Abstract
Platelet-derived growth factor receptor B (PDGFRB) gene rearrangements define a unique subgroup of myeloid and lymphoid neoplasms frequently associated with eosinophilia and characterized by high sensitivity to tyrosine kinase inhibition. To date, various PDGFRB/5q32 rearrangements, involving at least 40 fusion partners, have been reported. However, information on genomic and clinical features accompanying rearrangements of PDGFRB is still scarce. Here, we characterized a series of 14 cases with a myeloid neoplasm using cytogenetic, single nucleotide polymorphism array, and next-generation sequencing. We identified nine PDGFRB translocation partners, including the KAZN gene at 1p36.21 as a novel partner in a previously undescribed t(1;5)(p36;q33) chromosome change. In all cases, the PDGFRB recombination was the sole cytogenetic abnormality underlying the phenotype. Acquired somatic variants were mainly found in clinically aggressive diseases and involved epigenetic genes (TET2, DNMT3A, ASXL1), transcription factors (RUNX1 and CEBPA), and signaling modulators (HRAS). By using both cytogenetic and nested PCR monitoring to evaluate response to imatinib, we found that, in non-AML cases, a low dosage (100–200 mg) is sufficient to induce and maintain longstanding hematological, cytogenetic, and molecular remissions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The PDGFRB gene, at 5q32, encodes for the ß chain of the cell surface receptor for platelet-derived growth factor (PDGFRß), a class III receptor tyrosine kinase (RTK) that activates signaling pathways involved in cell growth and differentiation [1]. PDGFRB is a frequent target of chromosomal translocations in a subgroup of hematological malignancies recognized in the 2017 World Health Organization (WHO) as a stand-alone category under “Myeloproliferative neoplasms with eosinophilia and gene rearrangement” [2. These disorders are presenting as chronic myeloid neoplasms, frequently as chronic myelomonocytic leukemia with eosinophilia [2], although (hyper)-eosinophilia is not invariably present [3, 4]. Less frequently, PDGFRB is rearranged in lymphoid malignancies, including cases of both B- and T-cell acute lymphoblastic leukemia/lymphoma (ALL) [1, 4–6]. In addition, in the same patient, a PDGFRB rearrangement may underlie the occurrence of two different malignancies of both myeloid and lymphoid lineages, which may be diagnosed concomitantly or sequentially [7, 8]. To date, at least 40 fusion translocation partners of PDGFRB have been identified. Among them, ETV6 is the most frequently involved gene as a consequence of the t(5;12)(q32;p13) [4], while all other partners are rare and often found in single cases [9]. Whatever the partner, the PDGFRB gene always participates in the fusion with its 3′ end tyrosine kinase domain, resulting in the constitutive activation and in the deregulation of downstream signaling cascades, including Ras/mitogen-activated protein kinase, phosphatidylinositol 3′-kinase, and phospholipase-5γ pathways [4]. Information on somatic gene variants in cases with a PDGFRB rearrangement is still scarce mainly affecting ASXL1, TET2, STAG2, DNMT3A, NRAS, ZRSR2, BCOR, and STAT5B [4, 7, 10, 11]. Male predominance and a median age at onset in the late 40 s [2] have been established. Most patients have splenomegaly and/or hepatomegaly [2]. Rapid response and long-term remission are obtained with tyrosine kinase inhibitors. In particular, imatinib is the treatment of choice for this group of neoplasms [3, 4, 9]. Although this treatment is successful, few series of cases with clinical and molecular monitoring have been reported [3, 4, 12].
We carried out an in-depth characterization of genomic events accompanying the 5q32 rearrangement in a series of 14 PDGFRB-positive cases recruited in our center during the last 23 years. In addition, we evaluated the response to imatinib by cytogenetic and molecular monitoring.
Materials and methods
Patients
Patients with a myeloid neoplasm and a rearrangement of PDGFRB were recruited from the files of the Laboratory of Cytogenetics at the Department of Medicine and Surgery-Hematology section of the University of Perugia (Table 1). Data collection was done in accordance with the Declaration of Helsinki and its later amendments. The study was approved by the Bioethics Committee University of Perugia (number 2014–0259). All patients or their parents/guardians have provided informed consent for sample collection and use in approved research studies.
