like growth factor-I (IGF-I) stimulated breast cancer cell migration. Breast Cancer Res Treat. 2005;93:159–68.
Zhu C, Qi X, Chen Y, Sun B, Dai Y, Gu Y. PI3K/Akt and MAPK/ERK1/2 signaling pathways are involved in IGF-1-induced VEGF-C upregulation in breast cancer. J Cancer Res Clin Oncol. 2011;137:1587–94.
Hamelers IH, Steenbergh PH. Interactions between estrogen and insulin-like growth factor signaling pathways in human breast tumor cells. Endocr Relat Cancer. 2003;10:331–45.
Maor S, Mayer D, Yarden RI, Lee AV, Sarfstein R, Werner H, et al. Estrogen receptor regulates insulin-like growth factor-I receptor gene expression in breast tumor cells: involvement of transcription factor Sp1. J Endocrinol. 2006;191:605–12.
Heskamp S, Boerman OC, Molkenboer-Kuenen JD, Wauters CA, Strobbe LJ, Mandigers CM, et al. Upregulation of IGF-1R expression during neoadjuvant therapy predicts poor outcome in breast cancer patients. PLoS One. 2015;10:e0117745.
Diorio C, Brisson J, Berube S, Pollak M. Genetic polymorphisms involved in insulin-like growth factor (IGF) pathway in relation to mammographic breast density and IGF levels. Cancer Epidemiol Biomarkers Prev. 2008;17:880–8.
Gu F, Schumacher FR, Canzian F, Allen NE, Albanes D, Berg CD, et al. Eighteen insulin-like growth factor pathway genes, circulating levels of IGF-I and its binding protein, and risk of prostate and breast cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:2877–87.
Muendlein A, Lang AH, Geller-Rhomberg S, Winder T, Gasser K, Drexel H, et al. Association of a common genetic variant of the IGF-1 gene with event-free survival in patients with HER2-positive breast cancer. J Cancer Res Clin Oncol. 2013;139:491–8.
Winder T, Giamas G, Wilson PM, Zhang W, Yang D, Bohanes P, et al. Insulin-like growth factor receptor polymorphism defines clinical outcome in estrogen receptor-positive breast cancer patients treated with tamoxifen. Pharmacogenomics J. 2014;14:28–34.
Mauri D, Pavlidis N, Ioannidis JP. Neoadjuvant versus adjuvant systemic treatment in breast cancer: a meta-analysis. J Natl Cancer Inst. 2005;97:188–94.
Ogston KN, Miller ID, Payne S, Hutcheon AW, Sarkar TK, Smith I, et al. A new histological grading system to assess response of breast cancers to primary chemotherapy: prognostic significance and survival. Breast. 2003;12:320–7.
Romero A, Garcia-Saenz JA, Fuentes-Ferrer M, Lopez Garcia-Asenjo JA, Furio V, Roman JM, et al. Correlation between response to neoadjuvant chemotherapy and survival in locally advanced breast cancer patients. Ann Oncol. 2013;24:655–61.
Charehbili A, van de Ven S, Smit VT, Meershoek-Klein KE, Hamdy NA, Putter H, et al. Addition of zoledronic acid to neoadjuvant chemotherapy does not enhance tumor response in patients with HER2-negative stage II/III breast cancer: the NEOZOTAC trial (BOOG 2010-01). Ann Oncol. 2014;25:998–1004.
CTEP, CTCAE files 2009, National cancer institute, Bethesda USA. http://evs.nci.nih.gov/ftp1/CTCAE/Archive/CTCAE_4.0_2009-05-29_QuickReference_8.5x11.pdf.
NIH, National institute of Environmental Health Sciences, Research Triangle Park, USA. https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm. Accessed date July 2013.
Baak-Pablo R, Dezentje V, Guchelaar HJ, Van der Straaten T. Genotyping of DNA samples isolated from formalin-fixed paraffin-embedded tissues using preamplification. J Mol Diagn. 2010;12:746–9.
Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testingassociation with a clinical outcome. Bioinformatics. 2004;20:93-9.
Collecchi P, Giannessi PG, Baldini E, Naccarato AG, Passoni A, Bevilacqua G, et al. Effects of primary chemotherapy on biological parameters of locally advanced breast cancer. Ann N Y Acad Sci. 1996;784:419–26.
Chang CF, Pao JB, Yu CC, Huang CY, Huang SP, Yang YP, et al. Common variants in IGF1 pathway genes and clinical outcomes after radical prostatectomy. Ann Surg Oncol. 2013;20:2446–52.
Winder T, Zhang W, Yang D, Ning Y, Bohanes P, Gerger A, et al. Germline polymorphisms in genes involved in the IGF1 pathway predict efficacy of cetuximab in wild-type KRAS mCRC patients. Clin Cancer Res. 2010;16:5591–602.
Leung AK, Sharp PA. Function and localization of microRNAs in mammalian cells. Cold Spring Harb Symp Quant Biol. 2006;71:29–38.
