Abstract
Background
Heterogeneity of acute respiratory distress syndrome (ARDS) could be reduced by identification of biomarker-based phenotypes. The set of ARDS biomarkers to prospectively define these phenotypes remains to be established.
Objective
To provide an overview of the biomarkers that were multivariately associated with ARDS development or mortality.
Data sources
We performed a systematic search in Embase, MEDLINE, Web of Science, Cochrane CENTRAL, and Google Scholar from inception until 6 March 2020.
Study selection
Studies assessing biomarkers for ARDS development in critically ill patients at risk for ARDS and mortality due to ARDS adjusted in multivariate analyses were included.
Data extraction and synthesis
We included 35 studies for ARDS development (10,667 patients at risk for ARDS) and 53 for ARDS mortality (15,344 patients with ARDS). These studies were too heterogeneous to be used in a meta-analysis, as time until outcome and the variables used in the multivariate analyses varied widely between studies. After qualitative inspection, high plasma levels of angiopoeitin-2 and receptor for advanced glycation end products (RAGE) were associated with an increased risk of ARDS development. None of the biomarkers (plasma angiopoeitin-2, C-reactive protein, interleukin-8, RAGE, surfactant protein D, and Von Willebrand factor) was clearly associated with mortality.
Conclusions
Biomarker data reporting and variables used in multivariate analyses differed greatly between studies. Angiopoeitin-2 and RAGE in plasma were positively associated with increased risk of ARDS development. None of the biomarkers independently predicted mortality. Therefore, we suggested to structurally investigate a combination of biomarkers and clinical parameters in order to find more homogeneous ARDS phenotypes.
PROSPERO identifier
PROSPERO, CRD42017078957
Similar content being viewed by others
Introduction
The acute respiratory distress syndrome (ARDS) is a major problem in the intensive care unit (ICU) with a prevalence of 10% and an in-hospital mortality rate of 40% [1, 2]. ARDS pathophysiology is based on a triad of alveolar-capillary membrane injury, high permeability alveolar oedema, and migration of inflammatory cells [3]. This triad is not routinely measured in clinical practice. Therefore, arterial hypoxemia and bilateral opacities on chest imaging following various clinical insults are used as clinical surrogates in the American European Consensus Conference (AECC) definition and the newer Berlin definition of ARDS [4, 5].
Histologically, ARDS is characterized by diffuse alveolar damage (DAD). The correlation between a clinical and histological diagnosis of ARDS is poor [6]. Only half of clinically diagnosed patients with ARDS have histological signs of DAD at autopsy [7,8,9,10]. The number of risk factors for ARDS and consequently the heterogeneous histological substrates found in patients with clinical ARDS have been recognized as a major contributor to the negative randomized controlled trial results among patients with ARDS [11].
It has been suggested that the addition of biomarkers to the clinical definition of ARDS could reduce ARDS heterogeneity by the identification of subgroups [12,13,14,15]. A retrospective latent class analysis of large randomized controlled trials identified two ARDS phenotypes largely based on ARDS biomarkers combined with clinical parameters [16, 17]. These phenotypes responded differently to the randomly assigned intervention arms. Prospective studies are required to validate these ARDS phenotypes and their response to interventions. The set of ARDS biomarkers to prospectively define these phenotypes remains to be established.
Numerous biomarkers and their pathophysiological role in ARDS have been described [12, 18]. In an earlier meta-analysis, biomarkers for ARDS development and mortality were examined in univariate analysis [19]. However, pooling of univariate biomarker data may result in overestimation of the actual effect. For this reason, we conducted a systematic review and included all biomarkers that were multivariately associated with ARDS development or mortality. This study provides a synopsis of ARDS biomarkers that could be used for future research in the identification of ARDS phenotypes.
Methods
This systematic review was prospectively registered in PROSPERO International Prospective Register of Systematic Reviews (PROSPERO identifier CRD42017078957) and performed according to the Transparent Reporting of Systematic Reviews and Meta-analyses (PRISMA) Statement [20]. After the search strategy, two reviewers (PZ, PS, and/or WG) separately performed study eligibility criteria, data extraction, and quality assessment. Any discrepancies were resolved by consensus, and if necessary, a third reviewer was consulted.
We searched for studies that included biomarkers that were associated with ARDS development in critically ill patients at risk for ARDS and mortality in the ARDS population in multivariate analyses adjusted for background characteristics. We did not perform a meta-analysis, because the raw data in all studies was either not transformed or log transformed resulting in varying risk ratios and confidence intervals. In addition, the majority of studies used different biomarker concentration cut-offs, resulting in varying concentration increments for risk ratios. Lastly, the number of days until mortality and variables used in multivariate analysis differed between studies. For these reasons, we limited this study to a systematic review, as the multivariate odds ratios were not comparable and pooling would result in non-informative estimates [21].
Search strategy
We performed a systematic search in Embase, MEDLINE, Web of Science, Cochrane CENTRAL, and Google Scholar from inception until 30 July 2018 with assistance from the Erasmus MC librarian. The search was later updated to 6 March 2020. A detailed description of the systematic search string is presented in Additional file 1. In addition, the reference lists of included studies and recent systematic reviews were screened to identify additional eligible studies.
Study eligibility criteria
All retrieved studies were screened on the basis of title and abstract. Studies that did not contain adult patients at risk for ARDS or with ARDS and any biomarker for ARDS were excluded. The following eligibility criteria were used: human research, adult population, studies in which biomarkers were presented as odds ratios (OR) or risk ratios in multivariate analysis with ARDS development or mortality as outcome of interest, peer-reviewed literature only, and English language. Studies comparing ARDS with healthy control subjects, case series (< 10 patients included in the study), and studies presenting gene expression fold change were excluded.
Data extraction
A standardized form was used for data extraction from all eligible studies. Two clinical endpoints were evaluated in this study: development of ARDS in the at-risk population (patients that did develop ARDS versus critically ill patients that did not) and mortality in the ARDS population (survivors versus non-survivors). The following data were extracted: study design and setting, study population, sample size, the definition of ARDS used in the study, outcome, risk ratio with 95% confidence interval in multivariate analyses, and the variables used in the analyses. In addition, the role of the biomarker in ARDS pathophysiology as reported by the studies was extracted and divided into the following categories: increased endothelial permeability, alveolar epithelial injury, oxidative injury, inflammation, pro-fibrotic, myocardial strain, coagulation, and others. Subsequently, the relative frequency distribution of biomarker roles in ARDS pathophysiology was depicted in a bar chart.