FISH
Fluorescence in situ hybridization (FISH) was used to investigate PDGFRB involvement in our cohort, to identify translocation partners, and to assess cytogenetic response. A complete list of the genomic probes is reported in Supplementary Table 1. PDGFRB was studied by using either a commercial break-apart probe (LSI PDGFRB Break Apart, Abbott Molecular Diagnostics, Rome, Italy) or home-brew FISH assays with bac probes (RP11-759G10, 149,562,887–149,746,958, for the 5′, and RP11-100O5, 149,274,400–149,455,385, for the 3′) and even more specific cosmid clones: cosmid 4–1, for the 5′, and 9–4, for the 3′ of the gene [13]. The 1p36/KAZN breakpoint was characterized using a series of locus-specific probes mapping at 1p36–1p35 regions (Supplementary Table 1). Genomic coordinates are referred to GRCh37/hg19 assembly. One to two hundred nuclei and at least 7 abnormal metaphases were analyzed in each experiment. Follow-up analyses were carried out in one thousand nuclei. Cutoffs were assumed at the upper limit value obtained in 500 cells from a normal donor. FISH was used to assess the cytogenetic response in 13/14 cases (nos. 1–9, 11–14) with a monitoring range of 1–12 months.
Single nucleotide polymorphism array (SNPa)
Copy number variations (CNVs), gains, losses, and copy neutral loss of heterozygosity (cnLOH) were studied by SNP array in 9 cases with available material at diagnosis (nos. 1–4, 7–8, 11, 13–14) using a Cytoscan HD Array Kit (Affymetrix/Thermo Fisher Scientific, Santa Clara, CA, USA) following manufacturer’s instructions. Data were analyzed by Chromosome Analysis Suite (ChAS 4.1) software with hg19 build as reference. Filters were set at 400 kb, 50 markers for CNVs, and 10 Mb, 50 markers for cnLOH. Polymorphic copy number variants were excluded from the analysis by comparing with the Database of Genomic Variants (http://dgv.tcag.ca/dgv/app/home).
RT-PCR
Total RNA was extracted by Trizol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s protocol. One microgram was retrotranscribed using 50 U of SuperscriptII (Invitrogen). Nested PCR was performed to identify the fusion transcripts in 13/14 cases (nos. 1–9, 11–14) and for monitoring the minimal residual disease in 11 cases (nos. 1–4, 7–9, 11–14) with a monitoring range of 1–12 months (primer details are reported in Supplementary Table 2).
Treatment
Therapeutic regimens and responses for all patients are reported in Table 1 and .Table 2 Cytoreductive treatment was administered in eight cases (nos. 1–3, 5, 7, 9–11; Table 1) before the cytogenetic finding of a PDGFRB involvement was available. Thereafter, imatinib was administered as monotherapy in 13/14 cases (Table 2). Seven cases (nos. 1–2, 4, 6–8, 11) received a low dosage (100–200 mg/die), whereas 4 cases (nos. 3, 9, 13–14) received a high dosage (400 mg/die). In 2 cases (nos. 3 and 9), the dosage was reduced during follow-up. In 2 cases (nos. 5 and 12), the dosage was not available.
Targeted NGS
Mutational analysis of thirty genes mutated in myeloid malignancies and included in the Myeloid Solution SOPHiA GENETICS (Saint‐Sulpice, Switzerland) was performed following the manufacturer’s instructions on 13 cases at diagnosis (patient nos. 1–4, 6–14), in 8 cases (nos. 1, 4, 7–8, 11–14) also at cytogenetic and/or molecular remission, and in one case (no. 14) also after HSCT. The resulting captured libraries were further processed on a Miseq® sequencing platform (Illumina, San Diego, CA, USA). FASTQ sequencing files were uploaded to SOPHiA DDM® platform version 4 and, following adapter trimming and quality filtering, reads were aligned to the human reference genome (hg19 assembly) through the artificial SOPHiA™ intelligence. Variant calling of the resulted alignments was then performed using an in-house somatic variant caller, which takes into account the background noise level at each region. Only exonic and splice sites variants with MAF < 0.01 were taken into consideration and classified according to ACMG criteria [14] using the Varsome database [15]. Mutations with a variant allele frequency (VAF) of 40–60% or > 90% and/or associated with a clinical phenotype reported in OMIM (Online Mendelian Inheritance in Man, https://omim.org/) were defined as germline. All other variants were defined as somatic.
Results
Fourteen cases were collected. The median age at diagnosis was 38 years (range: 2–75) with a male/female ratio of 2.5 (Table 1). Eleven of fourteen patients were first observed in a non-aggressive phase (6 MPN/MDS, 2 CEL, 2 CMML, 1 aCML), 2 cases were first diagnosed as acute myeloid leukemia, and 1 case was diagnosed as myeloid sarcoma (Table 1). Thirteen of fourteen cases (93%) presented with leukocytosis (range: 11.2–106.8 × 109/L); eosinophilia (≥ 0.5 × 109/L) or hyper-eosinophilia (≥ 1.5 × 109/L) was demonstrated in 12 of them (range: 1.07–14.4 × 109/L) (Table 1). Six cases showed monocytosis (≥ 1.5 × 109/L). Anemia was seen in seven cases and thrombocytopenia in three. Hepatomegaly and/or splenomegaly was present in eleven cases (Table 1), and lymphadenopathy was noticed in three (Table 1). The serum lactate dehydrogenase level was available in seven cases and resulted increased in five (288–1604 U/L). Skin lesions have been noticed at diagnosis in two cases (nos. 7 and 9, Table 1).
