Abstract
Background
Myeloproliferative neoplasms (MPNs), typically defined by myeloid proliferation and eosinophilia, and are only rarely caused by platelet-derived growth factor receptor beta (PDGFRB) gene rearrangements.
Case presentation
Here, we report a unique case of MPN that is negative for eosinophilia and characterized by a novel PDGFRB rearrangement. After cytogenetic analysis revealed a karyotype of t(5;17) (q32;q11), we used fluorescence in situ hybridization to specifically identify the PDGFRB gene at 5q31-q33 as the gene that had been translocated. Subsequently, RNA sequencing identified a new MYO18A-PDGFRB gene fusion. This fusion presented a previously undescribed breakpoint composed of exon 37 of MYO18A and exon 13 of PDGFRB. Furthermore, both RT-PCR and Bi-directional Sanger sequencing confirmed this out-of-frame fusion. Interestingly, we simultaneously identified the presence of another three PDGFRB transcripts, all of which were in-frame fusions. After treating the patient with imatinib, the t(5;17) translocation was no longer detected by conventional cytogenetics or by FISH, and at the time of the last follow-up, the patient had been in complete remission for 26 months.
Conclusion
We prove that MYO18A-PDGFRB fusions are recurrent genetic aberrations involved in MPNs, and identify multiple fusion transcripts with novel breakpoints.
Similar content being viewed by others
Background
Constitutive activation of protein tyrosine kinases is a common feature of the pathogenesis of chronic myeloproliferative neoplasms (MPNs). The genes most commonly involved in these neoplasms are those encoding for the protein tyrosine kinases PDGFRA, PDGFRB, FGFR1, KIT, FLT3, JAK2 and ABL1. In 2008 however, the World Health Organization (WHO) classified rearrangements of PDGFRA, PDGFRB, and FGFR1 in a distinct disease category [1]. PDGFRB is a class III receptor tyrosine kinase located on chromosome 5 at position 5q31-q33. The most common type of PDGFRB aberration is a fusion translocation. To date, more than 30 different PDGFRB fusion partners have been identified [2, 3], with the majority of them only occurring in individual patients. Nonetheless, a few of these fusions - for example, ETV6-PDGFRB, H4-PDGFRB, and CCDC88C-PDGFRB [2, 4–6] – are in fact recurrent. Interestingly, each partner typically contains an oligomerization motif that contributes to protein dimerization and consequently to the constitutive activation of the PDGFRB kinase domain. Imatinib, a tyrosine kinase inhibitor typically used to treat those myeloid tumors characterized by PDGFRB fusions, is reported to produce sustained remission in nearly all cases [2, 3].
MYO18A, a member of the myosin superfamily originally identified in bone marrow stromal cells, is associated with the ability of these cells to support hematopoiesis [7]. In hematological malignancies, MYO18A has been found as fusions with FGFR1, PDGFRB, and in only a single case, with MLL, leading to the 8p11 myeloproliferative syndrome (EMS), eosinophilia-associated MPN (MPN-eo), and acute myeloid leukemia (AML) respectively [8–10].
Here, we present a novel case of MPN in which a unique MYO18A-PDGFRB fusion results in MPN without eosinophilia. To the best of our knowledge, this is the first time that fusion to exon 13 of PDGFB has been reported. Interestingly, this patient harbored multiple MYO18A-PDGFRB transcripts, with most of them being in-frame fusions. Therapeutically, this patient was sensitive to imatinib, and achieved both complete hematological remission (CHR) and complete cytogenetic remission (CCyR) in a sustained and rapid manner.
Case presentation
A 25-year-old man was admitted to our medical center after experiencing prolonged weakness and splenomegaly. A complete blood cell analysis indicated a white blood cell count of 81,190/μL, a platelet count of 206,000/μL, and a hemoglobin concentration of 10.5 g/dL. Peripheral blood analysis showed all stages of neutrophilic maturation, with 1% myeloblasts, 4% promyelocytes, 10% myelocytes, 17% metamyelocytes, 2% eosinophils and 9% basophils. Bone marrow aspirates revealed that the patient was in the chronic phase of chronic myeloid leukemia (CML), with 2.5% myeloblasts, 5.5% promyelocytes, 13% myelocytes, 13% metamyelocytes, 1.5% eosinophils and 9% basophils. A multiplex PCR screen for gene fusions typical of leukemia was negative. Chromosome analysis of the bone marrow showed the presence of 46,XY,t (5;17) (q32;q11) [10]. Subsequently, dual-color fluorescent in situ hybridization (FISH) proved positive for PDGFRB gene rearrangement. After one week of imatinib treatment at 400 mg/day, the patient achieved CHR. This dosage was then decreased to 200 mg/day, and the patient acquired CCyR three months later. Thereafter, the patient took 100 mg/day, and at the final follow-up (26 months later), the patient still remained in complete remission.