Quality assessment
Methodological quality of the included studies was assessed with the Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomized studies in systematic reviews and meta-analyses [22]. Items regarding patient selection, comparability, and outcome were assessed using a descriptive approach, and a risk-of-bias score, varying between 0 (high risk) and 9 (low risk), was assigned to each study.
Results
Literature search and study selection
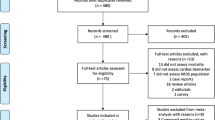
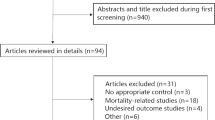
A total of 8125 articles were identified by the initial search and 972 by the updated search (Fig. 1). After removal of duplicates and reviewing titles and abstracts, we selected 438 articles for full-text review. A total of 86 studies was eligible for data extraction: 35 for ARDS development and 53 for ARDS mortality.
Study characteristics and quality assessment
The study characteristics of the 35 studies for ARDS development are presented in Table 1. A total of 10,667 critically ill patients was at risk for ARDS, of whom 2419 (24.6%) patients developed ARDS. The majority of studies used the Berlin definition of ARDS (21/35), followed by the AECC criteria of ARDS (13/35). The included biomarkers were measured in plasma, cerebrospinal fluid, and bronchoalveolar lavage fluid. In all studies, the first sample was taken within 72 h following ICU admission.
The study characteristics of the 53 studies for ARDS mortality are presented in Table 2. A total of 15,344 patients with ARDS were included with an observed mortality rate of 36.0%. The AECC definition of ARDS was used in the majority of included studies (39/53). The included biomarkers were measured in plasma, bronchoalveolar lavage fluid, and urine. All samples were taken within 72 h following the development of ARDS.
The median quality of the included publications according to the NOS was 7 (range 4–9) for ARDS development and 8 (range 5–9) for ARDS mortality (Additional file 2).
Biomarkers associated with ARDS development in the at-risk population
A total of 37 biomarkers in plasma, 7 in cerebrospinal fluid, and 1 in bronchoalveolar lavage fluid were assessed in multivariate analyses (Table 3). Five studies examined angiopoeitin-2 (Ang-2) and seven studies examined receptor for advanced glycation end products (RAGE). In all studies, high plasma levels of Ang-2 and RAGE were significantly associated with an increased risk of ARDS development in the at-risk population. Similar results were seen for surfactant protein D (SpD) in plasma in all three studies that assessed SpD. In contrast, biomarkers for inflammation as C-reactive protein (CRP), procalcitonin, interleukin-6, and interleukin-8 were not clearly associated with ARDS development. The majority of biomarkers in plasma are surrogates for inflammation in ARDS pathophysiology (Fig. 2).
Biomarkers associated with mortality in the ARDS population
A total of 49 biomarkers in plasma, 8 in bronchoalveolar lavage fluid, and 3 in urine were included in this study (Table 4). Ang-2, CRP, interleukin-8 (IL-8), RAGE, SpD, and Von Willebrand factor (VWF) in plasma were assessed in four or more studies. However, none of these biomarkers was associated with ARDS mortality in all four studies. Similarly to biomarkers in ARDS development, the majority of biomarkers for ARDS mortality in plasma had a pathophysiological role in inflammation (Fig. 2). The majority of biomarkers measured in bronchoalveolar lavage fluid had a pro-fibrotic role in ARDS pathophysiology.
Discussion
In the current systematic review, we present a synopsis of biomarkers for ARDS development and mortality tested in multivariate analyses. We did not perform a meta-analysis because of severe data heterogeneity between studies. Upon qualitative inspection, we found that high levels of Ang-2 and RAGE were associated with ARDS development in the at-risk population. None of the biomarkers assessed in four or more studies was associated with an increased mortality rate in all studies. The majority of plasma biomarkers for both ARDS development and mortality are surrogates for inflammation in ARDS pathophysiology.
Previously, Terpstra et al. [19] calculated univariate ORs from absolute biomarker concentrations and performed a meta-analysis. They found that 12 biomarkers in plasma were associated with mortality in patients with ARDS. However, a major limitation of their meta-analysis is that these biomarkers were tested in univariate analyses without considering confounders as disease severity scores. Given the high univariate ORs as compared to the multivariate ORs found in this systematic review, the performance of these biomarkers is likely to be overestimated. Jabaudon et al. [109] found in an individual patient data meta-analysis that high concentrations of plasma RAGE were associated with 90-day mortality independent of driving pressure or tidal volume. However, they could not correct for disease severity score as these differed between studies. Unfortunately, we were unable to perform a meta-analysis on multivariate data because of heterogeneity of the included studies, as transformation of raw data, biomarker concentration cut-offs, time until outcome, and the variables used in the multivariate analyses varied widely between studies. This could be an incentive to standardize the presentation of ARDS biomarker research in terms of statistics and outcome for future analyses or to make individual patient data accessible.
ARDS biomarkers are presumed to reflect the pathophysiology of ARDS, characterized by alveolar-capillary membrane injury, high permeability alveolar oedema, and migration of inflammatory cells [3]. Previously, Terpstra et al. [19] proposed that biomarkers for ARDS development were correlated with alveolar tissue injury, whereas biomarkers for ARDS mortality correlated more with inflammation. In this systematic review, we found that the majority of biomarkers tested for both ARDS development and mortality were surrogates for inflammation. However, following qualitative inspection, biomarkers for inflammation were not evidently associated with either ARDS development or mortality. In contrast, markers for alveolar epithelial injury (plasma RAGE and SpD) and endothelial permeability (plasma Ang-2) seem to be associated with ARDS development. Therefore, we should consider how we intend to use (a set of) biomarkers in patients with ARDS.