Cytogenetics and molecular findings
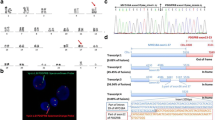
Involvement of 5q32 was detected by conventional cytogenetics in 13/14 patients while it was cryptic in case no. 11 (Table 1). In case no. 13, with t(5;12), hyperdiploidy was predominant. FISH revealed PDGFRB involvement and identified the translocation partner in all cases. Seven partners were already reported [16,17,18,19,20,21,22,23,24]. RT-PCR confirmed the presence of the corresponding fusion transcript in all cases with available material (12/14; nos. 1–4, 6–9, 11–14). Nine different partner genes were found (Table 1). Among them, ETV6 (n = 4), ERC1 (n = 2), and CCDC88C (n = 2) were recurrent partners. Moreover, in case no. 14, we identified KAZN at 1p36.21 as a novel partner gene of PDGFRB. Metaphase FISH identified the 1p breakpoint between RP11-1079F4, flanking the 5′, and RP11-317H5, covering the 3′ of the gene. Additional experiments, showing the splitting of WI2-0883F15 and WI2-0968K05 fosmids, narrowed the breakpoint between exons 4 and 5 of KAZN (Fig. 1a). RT-PCR confirmed an in-frame fusion between exon 4 of KAZN (NM_201628.3) and exon 12 of PDGFRB (NM_002609.3) (Fig. 1b)
Cytogenetic and molecular characterization of the novel KAZN::PDGFRB fusion. a FISH break-apart assay with fosmids WI2-0883F15 (spectrum green) and WI2-0968K05 (spectrum orange) for KAZN/1p36.21 showed a fusion signal on normal chromosome 1, a red signal on der(1), and a green signal on der(5) in case no. 14 (Table 1). b Direct sequencing showed an in-frame fusion joining exon 4 of KAZN to exon 12 of PDGFRB. GenBank accession numbers: NM_201628.3 for KAZN and NM_002609.3 for PDGFRB. nl, normal; ex, exon
SNParray
SNPa revealed a total of 13 events (3 gains, 8 losses, and 2 copy neutral LOH, Supplementary Table 3) in nine investigated cases. However, they were mainly concentrated in case no. 13 showing nine imbalances (gains at 3q and 8q, and losses at 1p, 2q, 4q, and 5q; Supplementary Table 3). Four events were distributed in case nos. 1, 3, 11, and 14 (Supplementary Table 3). The remaining four cases (nos. 2, 4, 7, and 8) were normal (Supplementary Table 3).
Mutational analysis
NGS was applied in 13 cases at diagnosis. Seven of them showed a total of 16 variants (range: 0–5 per case) (Fig. 2 and Table 3). One germline mutation at PTPN11 was confirmed in patient no. 9 (Fig. 2 and Table 3). A total of five somatic variants affecting HRAS (n = 2), TET2, DNMT3A, and CEBPA (n = 1 each) were found in 5 out of the 10 cases with chronic presentation (nos. 4, 8–11; 1 mutation/case). Longitudinal studies showed disappearance of HRAS and TET2 variants at remission in case nos. 4 and 8, respectively, whereas DNMT3A mutation load increased at cytogenetic remission in case no. 11 (Table 3). In the two cases with AML at presentation (nos. 13 and 14), we found a total of 10 somatic mutations (5 mutation/case) (Fig. 2 and Table 3). In both cases, RUNX1 was affected by multiple variants (n = 9), while one mutation at ASXL1 was found in case no. 14 (see Fig. 2, Table 3, and below for details).
Aggressive and unusual presentations
Two cases of our cohort presented as acute myeloid leukemia (nos. 13 and 14) and one case as myeloid sarcoma (no. 12).
Case no. 12
A 66-year-old man was diagnosed with myeloid sarcoma in another center based on a lymph node biopsy. At the same time, we found t(5;12)/PDGFRB::ETV6 and the absence of somatic mutations in bone marrow cells. Hematological, cytogenetic, and molecular response were achieved after 2 cycles of chemotherapy [25]. Remission was maintained by imatinib which was discontinued after 5 months because of iatrogenic interstitial infiltrates and pulmonary fibrosis. Four months later, the patient developed a B-ALL but material was not available for our cytogenetic-molecular studies. He died in aplasia after chemotherapy.