Results
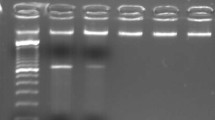
Through retrospective analysis, we identified an MPN patient with a karyotype of 46,XY,t(5;17) (q32;q11) (Fig. 1a). This suggested a rearrangement of the PDGFRB gene located at chromosome 5q31-q33. In order to confirm the suspected rearrangement, we performed FISH analysis using two distinct probes complementary to the 5’ and 3’ regions of the PDGFRB gene, and found separated signals in 75% of blast cells (Fig. 1b). A previous study by Walz et al. found that translocation events at 17q11 were associated with the MYO18A gene [9]. RT-PCR using MYO18A (exon 40) and PDGFRB (exon 10) primers was unable to detect this previously reported MYO18A-PDGFRB fusion transcript (data not show) [9]. Whole transcriptome sequencing, however, uncovered a fusion between exon 37 of MYO18A (NM_078471.3) and exon 13 of PDGFRB (NM_002609.3). Subsequently, RT-PCR and Bi-directional Sanger sequencing confirmed this out-of-frame fusion between MYO18A exon 37 and PDGFRB exon 13 (Fig. 1c). Given that the patient was highly sensitive to imatinib-mediated PDGFRB inhibition, but showed no other imatinib-related abnormalities, we hypothesized that there was some degree of splicing taking place within the MYO18A-PDGFRB fusion. In fact, we were able to amplify different products containing the same fusion point by RT-PCR, and a total of four types of MYO18A-PDGFRB fusion transcripts were identified by Sanger sequencing (Fig. 1d). Three out of four of these transcripts were actually in-frame PDGFRB fusions and accounted for a total of 90.9% of the identified transcripts (Fig. 1d). Two of the in-frame transcripts contained a deletion of MYO18A exon 30, with one of them also having parts of exons 36–37 deleted at the same time. Finally, the third in−frame fusion transcript had an insertion including part of MYO18A intron 39 and part of PDGFRB exon 12 (Fig. 1d).
Multiple MYO18A-PDGFRB fusion transcripts in a myeloproliferative neoplasm patient with t(5;17) (q32;q11) (a) The R-banded karyotype showing the translocation t(5;17) (q32;q11). The arrows indicate the structural aberrations of chromosomes 5 and 17; (b) FISH using Vysis LSI PDGFRB Spectrum Green and Orange Probes. A yellow signal denotes a normal PDGFRB gene (5’and 3’regions remain joined), whereas orange and green signals respectively denote the 5’ and 3’ regions of the PDGFRB gene after rearrangement; (c) Bi-directional Sanger sequencing of the PCR product confirms the fusion between MYO18A exon 37(NM_078471.3) and PDGFRB exon 13(NM_002609.3); (d) Four distinct types of MYO18A-PDGFRB fusion transcripts were detected in the patient
Discussion and Conclusion
Although PDGFRB fusions are rare, they can be observed in a diverse range of hematological malignancies including Ph-like acute lymphoblastic leukemia (ALL), AML, and atypical CML. These fusions have an adult male predominance, and are clinically defined by eosinophilia and splenomegaly [2]. To date, more than 30 partner genes of PDGFRB have been identified [2, 3]. However, only a minority of these are recurrent (the most common of which is ETV6-PDGFRB), with the vast majority being reported in only individual cases. Walz et al. reported the first eosinophilia-associated MPN attributed to a MYO18A-PDGFRB fusion in 2009 [9]. Here, we report another case of MPN that is characterized by unique MYO18A-PDGFRB fusions and an eosinophilia-free phenotype. In agreement with other studies, this suggests that eosinophilia is a prominent, but not invariable feature [11].