A biomarker for ARDS development should be specific for ARDS, i.e. a biomarker that reflects alveolar injury or alveolar-capillary injury. Half of plasma biomarkers for ARDS development included in this study reflected inflammation. An increase in inflammatory biomarkers is known to correlate with increased disease severity scores [71, 97, 110]. In turn, the majority of studies in this review found significantly higher disease severity scores in the critically ill patients that eventually developed ARDS. Thus, plasma biomarkers for inflammation rather represented an estimation of disease severity and its associated increased risk for the development of ARDS. In addition, biomarkers for inflammation in plasma lack the specificity to diagnose ARDS, as they are unlikely to differentiate sepsis with ARDS from sepsis without ARDS. In contrast, locally sampled biomarkers for inflammation, for example in the alveolar space, could potentially diagnose ARDS [111]. Biomarkers used for ARDS mortality or for the identification of less heterogeneous ARDS phenotypes do not require to be ARDS specific, provided that they adequately predict or stratify patients with ARDS.
The heterogeneity of ARDS has been recognized as a major contributor to the negative randomized controlled trial results among patients with ARDS [11]. Therefore, it is necessary to identify homogeneous ARDS phenotypes that are more likely to respond to an intervention. This is known as predictive enrichment [112]. Previously, patients with ARDS have been successfully stratified based on clinical parameters, such as ARDS risk factor (pulmonary or extra-pulmonary) or PaO2/FiO2 ratio [113]. ARDS biomarkers could be used to stratify patients with ARDS based on biological or pathophysiological phenotype. For example, trials of novel therapies designed to influence vascular permeability may benefit from preferentially enrolling patients with high Ang-2 concentrations. Recently, clinical parameters have been combined with a set of biomarkers in a retrospective latent class analysis. In three trials, two distinct phenotypes were found: hyperinflammatory and hypoinflammatory ARDS [16, 17]. Patients with the hyperinflammatory phenotype had reduced mortality rate with higher positive end-expiratory pressures and with liberal fluid treatment, whereas the trials themselves found no difference between the entire intervention groups. The next step is to validate the identification of ARDS phenotypes based on latent class analysis in prospective studies. An adequate combination of biomarkers and clinical parameters remains to be established. Until now, there is no list of biomarkers that are associated with ARDS development or mortality independently of clinical parameters. This systematic review may guide the selection of ARDS biomarkers used for predictive enrichment.
This systematic review has limitations. First, the intent of this systematic review was to perform a meta-analysis. However, we decided not to perform a meta-analysis, as the biomarker data handling and outcomes varied widely among studies, and pooling would have resulted in a non-informative estimate [21]. Arguably, this is a positive result, as it refrains us from focusing on the few biomarkers that could be pooled in a meta-analysis and guides us into a direction were multiple biomarkers combined with other parameters are of interest. In a heterogeneous syndrome as ARDS, the one biomarker probably does not exist. Second, the first sampling moment varied between sampling at ICU admission until 72 h following ICU admission. Initially, ARDS is characterized by an exudative phase followed by a second proliferative phase and late fibrotic phase [3]. The moment of sampling likely influences biomarker concentrations, as both alveolar membrane injury and inflammation increase during the exudative phase. This is also seen in six biomarkers that have been measured at separate days, resulting in a significant change in adjusted OR for four biomarkers (Table 4) [61, 98, 104, 105]. Third, the aim of this systematic review was to assess the independent risk effects of biomarkers measured in various bodily fluid compartments. However, the majority of studies assessed biomarkers in plasma. It remains to be answered whether other bodily fluid compartments, for example from the airways and alveolar space themselves, might outperform ARDS biomarkers in plasma, especially for ARDS development. Fourth, all studies found in this systematic review used a clinical definition of ARDS as standard for ARDS diagnosis. Given the poor correlation between a clinical diagnosis and a histopathological diagnosis of ARDS, these studies are diagnosing a very heterogeneous disease syndrome [7,8,9,10]. In order to actually evaluate ARDS development, biomarkers should be compared to a histopathological image of DAD, although acquiring histology poses great challenges by itself. Fifth, as only biomarkers assessed in multivariate analyses were included in this study, new promising biomarkers evaluated in univariate analyses were excluded from this study. Lastly, non-significant biomarkers in multivariate analyses were more likely not to be reported, although some studies report non-significant results nonetheless.
Conclusion
In here, we present a list of biomarkers for ARDS mortality and ARDS development tested in multivariate analyses. In multiple studies that assessed Ang-2 and RAGE, high plasma levels were associated with an increased risk of ARDS development. We did not find a biomarker that independently predicted mortality in all studies that assessed the biomarker. Furthermore, biomarker data reporting and variables used in multivariate analyses differed greatly between studies. Taken together, we should look for a combination of biomarkers and clinical parameters in a structured approach in order to find more homogeneous ARDS phenotypes. This systematic review may guide the selection of ARDS biomarkers for ARDS phenotyping.
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AECC:
-
American European Consensus Conference
- Ang-2:
-
Angiopoeitin-2
- ARDS:
-
Acute respiratory distress syndrome
- CRP:
-
C-reactive protein
- DAD:
-
Diffuse alveolar damage
- IL-8:
-
Interleukin-8
- NOS:
-
Newcastle-Ottawa Scale
- OR:
-
Odds ratio
- RAGE:
-
Receptor for advanced glycation end products
- SpD:
-
Surfactant protein D
- VWF:
-
Von Willebrand factor
References
Maca J, Jor O, Holub M, Sklienka P, Bursa F, Burda M, Janout V, Sevcik P. Past and present ARDS mortality rates: a systematic review. Respir Care. 2017;62(1):113–22.
Bellani G, Laffey JG, Pham T, Fan E, Brochard L, Esteban A, Gattinoni L, van Haren F, Larsson A, McAuley DF, et al. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. Jama. 2016;315(8):788–800.
Thompson BT, Chambers RC, Liu KD. Acute respiratory distress syndrome. N Engl J Med. 2017;377(19):1904–5.
Bernard GR, Artigas A, Brigham KL, Carlet J, Falke K, Hudson L, Lamy M, Legall JR, Morris A, Spragg R. The American-European consensus conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med. 1994;149(3 Pt 1):818–24.
Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, Fan E, Camporota L, Slutsky AS. Acute respiratory distress syndrome: the Berlin definition. Jama. 2012;307(23):2526–33.