Case no. 13
A 36-year-old man received imatinib only at relapse, occurring 4 years after the first identification of a t(5;12)-positive AML that underwent chemotherapy and autologous transplant. At relapse, in addition to the PDGFRB translocation, we found five somatic variants at the RUNX1 gene (Fig. 2 and Table 3). The patient was treated with imatinib (400 mg/day) followed by chemotherapy. The disappearance of hematological and cytogenetic remissions and RUNX1 mutations was observed, followed by an unrelated HSCT. The patient died because of transplant-related events.
Case no. 14
A 33-year-old man was first observed because of marked splenomegaly and leukocytosis (Table 1) with 22% of circulating myeloblasts and promonocytes, and bone marrow blasts < 20%. A t(1;5) translocation plus four somatic variants at RUNX1 and one at ASXL1 (Fig. 2 and Table 3) was documented. Cytogenetic remission was observed after 6 months of imatinib at 400 mg/die although increased blast cells (20–30%) were found in the bone marrow aspirate. At that time, a complex mutational landscape was identified. The ASXL1-positive cells persisted with unchanged VAF, while involvement of RUNX1 showed a decrease of the non-sense variant c.777dupT, p.Asn260*, but an increase of the three additional variants already found at diagnosis (Table 3). Moreover, a FLT3-ITD was identified in a small-size cell population. Treatment was standard chemotherapy and subsequent HSCT from an HLA-identical sibling donor. AML relapsed 1 year after HSCT, when bone marrow cells were negative for both PDGFRB and RUNX1 c.777dupT, p.Asn260*, but still positive for ASXL1 and for 1 out of the 3 RUNX1 variants identified at diagnosis (c.497G > A, p.Arg166Gln) (Table 3). Unfortunately, massive splenomegaly and CNS involvement occurred, requiring palliative splenic and cranio-spinal irradiation. The patient died of progressive disease 24 months after HSCT.
Imatinib dosage
In 10/11 cases first observed with a non-aggressive disease phase, imatinib was administered as sole treatment. Six cases (nos. 1–3, 5, 7, and 9) received a cytoreductive therapy before imatinib while four cases (nos. 4, 6, 8, and 11) received imatinib as front-line therapy (Table 1). In nine cases, precise information on dosage, response, and follow-up was available. A dosage of 100 mg/die induced complete cytogenetic and molecular remission in three cases (nos. 2, 4, and 7) while an increase to 200 mg/die was necessary in four other cases (nos. 1, 6, 8, and 11). In two cases (nos. 3 and 9), a higher dosage (400 mg/die) induced cytogenetic and molecular remission that was maintained by a dosage of 200 mg in case no. 3 and 100 mg in case no. 9. Case no. 3, still in monitoring, has the longest follow-up of 18 years, while case no. 9 died of sepsis of unknown origin in disease remission at + 129 months. A long-standing history was observed in our pediatric case (no. 7) who was started on a dosage of 100 mg/die and obtained a molecular remission after 32 months. However, after an additional 19 months, we detected molecular relapse and imatinib was increased to 200 mg/die, obtaining a second molecular remission after 10 months. The disease is stable with negative cytogenetic and molecular tests at + 44 months from the second remission and at + 105 months from the first imatinib administration. The median survival from the start of imatinib treatment in the nine chronic cases is 66.5 months (range: 30–222 months). Eight cases are still in follow-up.
Monitoring of PDGFRB rearrangements
Both FISH and nested or semi-nested RT-PCR were used for disease monitoring during imatinib treatment. FISH documented complete cytogenetic remission in all 13 cases with a median of 9 months after start of treatment. Molecular remission was also documented in 9 of 11 tested cases with a median of 24 months after start of treatment. In case no. 13, positivity disappeared only after HSCT, while case no. 11, who discontinued imatinib for 10 weeks due to kidney transplantation, was positive at last follow-up (+ 72 months from imatinib administration).
In four cases with complete monitoring (nos. 2–3, 7, and 9), the median time lapse between cytogenetic and molecular remission was 24 months (range: 14–32). The earliest cytogenetic remission was obtained in case no. 3 after 1 month of treatment, although the disease persisted at molecular level up to month 25. In case nos. 1 and 4, remission was simultaneously documented by FISH and RT-PCR after 24 and 9 months of treatment, respectively. The median overall survival after molecular remission in nine cases was 42 months (range: 6–197), with ongoing follow-up in seven cases.