Interestingly, in the present case exon 37 of MYO18A became fused in an out-of-frame fashion to exon 13 of PDGFRB. However, as the patient was highly sensitive to imatinib treatment - a pharmaceutical drug which inhibits activated receptor tyrosine kinases including PDGFRB - and displayed no other abnormalities, we suspected that the patient harbored different spliced versions of the fusion. Indeed, RT-PCR and sequencing enabled us to identify four types of fusion transcripts in our patient, with three of them being in-frame fusions. Interestingly, PDGFRB fusions principally involve exons 11 and 12, and more rarely exons 9 and 10 [9, 12]. In fact, there is no previous report of exon 13 being involved in the fusion point, possibly due to it being an out-of-frame fusion. The in-frame MYO18A-PDGFRB fusion proteins included the majority of the domains of MYO18A, including some of the coiled-coil motifs, and the entire tyrosine kinase domain of PDGFRB. It is known that only one, or a few of these coiled-coil motifs are enough to promote the dimerization or oligomerization of MYO18A-PDGFRB fusions. As in the case of the MYO18A-FGFR1 fusion, this leads to constitutive activation of the tyrosine kinase domain [8]. In addition, the lack of the PDGFRB transmembrane domain, which is found in exon 11, results in delocalization of the fusion protein to the cytoplasm. This may be another mechanism through which the kinase domain becomes constitutively activated. Furthermore, deletion of exon 12, which results in the disruption of the autoinhibitory WW-like domain of the juxtamembrane region, may also promote transformation properties of such fusion proteins [13–15]. Of course, however, loss of these domains is not mandatory for transformation because in previous cases, other MYO18A-PDGFRB fusions retained both the transmembrane and juxtamembrane domains.
Although PDGFRB fusions are rare, the identification of PDGFRB rearrangements is significant because of their sensitive response to imatinib, especially with respect to MPNs [16]. Moreover, both primary and secondary resistance to imatinib seems to be very rare [17, 18]. Due to its low incidence, a standard imatinib dose and medication time for patients with PDGFRB fusions has yet to be formally established in clinical treatment. With the standard dosage used for CML as a reference, patients are initially treated with 400 mg/day [16, 19]. In fact, PDGFR fusions are much more sensitive to imatinib than BCR-ABL1 fusions, at least in vitro. For instance, the IC50 of imatinib for inhibiting the proliferation of ETV6-PDGFRB transformed cells is 7.5 nM, a value significantly lower compared to the IC50 of 800 nM for BCR-ABL transformed cells [20, 21]. In our case, imatinib at 400 mg/day brought about CHR in only one week, and after decreasing the dosage, CCyR was also obtained within three months. Thereafter, complete remission remained stable, indicating that a low dose of imatinib is appropriate for patients with PDGFRB fusions.
Overall, our findings further emphasize the prominent role of PDGFRB in the pathogenesis of MPNs, and highlight the importance of accurate diagnosis and targeted therapy.
Methods
R-banding and karyotyping
At the time of diagnosis, the bone marrow cells were cultured for 24 h and analyzed for standard cytogenetic R-banding. The karyotype was described according to the International System for Human Cytogenetic Nomenclature (ISCN 2013).
Fluorescence in situ hybridization (FISH) analysis
FISH analysis was performed on metaphase spreads, using the Vysis LSI PDGFRB Spectrum Orange and Green Probes (Abbott Molecular, Illinois, USA) according to the manufacturer’s instructions.
RNA sequencing
Whole transcriptome sequencing of bone marrow mononuclear cells was performed, and transcription sequence data were generated by high-throughput RNA sequencing (Illumina HiSeq 2500). The online software deFuse was used for the discovery of fusion transcripts.
RT-PCR and Sanger sequencing
RNA was reverse transcribed with random hexamers using standard techniques (Thermo Fisher), and the following primer sets were used to detect MYO18A-PDGFRB transcripts: MYO18A(c.5165) Forward (5′-ACATCGCCAAAGCCAAGA-3′) and PDGFRB(c.2014) Reverse (5′-GATGGGTCCTCCTTTGGT-3′) at an annealing temperature of 58 °C over 32 cycles; MYO18A(c.2524) Forward (5′-CTGGCGTTTGACGACTTG-3′) and PDGFRB(c.2739) Reverse (5′-CCGTTTGATGGCATTGTAG-3′); and MYO18A(c.5436) Forward (5′-GGACAAGTCCCTGGTGAGC-3′) and PDGFRB(c.3301) Reverse (5′-CTACAGGAAGCTATCCTCTGC-3′) at an annealing temperature of 55 °C over 35 cycles. PCR products were analyzed by Bi-directional Sanger sequencing.