Tomashefski JF Jr. Pulmonary pathology of acute respiratory distress syndrome. Clin Chest Med. 2000;21(3):435–66.
de Hemptinne Q, Remmelink M, Brimioulle S, Salmon I, Vincent JL. ARDS: a clinicopathological confrontation. Chest. 2009;135(4):944–9.
Kao KC, Hu HC, Chang CH, Hung CY, Chiu LC, Li SH, Lin SW, Chuang LP, Wang CW, Li LF, et al. Diffuse alveolar damage associated mortality in selected acute respiratory distress syndrome patients with open lung biopsy. Crit Care. 2015;19:228.
Lorente JA, Cardinal-Fernandez P, Munoz D, Frutos-Vivar F, Thille AW, Jaramillo C, Ballen-Barragan A, Rodriguez JM, Penuelas O, Ortiz G, et al. Acute respiratory distress syndrome in patients with and without diffuse alveolar damage: an autopsy study. Intensive Care Med. 2015;41(11):1921–30.
Thille AW, Esteban A, Fernandez-Segoviano P, Rodriguez JM, Aramburu JA, Penuelas O, Cortes-Puch I, Cardinal-Fernandez P, Lorente JA, Frutos-Vivar F. Comparison of the Berlin definition for acute respiratory distress syndrome with autopsy. Am J Respir Crit Care Med. 2013;187(7):761–7.
Matthay MA, McAuley DF, Ware LB. Clinical trials in acute respiratory distress syndrome: challenges and opportunities. Lancet Respir Med. 2017;5(6):524–34.
Garcia-Laorden MI, Lorente JA, Flores C, Slutsky AS, Villar J. Biomarkers for the acute respiratory distress syndrome: how to make the diagnosis more precise. Ann Transl Med. 2017;5(14):283.
Villar J, Slutsky AS. GOLDEN anniversary of the acute respiratory distress syndrome: still much work to do! Curr Opin Crit Care. 2017;23(1):4–9.
Frohlich S, Murphy N, Boylan JF. ARDS: progress unlikely with non-biological definition. Br J Anaesth. 2013;111(5):696–9.
Ware LB, Koyama T, Billheimer DD, Wu W, Bernard GR, Thompson BT, Brower RG, Standiford TJ, Martin TR, Matthay MA, et al. Prognostic and pathogenetic value of combining clinical and biochemical indices in patients with acute lung injury. Chest. 2010;137(2):288–96.
Calfee CS, Delucchi K, Parsons PE, Thompson BT, Ware LB, Matthay MA, Network NA. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med. 2014;2(8):611–20.
Famous KR, Delucchi K, Ware LB, Kangelaris KN, Liu KD, Thompson BT, Calfee CS, Network A. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am J Respir Crit Care Med. 2017;195(3):331–8.
Blondonnet R, Constantin JM, Sapin V, Jabaudon M. A pathophysiologic approach to biomarkers in acute respiratory distress syndrome. Dis Markers. 2016;2016:3501373.
Terpstra ML, Aman J, van Nieuw Amerongen GP, Groeneveld AB. Plasma biomarkers for acute respiratory distress syndrome: a systematic review and meta-analysis*. Crit Care Med. 2014;42(3):691–700.
Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J, Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6(7):e1000100.
Egger MS, GD., Altman DG. Systematic reviews in health care: meta-analysis in context. 2nd ed. London: BMJ Publishing Group; 2001.
Wells GA SB, O'Connell D, Peterson J, Welch V, Losos M, Tugwell P: The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Online http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp (cited 04-07-2018).
Agrawal A, Matthay MA, Kangelaris KN, Stein J, Chu JC, Imp BM, Cortez A, Abbott J, Liu KD, Calfee CS. Plasma angiopoietin-2 predicts the onset of acute lung injury in critically ill patients. Am J Respir Crit Care Med. 2013;187(7):736–42.
Ahasic AM, Zhai R, Su L, Zhao Y, Aronis KN, Thompson BT, Mantzoros CS, Christiani DC. IGF1 and IGFBP3 in acute respiratory distress syndrome. Eur J Endocrinol. 2012;166(1):121–9.
Aisiku IP, Yamal JM, Doshi P, et al. Plasma cytokines IL-6, IL-8, and IL-10 are associated with the development of acute respiratory distress syndrome in patients with severe traumatic brain injury. Crit Care. 2016;20:288. Published 2016 Sep 15. https://doi.org/10.1186/s13054-016-1470-7.
Amat M, Barcons M, Mancebo J, Mateo J, Oliver A, Mayoral JF, Fontcuberta J, Vila L. Evolution of leukotriene B4, peptide leukotrienes, and interleukin-8 plasma concentrations in patients at risk of acute respiratory distress syndrome and with acute respiratory distress syndrome: mortality prognostic study. Crit Care Med. 2000;28(1):57–62.
Bai W, Zhu WL, Ning YL, Li P, Zhao Y, Yang N, Chen X, Jiang YL, Yang WQ, Jiang DP, et al. Dramatic increases in blood glutamate concentrations are closely related to traumatic brain injury-induced acute lung injury. Sci Rep. 2017;7(1):5380.
Bai W, Li W, Ning YL, et al. Blood Glutamate Levels Are Closely Related to Acute Lung Injury and Prognosis after Stroke. Front Neurol. 2018;8:755. Published 2018 Jan 19. https://doi.org/10.3389/fneur.2017.00755.
Chen D, Wu X, Yang J, Yu L. Serum plasminogen activator urokinase receptor predicts elevated risk of acute respiratory distress syndrome in patients with sepsis and is positively associated with disease severity, inflammation and mortality. Exp Ther Med. 2019;18(4):2984–92.
Du S, Ai J, Zeng X, Wan J, Wu X, He J. Plasma level of advanced oxidation protein products as a novel biomarker of acute lung injury following cardiac surgery. Springerplus. 2016;5:231. Published 2016 Feb 29. https://doi.org/10.1186/s40064-016-1899-9.
Faust HE, Reilly JP, Anderson BJ, Ittner CAG, Forker CM, Zhang P, Weaver BA, Holena DN, Lanken PN, Christie JD, et al. Plasma mitochondrial DNA levels are associated with ARDS in trauma and sepsis patients. Chest. 2020;157(1):67–76.