Discussion
Chromosomal rearrangements involving PDGFRB in myeloid malignancies are rare events. Here, we characterized a series of 14 cases with a myeloid neoplasm at diagnosis and a rearrangement of PDGFRB, providing data from in-depth genomic characterization by SNPa and NGS analysis. At cytogenetics, the 5q32/PDGFRB rearrangement involved multiple partners identified by banding in all cases but one with a cryptic change. Notably, in one case, complex chromosome rearrangements corresponded to a PDGFRB::ETV6 fusion underlying a four-break translocation. These results confirm the clinical relevance of FISH studies for PDGFRB gene rearrangements in patients with myeloid neoplasms, especially when associated with eosinophilia, even in the absence of karyotypic abnormalities. Notably, the 5q32/PDGFRB rearrangement was the sole cytogenetic abnormality in our cohort and SNPa confirmed a low burden of co-occurring abnormalities in all cases with chronic disease, strongly supporting the driver role of PDGFRß tyrosine kinase activation to address the clinical phenotype.
This study first identified the KAZN gene at 1p36 as a novel partner of PDGFRB in one case with a t(1;5)(p36;q33). The KAZN gene is involved in intercellular adhesion, cell differentiation, signal transduction, cytoskeletal organization, and apoptosis [26]. To date, PTBP2, TMEM51, and MTOR have been found as fusion partners of KAZN by RNA sequencing studies in solid tumors [27, 28], while, as far as we know, rearrangements of KAZN have never been described in hematological malignancies. Recurrent KAZN variants have not been specifically associated with human tumors (COSMIC, Catalogue of Somatic Mutation in Cancer [29]).
In a literature review, we found only 8 cases in chronic phase with information on the mutational background that showed variants at TET2, ASXL1, and STAG2 genes [4, 7, 10]. In our cases presenting with a chronic clinical phenotype, we found a low burden of somatic mutations involving TET2, DNMT3A, HRAS, and CEBPA. Notably, five and four variants affected the RUNX1 gene in our AML case no. 13 and no. 14, respectively. These findings suggest that instability at RUNX1 is a nonrandom event accompanying an acute clinical phenotype in PDGFRB + malignancies. This hypothesis is supported by the work of Stengel et al. [30], who rarely (< 1%) found more than three variants of the RUNX1 gene in a large series of mutated de novo AML cases. With respect to the nature of RUNX1 mutations in our acute cases, they are all predicted to be pathogenic. Six of them fell in the Runt-Homology DNA binding Domain (RHD), one fell in the Trans-Activation Domain (TAD), and two non-sense mutations truncated the protein between RHD and TAD.
Longitudinal studies helped us to characterize the behavior of somatic mutations over the disease course. In particular, both HRAS and TET2 gene variants disappeared concomitantly to cytogenetic and molecular remission of the PDGFRB abnormality. These cases, in addition to one case with somatic mutation at the BCOR gene that disappeared at remission of the PDGFRB rearrangement [11], suggest that all these acquired mutations were present in the PDGFRB + clonal population sensitive to imatinib. Instead, a peculiar dynamics of somatic gene variants, as compared to the PDGFRB rearrangement, was seen in our case no. 14, in which one of the RUNX1 variants (c.777dupT, p.Asn260*) decreased concomitantly to cytogenetic remission, whereas both the ASXL1 and the c.497G > A RUNX1 variants appeared as the leukemic fil rouge over the whole disease course in this case, from diagnosis till post-transplant relapse, when PDGFRB was absent. Altogether, these results raise the question whether AML in the last case was a second malignancy instead of an acute phase originated by linear clonal evolution [31] of the PDGFRB-positive cells. Single-cell analysis will be helpful to better understand the origin of AML in PDGFRB-positive cases. Finally, the increased clonal size of DNMT3A in another responder (case no. 11) with cytogenetic disappearance of the PDGFRB rearrangement is consistent with clonal hematopoiesis, similar to that observed in other myeloid malignancies [32].
Our study confirmed that prognosis in cases with PDGFRB rearrangement is deeply influenced by the clinical presentation [3], as our cases first seen with acute disease had a more unfavorable outcome compared to cases with less aggressive hematological phenotypes at diagnosis.
A consensus on the posology of the TKI inhibitor in PDGFRB-related disorders presenting in chronic phase is still not available. The present series underlines the low dosage of 100–200 mg as a successful approach to inducing and maintaining cytogenetic and molecular remission. Moreover, molecular analysis is essential to carefully monitoring response to therapy, to evaluate the amount of residual disease, and to address changes of imatinib dosage over the disease course.