Abbreviations
- ALL:
-
acute lymphoblastic leukemia
- AML:
-
acute myeloid leukemia
- CCyR:
-
complete cytogenetic remission
- CHR:
-
complete hematological remission
- CML:
-
chronic myeloid leukemia
- EMS:
-
the 8p11 myeloproliferative syndrome
- FISH:
-
fluorescence in situ hybridization
- MPN-eo:
-
eosinophilia-associated MPN
- MPNs:
-
myeloproliferative neoplasms
- PDGFRB:
-
platelet-derived growth factor receptor beta
- WHO:
-
World Health Organization
References
Bain BJ. Myeloid and lymphoid neoplasms with eosinophilia and abnormalities of PDGFRA, PDGFRB or FGFR1. Haematologica. 2010;95(5):696–8.
Naumann N, Schwaab J, Metzgeroth G, Jawhar M, Haferlach C, Gohring G, Schlegelberger B, Dietz CT, Schnittger S, Lotfi S, et al. Fusion of PDGFRB to MPRIP, CPSF6, and GOLGB1 in three patients with eosinophilia-associated myeloproliferative neoplasms. Genes Chromosomes Cancer. 2015;54(12):762–70.
Vega F, Medeiros LJ, Bueso-Ramos CE, Arboleda P, Miranda RN. Hematolymphoid neoplasms associated with rearrangements of PDGFRA, PDGFRB, and FGFR1. Am J Clin Pathol. 2015;144(3):377–92.
Golub TR, Barker GF, Lovett M, Gilliland DG. Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell. 1994;77(2):307–16.
Kulkarni S, Heath C, Parker S, Chase A, Iqbal S, Pocock CF, Kaeda J, Cwynarski K, Goldman JM, Cross NC. Fusion of H4/D10S170 to the platelet-derived growth factor receptor beta in BCR-ABL-negative myeloproliferative disorders with a t(5;10) (q33;q21). Cancer Res. 2000;60(13):3592–8.
Levine RL, Wadleigh M, Sternberg DW, Wlodarska I, Galinsky I, Stone RM, DeAngelo DJ, Gilliland DG, Cools J. KIAA1509 is a novel PDGFRB fusion partner in imatinib-responsive myeloproliferative disease associated with a t(5;14) (q33;q32). Leukemia. 2005;19(1):27–30.
Furusawa T, Ikawa S, Yanai N, Obinata M. Isolation of a novel PDZ-containing myosin from hematopoietic supportive bone marrow stromal cell lines. Biochem Biophys Res Commun. 2000;270(1):67–75.
Walz C, Chase A, Schoch C, Weisser A, Schlegel F, Hochhaus A, Fuchs R, Schmitt-Graff A, Hehlmann R, Cross NC, et al. The t(8;17) (p11;q23) in the 8p11 myeloproliferative syndrome fuses MYO18A to FGFR1. Leukemia. 2005;19(6):1005–9.
Walz C, Haferlach C, Hanel A, Metzgeroth G, Erben P, Gosenca D, Hochhaus A, Cross NC, Reiter A. Identification of a MYO18A-PDGFRB fusion gene in an eosinophilia-associated atypical myeloproliferative neoplasm with a t(5;17) (q33-34;q11.2). Genes Chromosomes Cancer. 2009;48(2):179–83.
Ussowicz M, Jaskowiec A, Meyer C, Marschalek R, Chybicka A, Szczepanski T, Haus O. A three-way translocation of MLL, MLLT11, and the novel reciprocal partner gene MYO18A in a child with acute myeloid leukemia. Cancer Genet. 2012;205(5):261–5.
Nadal N, Flandrin P, Cornillon J, Delabesse E, Mauvieux L, Olaru D, Morel S, Campos L. Successful treatment with imatinib mesylate in a case of chronic myeloproliferative disorder with a t(5;12) (q33;p13.1) without eosinophilia. Cancer Genet Cytogenet. 2006;169(2):174–5.
Curtis CE, Grand FH, Waghorn K, Sahoo TP, George J, Cross NC. A novel ETV6-PDGFRB fusion transcript missed by standard screening in a patient with an imatinib responsive chronic myeloproliferative disease. Leukemia. 2007;21(8):1839–41.