Fremont RD, Koyama T, Calfee CS, Wu W, Dossett LA, Bossert FR, Mitchell D, Wickersham N, Bernard GR, Matthay MA, et al. Acute lung injury in patients with traumatic injuries: utility of a panel of biomarkers for diagnosis and pathogenesis. J Trauma Inj Infect Crit Care. 2010;68(5):1121–7.
Gaudet A, Parmentier E, Dubucquoi S, Poissy J, Duburcq T, Lassalle P, De Freitas CN, Mathieu D. Low endocan levels are predictive of acute respiratory distress syndrome in severe sepsis and septic shock. J Crit Care. 2018;47:121–6.
Hendrickson CM, Gibb SL, Miyazawa BY, Keating SM, Ross E, Conroy AS, Calfee CS, Pati S, Cohen MJ. Elevated plasma levels of TIMP-3 are associated with a higher risk of acute respiratory distress syndrome and death following severe isolated traumatic brain injury. Trauma Surg Acute Care Open. 2018;3(1):e000171.
Huang X, Zhao M. High expression of long non-coding RNA MALAT1 correlates with raised acute respiratory distress syndrome risk, disease severity, and increased mortality in sepstic patients. Int J Clin Exp Pathol. 2019;12(5):1877–87.
Huang Y, Xiao J, Cai T, Yang L, Shi F, Wang Y, Li Y, Shi T, Li C, Peng Y, et al. Immature granulocytes: a novel biomarker of acute respiratory distress syndrome in patients with acute pancreatitis. J Crit Care. 2019;50:303–8.
Jabaudon M, Berthelin P, Pranal T, Roszyk L, Godet T, Faure JS, Chabanne R, Eisenmann N, Lautrette A, Belville C, et al. Receptor for advanced glycation end-products and ARDS prediction: a multicentre observational study. Sci Rep. 2018;8(1):2603.
Jensen JS, Itenov TS, Thormar KM, et al. Prediction of non-recovery from ventilator-demanding acute respiratory failure, ARDS and death using lung damage biomarkers: data from a 1200-patient critical care randomized trial. Ann Intensive Care. 2016;6(1):114. https://doi.org/10.1186/s13613-016-0212-y.
Jones TK, Feng R, Kerchberger VE, Reilly JP, Anderson BJ, Shashaty MGS, Wang F, Dunn TG, Riley TR, Abbott J, et al. Plasma sRAGE acts as a genetically regulated causal intermediate in sepsis-associated acute respiratory distress syndrome. Am J Respir Crit Care Med. 2020;201(1):47–56.
Komiya K, Ishii H, Teramoto S, et al. Diagnostic utility of C-reactive protein combined with brain natriuretic peptide in acute pulmonary edema: a cross sectional study. Respir Res. 2011;12(1):83. Published 2011 Jun 22. https://doi.org/10.1186/1465-9921-12-83.
Lee KA, Gong MN. Pre-B-cell colony-enhancing factor and its clinical correlates with acute lung injury and sepsis. Chest. 2011;140(2):382–90.
Lin J, Zhang W, Wang L, Tian F. Diagnostic and prognostic values of Club cell protein 16 (CC16) in critical care patients with acute respiratory distress syndrome. J Clin Lab Anal. 2018;32(2):e22262. https://doi.org/10.1002/jcla.22262.
Liu XW, Ma T, Cai Q, Wang L, Song HW, Liu Z. Elevation of Serum PARK7 and IL-8 Levels Is Associated With Acute Lung Injury in Patients With Severe Sepsis/Septic Shock. J Intensive Care Med. 2019;34(8):662‐668. https://doi.org/10.1177/0885066617709689.
Luo J, Yu H, Hu YH, Liu D, Wang YW, Wang MY, Liang BM, Liang ZA. Early identification of patients at risk for acute respiratory distress syndrome among severe pneumonia: a retrospective cohort study. J Thorac Dis. 2017;9(10):3979–95.
Meyer NJ, Reilly JP, Feng R, et al. Myeloperoxidase-derived 2-chlorofatty acids contribute to human sepsis mortality via acute respiratory distress syndrome. JCI Insight. 2017;2(23):e96432. Published 2017 Dec 7. https://doi.org/10.1172/jci.insight.96432.
Mikkelsen ME, Shah CV, Scherpereel A, et al. Lower serum endocan levels are associated with the development of acute lung injury after major trauma. J Crit Care. 2012;27(5):522.e11‐522.e5.22E17. https://doi.org/10.1016/j.jcrc.2011.07.077.
Osaka D, Shibata Y, Kanouchi K, Nishiwaki M, Kimura T, Kishi H, Abe S, Inoue S, Tokairin Y, Igarashi A, et al. Soluble endothelial selectin in acute lung injury complicated by severe pneumonia. Int J Med Sci. 2011;8(4):302–8.
Palakshappa JA, Anderson BJ, Reilly JP, et al. Low Plasma Levels of Adiponectin Do Not Explain Acute Respiratory Distress Syndrome Risk: a Prospective Cohort Study of Patients with Severe Sepsis. Crit Care. 2016;20:71. Published 2016 Mar 16. https://doi.org/10.1186/s13054-016-1244-2.
Reilly JP, Wang F, Jones TK, Palakshappa JA, Anderson BJ, Shashaty MGS, Dunn TG, Johansson ED, Riley TR, Lim B, et al. Plasma angiopoietin-2 as a potential causal marker in sepsis-associated ARDS development: evidence from Mendelian randomization and mediation analysis. Intensive Care Med. 2018;44(11):1849–58.
Shashaty MGS, Reilly JP, Faust HE, et al. Plasma receptor interacting protein kinase-3 levels are associated with acute respiratory distress syndrome in sepsis and trauma: a cohort study. Crit Care. 2019;23(1):235. Published 2019 Jun 28. https://doi.org/10.1186/s13054-019-2482-x.
Shaver CM, Woods J, Clune JK, et al. Circulating microparticle levels are reduced in patients with ARDS. Crit Care. 2017;21(1):120. Published 2017 May 25. https://doi.org/10.1186/s13054-017-1700-7.
Suzuki A, Taniguchi H, Kondoh Y, Ando M, Watanabe N, Kimura T, Kataoka K, Yokoyama T, Sakamoto K, Hasegawa Y. Soluble thrombomodulin in bronchoalveolar lavage fluid is an independent predictor of severe drug-induced lung injury. Respirology. 2017;22(4):744–9.