Data availability
All datasets generated during the current study were deposited in NCBI Gene Expression Omnibus (GEO) under accession number GSE182820 (GSE182785 for SNParray and GSE182817 for Sequencing data).
References
Ondrejka SL, Jegalian AG, Kim AS, Chabot-Richards DS, Giltnane J, Czuchlewski DR, Shetty S, Sekeres MA, Yenamandra A, Head D, Jagasia M, His ED (2014) PDGFRB-rearranged T-lymphoblastic leukemia/lymphoma occurring with myeloid neoplasms: the missing link supporting a stem cell origin. Haematologica 99:e148–e151. https://doi.org/10.3324/haematol.2014.105452
Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J (2017) WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon, France
Jawhar M, Naumann N, Schwaab J, Baurmann H, Casper J, Dang TA, Dietze L, Döhner K, Hänel A, Lathan B, Link H, Lotfi S, Maywald O, Mielke S, Müller L, Platzbecker U, Prümmer O, Thomssen H, Töpelt K, Panse J, Vieler T, Hofmann WK, Haferlach T, Haferlach C, Fabarius A, Hochhaus A, Cross NCP, Reiter A, Metzgeroth G (2017) Imatinib in myeloid/lymphoid neoplasms with eosinophilia and rearrangement of PDGFRB in chronic or blast phase. Ann Hematol 96:1463–1470. https://doi.org/10.1007/s00277-017-3067-x
Fang H, Tang G, Loghavi S, Greipp P, Wang W, Verstovsek S, Medeiros LJ, Reichard KK, Miranda RN, Wang SA (2020) Systematic use of fluorescence in-situ hybridisation and clinicopathological features in the screening of PDGFRB rearrangements of patients with myeloid/lymphoid neoplasms. Histopathology 76:1042–1054. https://doi.org/10.1111/his.14097
Zabriskie MS, Antelope O, Verma AR, Draper LR, Eide CA, Pomicter AD, Tran TH, Druker BJ, Tyner JW, Miles RR, Graham JM, Hwang JH, Varley KE, Toydemir RM, Deininger MW, Raetz EA, O’Hare T (2018) A novel AGGF1-PDGFRb fusion in pediatric T-cell acute lymphoblastic leukemia. Haematologica 103:e87–e91. https://doi.org/10.3324/haematol.2017.165282
Heilmann AM, Schrock AB, He J, Nahas M, Curran K, Shukla N, Cramer S, Draper L, Verma A, Erlich R, Ross J, Stephens P, Miller VA, Ali SM, Verglio JA, Tallman MS, Mughal TI (2017) Novel PDGFRB fusions in childhood B- and T-acute lymphoblastic leukemia. Leukemia 31:1989–1992. https://doi.org/10.1038/leu.2017.161
Pozdnyakova O, Orazi A, Kelemen K, King R, Reichard KK, Craig FE, Quintanilla-Martinez L, Rimsza L, George TI, Horny HP, Wang SA (2021) Myeloid/lymphoid neoplasms associated with eosinophilia and rearrangements of PDGFRA, PDGFRB, or FGFR1 or with PCM1-JAK2. Am J Clin Pathol 155:160–178. https://doi.org/10.1093/ajcp/aqaa208
Chen X, Wang F, Wang T, Zhang Y, Ma X, Yuan L, Teng W, Guo L, Liu M, Liu M, Chen J, Nie D, Zhang Y, Zhou X, Wang M, Chen KN, Zhu P, Liu H (2020) The incidence, genetic characteristics, and prognosis of leukemia with concurrent pathogenic fusion genes: a series of 25 cases from a large cohort of leukemia patients. Cancer Gene Ther 27:89–97. https://doi.org/10.1038/s41417-019-0147-1
Appiah-Kubi K, Lan T, Wang Y, Qian H, Wu M, Yao X, Wu Y, Chen Y (2017) Platelet- derived growth factor receptors (PDGFRs) fusion genes involvement in hematological malignancies. Crit Rev Oncol Hematol 109:20–34. https://doi.org/10.1016/j.critrevonc.2016.11.008
Zimmermann N, Nassiri M, Zhou J, Miller AM, Zhang S (2020) Myeloid neoplasm with a novel cryptic PDGFRB rearrangement detected by next-generation sequencing. Cancer Genet 244:55–59. https://doi.org/10.1016/j.cancergen.2020.03.002
Baer C, Muehlbacher V, Kern W, Haferlach C, Haferlach T (2018) Molecular genetic characterization of myeloid/lymphoid neoplasms associated with eosinophilia and rearrangement of PDGFRA, PDGFRB, FGFR1 or PCM1-JAK2. Haematologica 103:e348–e350. https://doi.org/10.3324/haematol.2017.187302