Irusta PM, Luo Y, Bakht O, Lai CC, Smith SO, DiMaio D. Definition of an inhibitory juxtamembrane WW-like domain in the platelet-derived growth factor beta receptor. J Biol Chem. 2002;277(41):38627–34.
Chen J, Williams IR, Kutok JL, Duclos N, Anastasiadou E, Masters SC, Fu H, Gilliland DG. Positive and negative regulatory roles of the WW-like domain in TEL-PDGFbetaR transformation. Blood. 2004;104(2):535–42.
Stover EH, Chen J, Folens C, Lee BH, Mentens N, Marynen P, Williams IR, Gilliland DG, Cools J. Activation of FIP1L1-PDGFRalpha requires disruption of the juxtamembrane domain of PDGFRalpha and is FIP1L1-independent. Proc Natl Acad Sci U S A. 2006;103(21):8078–83.
Apperley JF, Gardembas M, Melo JV, Russell-Jones R, Bain BJ, Baxter EJ, Chase A, Chessells JM, Colombat M, Dearden CE, et al. Response to imatinib mesylate in patients with chronic myeloproliferative diseases with rearrangements of the platelet-derived growth factor receptor beta. N Engl J Med. 2002;347(7):481–7.
Bastie JN, Garcia I, Terre C, Cross NC, Mahon FX, Castaigne S. Lack of response to imatinib mesylate in a patient with accelerated phase myeloproliferative disorder with rearrangement of the platelet-derived growth factor receptor beta-gene. Haematologica. 2004;89(10):1263–4.
Grand FH, Burgstaller S, Kuhr T, Baxter EJ, Webersinke G, Thaler J, Chase AJ, Cross NC. p53-Binding protein 1 is fused to the platelet-derived growth factor receptor beta in a patient with a t(5;15) (q33;q22) and an imatinib-responsive eosinophilic myeloproliferative disorder. Cancer Res. 2004;64(20):7216–9.
David M, Cross NC, Burgstaller S, Chase A, Curtis C, Dang R, Gardembas M, Goldman JM, Grand F, Hughes G, et al. Durable responses to imatinib in patients with PDGFRB fusion gene-positive and BCR-ABL-negative chronic myeloproliferative disorders. Blood. 2007;109(1):61–4.
Gunby RH, Cazzaniga G, Tassi E, Le Coutre P, Pogliani E, Specchia G, Biondi A, Gambacorti-Passerini C. Sensitivity to imatinib but low frequency of the TEL/PDGFRbeta fusion protein in chronic myelomonocytic leukemia. Haematologica. 2003;88(4):408–15.
Roumiantsev S, Shah NP, Gorre ME, Nicoll J, Brasher BB, Sawyers CL, Van Etten RA. Clinical resistance to the kinase inhibitor STI-571 in chronic myeloid leukemia by mutation of Tyr-253 in the Abl kinase domain P-loop. Proc Natl Acad Sci U S A. 2002;99(16):10700–5.
Acknowledgements
Not applicable.
Funding
This work was supported in part by grants from the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), Jiangsu Province Natural Science Foundation for Distinguished Young Scholars (BK2012006), Jiangsu Provincial Special Program of Medical Science (BL2012005), Jiangsu Province’s Key Medical Center (ZX201102), Jiangsu Provincial Postdoctoral Science Foundation(1501059C), China Natural Youth Science Foundation(81300424 and 81600116), China Natural Science Foundation (81370626), and China Postdoctoral Science Foundation (2015 M580467).
Availability of data and materials
Data sharing is not applicable for this article as no datasets were generated or analyzed during the current study.
Authors’ contributions
SC, GS and ZZ designed and performed the research, and drafted the manuscript. JP and LK performed the bone marrow karyotype and FISH analysis. QW, HY and LW participated in the RNA-sequencing data analysis. GS and ZZ confirmed the multiple fusion transcripts by PCR. LM, HQ and DW assessed the clinical data of the patient. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Informed written consent for publication of the clinical details was obtained from the patient. A copy of the consent form is available for inspection by the editors.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the First Affiliated Hospital of the Soochow University according to the conditions of the Declaration of Helsinki. The patient has given his consent for participation in this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Sheng, G., Zeng, Z., Pan, J. et al. Multiple MYO18A-PDGFRB fusion transcripts in a myeloproliferative neoplasm patient with t(5;17)(q32;q11). Mol Cytogenet 10, 4 (2017). https://doi.org/10.1186/s13039-017-0306-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-017-0306-8