Wang Y, Fu X, Yu B, Ai F. Long non-coding RNA THRIL predicts increased acute respiratory distress syndrome risk and positively correlates with disease severity, inflammation, and mortality in sepsis patients. J Clin Lab Anal. 2019;33(6):e22882. https://doi.org/10.1002/jcla.22882.
Ware LB, Zhao Z, Koyama T, Brown RM, Semler MW, Janz DR, May AK, Fremont RD, Matthay MA, Cohen MJ, et al. Derivation and validation of a two-biomarker panel for diagnosis of ARDS in patients with severe traumatic injuries. Trauma Surg Acute Care Open. 2017;2(1):e000121.
Xu Z, Wu GM, Li Q, Ji FY, Shi Z, Guo H, Yin JB, Zhou J, Gong L, Mei CX, et al. Predictive value of combined LIPS and ANG-2 level in critically ill patients with ARDS risk factors. Mediat Inflamm. 2018;2018:1739615.
Yeh LC, Huang PW, Hsieh KH, Wang CH, Kao YK, Lin TH, Lee XL. Elevated plasma levels of Gas6 are associated with acute lung injury in patients with severe sepsis. Tohoku J Exp Med. 2017;243(3):187–93.
Ying J, Zhou D, Gu T, Huang J. Endocan, a Risk Factor for Developing Acute Respiratory Distress Syndrome among Severe Pneumonia Patients. Can Respir J. 2019;2019:2476845. Published 2019 Apr 1. https://doi.org/10.1155/2019/2476845.
Adamzik M, Broll J, Steinmann J, Westendorf AM, Rehfeld I, Kreissig C, Peters J. An increased alveolar CD4 + CD25 + Foxp3 + T-regulatory cell ratio in acute respiratory distress syndrome is associated with increased 30-day mortality. Intensive Care Med. 2013;39(10):1743–51.
Bajwa EK, Januzzi JL, Gong MN, Thompson BT, Christiani DC. Prognostic value of plasma N-terminal probrain natriuretic peptide levels in the acute respiratory distress syndrome. Crit Care Med. 2008;36(8):2322–7.
Bajwa EK, Khan UA, Januzzi JL, Gong MN, Thompson BT, Christiani DC. Plasma C-reactive protein levels are associated with improved outcome in ARDS. Chest. 2009;136(2):471–80.
Bajwa EK, Volk JA, Christiani DC, Harris RS, Matthay MA, Thompson BT, Januzzi JL. Prognostic and diagnostic value of plasma soluble suppression of tumorigenicity-2 concentrations in acute respiratory distress syndrome. Crit Care Med. 2013;41(11):2521–31.
Calfee CS, Ware LB, Eisner MD, Parsons PE, Thompson BT, Wickersham N, Matthay MA. Plasma receptor for advanced glycation end products and clinical outcomes in acute lung injury. Thorax. 2008;63(12):1083–9.
Calfee CS, Eisner MD, Parsons PE, Thompson BT, Conner ER Jr, Matthay MA, Ware LB. Soluble intercellular adhesion molecule-1 and clinical outcomes in patients with acute lung injury. Intensive Care Med. 2009;35(2):248–57.
Calfee CS, Ware LB, Glidden DV, Eisner MD, Parsons PE, Thompson BT, Matthay MA. Use of risk reclassification with multiple biomarkers improves mortality prediction in acute lung injury. Crit Care Med. 2011;39(4):711–7.
Calfee CS, Gallagher D, Abbott J, Thompson BT, Matthay MA. Plasma angiopoietin-2 in clinical acute lung injury: prognostic and pathogenetic significance. Crit Care Med. 2012;40(6):1731–7.
Calfee CS, Janz DR, Bernard GR, May AK, Kangelaris KN, Matthay MA, Ware LB. Distinct molecular phenotypes of direct vs indirect ARDS in single-center and multicenter studies. Chest. 2015;147(6):1539–48.
Cartin-Ceba R, Hubmayr RD, Qin R, Peters S, Determann RM, Schultz MJ, Gajic O. Predictive value of plasma biomarkers for mortality and organ failure development in patients with acute respiratory distress syndrome. J Crit Care. 2015;30(1):219.e211–7.
Chen CY, Yang KY, Chen MY, Chen HY, Lin MT, Lee YC, Perng RP, Hsieh SL, Yang PC, Chou TY. Decoy receptor 3 levels in peripheral blood predict outcomes of acute respiratory distress syndrome. Am J Respir Crit Care Med. 2009;180(8):751–60.
Clark JG, Milberg JA, Steinberg KP, Hudson LD. Type III procollagen peptide in the adult respiratory distress syndrome. Association of increased peptide levels in bronchoalveolar lavage fluid with increased risk for death. Ann Intern Med. 1995;122(1):17–23.
Clark BJ, Bull TM, Benson AB, et al. Growth differentiation factor-15 and prognosis in acute respiratory distress syndrome: a retrospective cohort study. Crit Care. 2013;17(3):R92. Published 2013 May 24. https://doi.org/10.1186/cc12737.
Dolinay T, Kim YS, Howrylak J, Hunninghake GM, An CH, Fredenburgh L, Massaro AF, Rogers A, Gazourian L, Nakahira K, et al. Inflammasome-regulated cytokines are critical mediators of acute lung injury. Am J Respir Crit Care Med. 2012;185(11):1225–34.
Eisner MD, Parsons P, Matthay MA, Ware L, Greene K. Plasma surfactant protein levels and clinical outcomes in patients with acute lung injury. Thorax. 2003;58(11):983–8.
Forel JM, Guervilly C, Hraiech S, Voillet F, Thomas G, Somma C, Secq V, Farnarier C, Payan MJ, Donati SY, et al. Type III procollagen is a reliable marker of ARDS-associated lung fibroproliferation. Intensive Care Med. 2015;41(1):1–11.
Forel JM, Guervilly C, Farnarier C, et al. Transforming Growth Factor-β1 in predicting early lung fibroproliferation in patients with acute respiratory distress syndrome. PLoS One. 2018;13(11):e0206105. Published 2018 Nov 5. https://doi.org/10.1371/journal.pone.0206105.