Cheah CY, Burbury K, Apperley JF, Huguet F, Pitini V, Gardembas M, Ross DM, Forrest D, Genet P, Rousselot P, Patton N, Smith G, Dunbar CE, Ito S, Aguiar RCT, Odenike O, Gimelfarb A, Cross NCP, Seymour JF (2014) Patients with myeloid malignancies bearing PDGFRB fusion genes achieve durable long-term remissions with imatinib. Blood 123:3574–3577. https://doi.org/10.1182/blood-2014-02-555607
Baxter EJ, Kulkarni S, Vizmanos J-L, Jaju R, Martinelli G, Testoni N, Hughes G, Salamanchuk Z, Calasanz MJ, Lahortiga I, Pocock CF, Dang R, Fidler C, Wainscoat JS, Boultwood J, Cross NCP (2003) Novel translocations that disrupt the platelet-derived growth factor receptor β (PDGFRB) gene in BCR-ABL-negative chronic myeloproliferative disorders. Br J Haematol 120:251–256. https://doi.org/10.1046/j.1365-2141.2003.04051.x
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–423. https://doi.org/10.1038/gim.2015.30
Kopanos C, Tsiolkas V, Kouris A et al (2019) VarSome: the human genomic variant search engine. Bioinformatics 35:1978–1980. https://doi.org/10.1093/bioinformatics/bty897
Siena S, Sammarelli G, Grimoldi MG, Schiavo R, Nozza A, Roncalli M, Mecucci C, Santoro A, Carlo-Stella C (1999) New reciprocal translocation t(5;10)(q33;q22) associated with atypical chronic myeloid leukemia. Haematologica 84:369–372
Luciano L, Catalano L, Sarrantonio C, Guerriero A, Califano C, Rotoli B (1999) AlphaIFN- induced hematologic and cytogenetic remission in chronic eosinophilic leukemia with t(1;5). Haematologica 84:651–653
Rosati R, La Starza R, Luciano L, Gorello P, Matteucci C, Pierini V, Romoli S, Crescenzi B, Rotoli B, Martelli MF, Pane F, Mecucci C (2006) TPM3/PDGFRB fusion transcript and its reciprocal in chronic eosinophilic leukemia. Leukemia 20:1623–1624. https://doi.org/10.1038/sj.leu.2404307
Albano F, Anelli L, Zagaria A, Lonoce A, La Starza R, Liso V, Rocchi M, Specchia G (2008) Bosi A, Martelli MF, Marynen P, Mecucci C (2007) Molecular cytogenetic findings in a four- way t(1;12;5;12)(p36;p13;q33;q24) underlying the ETV6-PDGFRB fusion gene in chronic myelomonocytic leukemia. Cancer Genet Cytogenet 176:67–71. https://doi.org/10.1016/j.cancergencyto.2007.03.004
Crescenzi B, La Starza R, Nozzoli C, Ciolli S, Matteucci C, Romoli S, Rigacci L, Gorello P, Bosi A, Martelli MF, Marynen P, Mecucci C (2007) Molecular cytogenetic findings in a four-way t(1;12;5;12)(p36;p13;q33;q24) underlying the ETV6-PDGFRB fusion gene in chronic myelomonocytic leukemia. Cancer Genet Cytogenet 176:67–71. https://doi.org/10.1016/j.cancergencyto.2007.03.004
La Starza R, Rosati R, Roti G, Gorello P, Bardi A, Crescenzi B, Pierini V, Calabrese O, Baens M, Folens C, Cools J, Marynen P, Martelli MF, Mecucci C, Cuneo A (2007) A new NDE1/PDGFRB fusion transcript underlying chronic myelomonocytic leukaemia in Noonan Syndrome. Leukemia 21:830–833. https://doi.org/10.1038/sj.leu.2404541
Gorello P, La Starza R, Brandimarte L, Trisolini SM, Pierini V, Crescenzi B, Limongi MZ, Nanni M, Belloni E, Tapinassi C, Gerbino E, Martelli MF, Foà R, Meloni G, Pelicci PG, Mecucci C (2008) A PDGFRB-positive acute myeloid malignancy with a new t(5;12)(q33;p13.3) involving the ERC1 gene. Leukemia 22:216–218. https://doi.org/10.1038/sj.leu.2404894
Cavazzini F, Bardi A, Ciccone M, Rigolin GM, Gorello P, La Starza R, Mecucci C, Cuneo A (2009) Trisomy 8 in PDGFRB-negative cells in a patient with imatinib-sensitive chronic myelomonocytic leukemia and t(5;16)(q33;p13), PDGFRB–NDE1 fusion. Cancer Genet Cytogenet 194:67–69. https://doi.org/10.1016/j.cancergencyto.2009.04.026
Maccaferri M, Pierini V, Di Giacomo D, Zucchini P, Forghieri F, Bonacorsi G, Paolini A, Quadrelli C, Giacobbi F, Fontana F, Cappelli G, Potenza L, Marasca R, Luppi M, Mecucci C (2017) The importance of cytogenetic and molecular analyses in eosinophilia-associated myeloproliferative neoplasms: an unusual case with normal karyotype and TNIP1- PDGFRB rearrangement and overview of PDGFRB partner genes. Leuk Lymphoma 58:489–493. https://doi.org/10.1080/10428194.2016.1197396