Patel AV, Cheng I, Canzian F, Le ML, Thun MJ, Berg CD, et al. IGF-1, IGFBP-1, and IGFBP-3 polymorphisms predict circulating IGF levels but not breast cancer risk: findings from the Breast and Prostate Cancer Cohort Consortium (BPC3). PLoS One. 2008;3:e2578.
D’Aloisio AA, Schroeder JC, North KE, Poole C, West SL, Travlos GS, et al. IGF-I and IGFBP-3 polymorphisms in relation to circulating levels among African American and Caucasian women. Cancer Epidemiol Biomarkers Prev. 2009;18:954–66.
Zhang GC, Zhang YF, Xu FP, Qian XK, Guo ZB, Ren CY, et al. Axillary lymph node status, adjusted for pathologic complete response in breast and axilla after neoadjuvant chemotherapy, predicts differential disease-free survival in breast cancer. Curr Oncol. 2013;20:e180–92.
Ma CX, Suman VJ, Goetz M, Haluska P, Moynihan T, Nanda R, et al. A phase I trial of the IGF-1R antibody cixutumumab in combination with temsirolimus in patients with metastatic breast cancer. Breast Cancer Res Treat. 2013;139:145–53.
Murakami H, Doi T, Yamamoto N, Watanabe J, Boku N, Fuse N, et al. Phase 1 study of ganitumab (AMG 479), a fully human monoclonal antibody against the insulin-like growth factor receptor type I (IGF1R), in Japanese patients with advanced solid tumors. Cancer Chemother Pharmacol. 2012;70:407–14.
Robertson JF, Ferrero JM, Bourgeois H, Kennecke H, de Boer RH, Jacot W, et al. Ganitumab with either exemestane or fulvestrant for postmenopausal women with advanced, hormone-receptor-positive breast cancer: a randomised, controlled, double-blind, phase 2 trial. Lancet Oncol. 2013;14:228–35.
Verheus M, McKay JD, Kaaks R, Canzian F, Biessy C, Johansson M, et al. Common genetic variation in the IGF-1 gene, serum IGF-I levels and breast density. Breast Cancer Res Treat. 2008;112:109–22.
Al-Zahrani A, Sandhu MS, Luben RN, Thompson D, Baynes C, Pooley KA, et al. IGF1 and IGFBP3 tagging polymorphisms are associated with circulating levels of IGF1, IGFBP3 and risk of breast cancer. Hum Mol Genet. 2006;15:1–10.
Cheng I, DeLellis HK, Haiman CA, Kolonel LN, Henderson BE, Freedman ML, et al. Genetic determinants of circulating insulin-like growth factor (IGF)-I, IGF binding protein (BP)-1, and IGFBP-3 levels in a multiethnic population. J Clin Endocrinol Metab. 2007;92:3660–6.
Graziano F, Ruzzo A, Canestrari E, Catalano V, Santini D, Galluccio N, et al. Host genetic variants in the IGF binding protein-3 impact on survival of patients with advanced gastric cancer treated with palliative chemotherapy. Pharmacogenomics. 2010;11:1247–56.
Lu L, Risch E, Deng Q, Biglia N, Picardo E, Katsaros D, et al. An insulin-like growth factor-II intronic variant affects local DNA conformation and ovarian cancer survival. Carcinogenesis. 2013;34:2024–30.
Acknowledgments
The authors thank all of the participating centers and are greatly indebted to the patients for participating in this study. They thank the LUMC Datacenter, Department of Surgery, for trial coordination and data collection. This work was supported by grants from the Dutch Cancer Society (2010-4682), Amgen, Novartis, and Sanofi Aventis. The Dutch Breast Cancer Research Group (BOOG), Amsterdam, the Netherlands is sponsor of this study.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conception and design: SdG, AC, HWMvL, SvdV, LGMJ, HG, H-JG, HP, and JRK. Study coordination: AC, SvdV, JRK, and Dutch breast cancer group. Development of experiments: SdG, AC, HWMvL, ALM, NGD-E, LGMJ, TvdS, HP, and JRK. Acquisition of data: SdG, AC, HWMvL, ALM, NGD-E, LGMJ, VTHBMS, JBH, LWK, TvdS, and JRK. Analysis and interpretation of data: SdG, AC, ALM, LGMJ, JJS, TvdS, SB, JJMvH, H-JG, HP, and JRK. Study supervision: HG, JJMvdH, HP, and JRK. Writing of the manuscript: SdG, NGD-E, SB, HP, and JRK. All authors critically revised and approved the final manuscript and agree to be accountable for all aspects of the work.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
de Groot, S., Charehbili, A., van Laarhoven, H.W.M. et al. Insulin-like growth factor 1 receptor expression and IGF1R 3129G > T polymorphism are associated with response to neoadjuvant chemotherapy in breast cancer patients: results from the NEOZOTAC trial (BOOG 2010-01). Breast Cancer Res 18, 3 (2016). https://doi.org/10.1186/s13058-015-0663-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13058-015-0663-3