Guervilly C, Lacroix R, Forel JM, et al. High levels of circulating leukocyte microparticles are associated with better outcome in acute respiratory distress syndrome. Crit Care. 2011;15(1):R31. https://doi.org/10.1186/cc9978.
Kim JM, Lee JK, Choi SM, et al. Diagnostic and prognostic values of serum activin-a levels in patients with acute respiratory distress syndrome. BMC Pulm Med. 2019;19(1):115. Published 2019 Jun 25. https://doi.org/10.1186/s12890-019-0879-6.
Lee HW, Choi SM, Lee J, Park YS, Lee CH, Yim JJ, Yoo CG, Kim YW, Han SK, Lee SM. Serum uric acid level as a prognostic marker in patients with acute respiratory distress syndrome. J Intensive Care Med. 2019;34(5):404–10.
Lesur O, Langevin S, Berthiaume Y, Légaré M, Skrobik Y, Bellemare JF, Lévy B, Fortier Y, Lauzier F, Bravo G, et al. Outcome value of Clara cell protein in serum of patients with acute respiratory distress syndrome. Intensive Care Med. 2006;32(8):1167–74.
Li W, Ai X, Ni Y, Ye Z, Liang Z. The association between the neutrophil-to-lymphocyte ratio and mortality in patients with acute respiratory distress syndrome: a retrospective cohort study. Shock (Augusta). 2019;51(2):161–7.
Lin MT, Wei YF, Ku SC, Lin CA, Ho CC, Yu CJ. Serum soluble triggering receptor expressed on myeloid cells-1 in acute respiratory distress syndrome: a prospective observational cohort study. J Formos Med Assoc. 2010;109(11):800–9.
Lin Q, Fu F, Chen H, Zhu B. Copeptin in the assessment of acute lung injury and cardiogenic pulmonary edema. Respir Med. 2012;106(9):1268–77.
Lin Q, Shen J, Shen L, Zhang Z, Fu F. Increased plasma levels of heparin-binding protein in patients with acute respiratory distress syndrome. Crit Care. 2013;17(4):R155. Published 2013 Jul 24. https://doi.org/10.1186/cc12834.
Madtes DK, Rubenfeld G, Klima LD, Milberg JA, Steinberg KP, Martin TR, Raghu G, Hudson LD, Clark JG. Elevated transforming growth factor-α levels in bronchoalveolar lavage fluid of patients with acute respiratory distress syndrome. Am J Respir Crit Care Med. 1998;158(2):424–30.
McClintock DE, Starcher B, Eisner MD, Thompson BT, Hayden DL, Church GD, Matthay MA, Wiedemann HP, Arroliga AC, Fisher CJ Jr, et al. Higher urine desmosine levels are associated with mortality in patients with acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2006;291(4):L566–71.
McClintock DE, Ware LB, Eisner MD, Wickersham N, Thompson BT, Matthay MA, Wiedemann HP, Arroliga AC, Fisher CJ Jr, Komara JJ Jr, et al. Higher urine nitric oxide is associated with improved outcomes in patients with acute lung injury. Am J Respir Crit Care Med. 2007;175(3):256–62.
McClintock D, Zhuo H, Wickersham N, Matthay MA, Ware LB. Biomarkers of inflammation, coagulation and fibrinolysis predict mortality in acute lung injury. Crit Care. 2008;12(2):R41.
Menk M, Giebelhäuser L, Vorderwülbecke G, et al. Nucleated red blood cells as predictors of mortality in patients with acute respiratory distress syndrome (ARDS): an observational study. Ann Intensive Care. 2018;8(1):42. Published 2018 Mar 27. https://doi.org/10.1186/s13613-018-0387-5.
Metkus TS, Guallar E, Sokoll L, Morrow D, Tomaselli G, Brower R, Schulman S, Korley FK. Prevalence and prognostic association of circulating troponin in the acute respiratory distress syndrome. Crit Care Med. 2017;45(10):1709–17.
Mrozek S, Jabaudon M, Jaber S, Paugam-Burtz C, Lefrant JY, Rouby JJ, Asehnoune K, Allaouchiche B, Baldesi O, Leone M, et al. Elevated plasma levels of sRAGE are associated with nonfocal CT-based lung imaging in patients with ARDS: a prospective multicenter study. Chest. 2016;150(5):998–1007.
Ong T, McClintock DE, Kallet RH, Ware LB, Matthay MA, Liu KD. Ratio of angiopoietin-2 to angiopoietin-1 as a predictor of mortality in acute lung injury patients. Crit Care Med. 2010;38(9):1845–51.
Parsons PE, Matthay MA, Ware LB, Eisner MD. Elevated plasma levels of soluble TNF receptors are associated with morbidity and mortality in patients with acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2005;288(3 32–3):L426–31.
Parsons PE, Eisner MD, Thompson BT, Matthay MA, Ancukiewicz M, Bernard GR, Wheeler AP. Lower tidal volume ventilation and plasma cytokine markers of inflammation in patients with acute lung injury. Crit Care Med. 2005;33(1):1–6.
Quesnel C, Piednoir P, Gelly J, Nardelli L, Garnier M, Leçon V, Lasocki S, Bouadma L, Philip I, Elbim C, et al. Alveolar fibrocyte percentage is an independent predictor of poor outcome in patients with acute lung injury. Crit Care Med. 2012;40(1):21–8.
Rahmel T, Rump K, Adamzik M, Peters J, Frey UH. Increased circulating microRNA-122 is associated with mortality and acute liver injury in the acute respiratory distress syndrome. BMC Anesthesiol. 2018;18(1):75. Published 2018 Jun 23. https://doi.org/10.1186/s12871-018-0541-5.
Reddy R, Asante I, Liu S, et al. Circulating angiotensin peptides levels in Acute Respiratory Distress Syndrome correlate with clinical outcomes: A pilot study. PLoS One. 2019;14(3):e0213096. Published 2019 Mar 7. https://doi.org/10.1371/journal.pone.0213096.
Rivara MB, Bajwa EK, Januzzi JL, Gong MN, Thompson BT, Christiani DC. Prognostic significance of elevated cardiac troponin-T levels in acute respiratory distress syndrome patients. PLoS One. 2012;7(7):e40515. https://doi.org/10.1371/journal.pone.0040515.