Caprioli C, Lussana F, Salmoiraghi S, et al (2020) Clinical significance of chromatin- spliceosome acute myeloid leukemia: a report from the Northern Italy Leukemia Group (NILG) randomized trial 02/06. Haematologica haematol 2020 252825. https://doi.org/10.3324/haematol.2020.252825
Le Pennec S, Konopka T, Gacquer D, Fimereli D, Tarabichi M, Tomás G, Savagner F, Decaussin-Petrucci M, Trésallet C, Andry G, Larsimont D, Detours V, Maenhaut C (2015) Intratumor heterogeneity and clonal evolution in an aggressive papillary thyroid cancer and matched metastases. Endocr Relat Cancer 22:205–216. https://doi.org/10.1530/ERC-14-0351
Yoshihara K, Wang Q, Torres-Garcia W, Zheng S, Vegesna R, Kim H, Verhaak RGW (2015) The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene 34:4845–4854. https://doi.org/10.1038/onc.2014.406
Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C (2014) The landscape of kinase fusions in cancer. Nat Commun 5:4846. https://doi.org/10.1038/ncomms5846
Tate JG, Bamford S, Jubb HC et al (2019) COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 47:D941–D947. https://doi.org/10.1093/nar/gky1015
Stengel A, Kern W, Meggendorfer M, Nadarajah N, Perglerovà K, Haferlach T, Haferlach C (2018) Number of RUNX1 mutations, wild-type allele loss and additional mutations impact on prognosis in adult RUNX1-mutated AML. Leukemia 32:295–302. https://doi.org/10.1038/leu.2017.239
Vosberg S, Greif PA (2019) Clonal evolution of acute myeloid leukemia from diagnosis to relapse. Genes, Chromosom Cancer 58:839–849. https://doi.org/10.1002/gcc.22806
Pløen GG, Nederby L, Guldberg P, Hansen M, Ebbesen LH, Jensen UB, Hokland P, Aggerholm A (2014) Persistence of DNMT3A mutations at long-term remission in adult patients with AML. Br J Haematol 167:478–486. https://doi.org/10.1111/bjh.13062
Funding
This work was supported by AIRC 5 × 1000, MYNERVA project, #21267 (MYeloid NEoplasms Research Venture Airc, http://www.progettoagimm.it) (CM); Sergio Luciani Association, Fabriano, Italy (CM); Consorzio Internazionale per le Biotecnologie, CIB (CM); Roche per la Medicina di Precisione, bando 2020 (DDG)., #21267 (MYeloid NEoplasms Research Venture Airc, http://www.progettoagimm.it) (CM); Sergio Luciani Association, Fabriano, Italy (CM); Consorzio Internazionale per le Biotecnologie, CIB (CM); Roche per la Medicina di Precisione, bando 2020 (DDG).
Author information
Authors and Affiliations
Contributions
D.D.G. and P.G. performed molecular studies and monitoring. V.P., R.L.S., B.C., and C.M. performed and analyzed cytogenetics, FISH, and SNP arrays. F.P. and Ca. Ma. performed and analyzed NGS data. M.Q., R.L.S., P.F.F., M.V., E.B., F.A., F.F. M.M., F.B., M.L., A.C., G.R., and C.M. collected samples and patient data. C.M. conceived the study. C.M. and D.D.G. collected all of the data and wrote the manuscript. All of the authors read and approved the final paper.
Corresponding author
Ethics declarations
Ethics approval
The study was approved by the Bioethics Committee of the University of Perugia (number 2014–0259). The study was performed in accordance with the Declaration of Helsinki and its later amendments.
Consent to participate
Informed consent was obtained for all individual participants included in the study.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Di Giacomo, D., Quintini, M., Pierini, V. et al. Genomic and clinical findings in myeloid neoplasms with PDGFRB rearrangement. Ann Hematol 101, 297–307 (2022). https://doi.org/10.1007/s00277-021-04712-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00277-021-04712-8