Rogers AJ, Guan J, Trtchounian A, Hunninghake GM, Kaimal R, Desai M, Kozikowski LA, DeSouza L, Mogan S, Liu KD, et al. Association of elevated plasma interleukin-18 level with increased mortality in a clinical trial of statin treatment for acute respiratory distress syndrome. Crit Care Med. 2019;47(8):1089–96.
Sapru A, Calfee CS, Liu KD, Kangelaris K, Hansen H, Pawlikowska L, Ware LB, Alkhouli MF, Abbot J, Matthay MA. Plasma soluble thrombomodulin levels are associated with mortality in the acute respiratory distress syndrome. Intensive Care Med. 2015;41(3):470–8.
Suratt BT, Eisner MD, Calfee CS, Allard JB, Whittaker LA, Engelken DT, Petty JM, Trimarchi T, Gauthier L, Parsons PE. Plasma granulocyte colony-stimulating factor levels correlate with clinical outcomes in patients with acute lung injury. Crit Care Med. 2009;37(4):1322–8.
Tang L, Zhao Y, Wang D, et al. Endocan levels in peripheral blood predict outcomes of acute respiratory distress syndrome. Mediators Inflamm. 2014;2014:625180. https://doi.org/10.1155/2014/625180.
Tsangaris I, Tsantes A, Bonovas S, Lignos M, Kopterides P, Gialeraki A, Rapti E, Orfanos S, Dimopoulou I, Travlou A, et al. The impact of the PAI-1 4G/5G polymorphism on the outcome of patients with ALI/ARDS. Thromb Res. 2009;123(6):832–6.
Tsangaris I, Tsantes A, Vrigkou E, et al. Angiopoietin-2 Levels as Predictors of Outcome in Mechanically Ventilated Patients with Acute Respiratory Distress Syndrome. Dis Markers. 2017;2017:6758721. https://doi.org/10.1155/2017/6758721.
Tsantes AE, Kopterides P, Bonovas S, Bagos P, Antonakos G, Nikolopoulos GK, Gialeraki A, Kapsimali V, Kyriakou E, Kokori S, et al. Effect of angiotensin converting enzyme gene I/D polymorphism and its expression on clinical outcome in acute respiratory distress syndrome. Minerva Anestesiol. 2013;79(8):861–70.
Tseng CC, Fang WF, Leung SY, et al. Impact of serum biomarkers and clinical factors on intensive care unit mortality and 6-month outcome in relatively healthy patients with severe pneumonia and acute respiratory distress syndrome. Dis Markers. 2014;2014:804654. https://doi.org/10.1155/2014/804654.
Wang T, Zhu Z, Liu Z, Yi L, Yang Z, Bian W, Chen W, Wang S, Li G, Li A, et al. Plasma neutrophil elastase and Elafin as prognostic biomarker for acute respiratory distress syndrome: a multicenter survival and longitudinal prospective observation study. Shock (Augusta). 2017;48(2):168–74.
Wang Y, Ju M, Chen C, Yang D, Hou D, Tang X, Zhu X, Zhang D, Wang L, Ji S, et al. Neutrophil-to-lymphocyte ratio as a prognostic marker in acute respiratory distress syndrome patients: a retrospective study. J Thorac Dis. 2018;10(1):273–82.
Ware LB, Eisner MD, Thompson BT, Parsons PE, Matthay MA. Significance of Von Willebrand factor in septic and nonseptic patients with acute lung injury. Am J Respir Crit Care Med. 2004;170(7):766–72.
Xu Z, Li X, Huang Y, Mao P, Wu S, Yang B, Yang Y, Chen K, Liu X, Li Y. The predictive value of plasma galectin-3 for ARDS severity and clinical outcome. Shock (Augusta). 2017;47(3):331–6.
Jabaudon M, Blondonnet R, Pereira B, Cartin-Ceba R, Lichtenstern C, Mauri T, Determann RM, Drabek T, Hubmayr RD, Gajic O, et al. Plasma sRAGE is independently associated with increased mortality in ARDS: a meta-analysis of individual patient data. Intensive Care Med. 2018;44(9):1388–99.
Gardlund B, Sjolin J, Nilsson A, Roll M, Wickerts CJ, Wretlind B. Plasma levels of cytokines in primary septic shock in humans: correlation with disease severity. J Infect Dis. 1995;172(1):296–301.
van der Zee P, van Walree I, Fijen JW, van Houte AJ, van Velzen-Blad H, Rijkers G, Gommers D, Endeman H. Cytokines and chemokines are detectable in swivel-derived exhaled breath condensate (SEBC): a pilot study in mechanically ventilated patients. Dis Markers. 2020;2020:2696317.
Prescott HC, Calfee CS, Thompson BT, Angus DC, Liu VX. Toward smarter lumping and smarter splitting: rethinking strategies for sepsis and acute respiratory distress syndrome clinical trial design. Am J Respir Crit Care Med. 2016;194(2):147–55.
Sinha P, Calfee CS. Phenotypes in acute respiratory distress syndrome: moving towards precision medicine. Curr Opin Crit Care. 2019;25(1):12–20.
Acknowledgements
We thank Wan-Jie Gu (abbreviated in the text as WG) for his support in study eligibility evaluation (Nanjing University, China).
We thank Wichor Bramer and Elise Krabbendam (Biomedical Information Specialists Medical Library Erasmus MC) for their support in the literature search.
Funding
None
Author information
Authors and Affiliations
Contributions
PZ collected and analysed the data and drafted the manuscript. WR analysed the data and substantially revised the manuscript. PS collected the data and substantially revised the manuscript. HE and DG substantially revised the manuscript. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
PZ, WR, PS, and HE have no conflict of interest. DG received speaker’s fee and travel expenses from Dräger, GE Healthcare (medical advisory board 2009–2012), Maquet, and Novalung (medical advisory board).
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Literature search.
Additional file 2.
Quality assessment
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
van der Zee, P., Rietdijk, W., Somhorst, P. et al. A systematic review of biomarkers multivariately associated with acute respiratory distress syndrome development and mortality. Crit Care 24, 243 (2020). https://doi.org/10.1186/s13054-020-02913-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13054-020-02913-7