Abstract
Background
Therapy-related myeloid neoplasm after treatment for acute promyelocytic leukemia (APL) is a relatively infrequent but severe complication. Most therapy-related myeloid neoplasms after treatment for APL are classified as therapy-related myelodysplastic syndrome or therapy-related acute myeloid leukemia. Translocation of 5q31-33, PDGFRB occur rarely in therapy-related myeloid neoplasm and there has been two identified PDGFRB partner genes located at 14q32, TRIP11 and KIAA1509.
Results
The TRIP11-PDGFRB fusion was identified in a patient with therapy-related myeloid neoplasm with t(5;14)(q33;q32) after treatment of APL using conventional cytogenetics, fluorescence in situ hybridization (FISH) and molecular methods. Cytogenetic analysis of the bone marrow aspirate revealed 46, XY, t(5;14)(q33;q32) in all 20 analyzed cells. No other cytogenetic abnormalities were observed. Break-apart FISH analysis demonstrated that rearrangement of PDGFRB at 5q33 was positive in 460 of 500 cells analyzed, while the PML-RARA rearrangement remained undetectable by RT-PCR. Sequencing of RT-PCR products revealed fusion between exon 16 of TRIP11 and exon 11 of PDGFRB. However, the KIAA1509-PDGFRB fusion was not detected by RT-PCR.
Conclusion
We firstly demonstrated that therapy-related myeloid neoplasm with TRIP11-PDGFRB fusion was identified after treatment of APL.
Similar content being viewed by others
Background
Therapy-related myeloid neoplasms (t-MNs) are late complications of cytotoxic therapies used to treat malignant and non-malignant conditions. The incidence of t-MNs is increasing worldwide, due to improved survival rates following treatment of primary malignancies [1]. t-MNs account for 10-20% of all malignant myeloid diagnoses [2].
The introduction of all trans retinoic acid (ATRA) has been a major breakthrough in the treatment of acute promyelocytic leukemia (APL), characterized by t(15;17)(q22;q12). The combination of ATRA and anthracycline-based chemotherapy results in high rates of complete remission and survival. However, ATRA combined with chemotherapeutic drugs increases the risk of t-MN. Most t-MNs after treatment for APL are classified as therapy related myelodysplastic syndrome (t-MDS) or therapy related acute myeloid leukemia (t-AML). Rearrangement of platelet-derived growth factor receptor beta (PDGFRB) is a distinctive type of myeloid neoplasm which occurs rarely in t-MN. More than 20 different partner genes of PDGFRB have been described [3]. The most common abnormality is the t(5;12)(q31 ~ 33;p12), which forms an ETV6-PDGFRB fusion gene. The TRIP11-PDGFRB fusion gene and the KIAA1509 (HGNC approved gene symbol; CCDC88C)-PDGFRB fusion gene were identified in a patient with myeloproliferative neoplasm (MPN) and t(5;14)(q33;q32) [4],[5]. t-MN has an extremely poor clinical outcome. However, t-MNs with PDGFRB rearrangement are very sensitive to imatinib methylate, a tyrosine kinase inhibitor with activity against ABL, c-KIT, and PDGFR[6].
Here, we report the detection of a TRIP11-PDGFRB fusion in a patient with t-MN with t(5;14)(q33;q32) after treatment of APL.
Case presentation
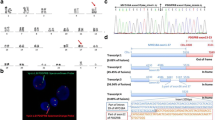
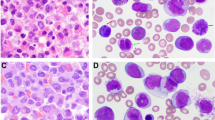
A 54-year-old Korean male was referred to our hospital due to hematemesis in July of 2010. Peripheral blood results showed a hemoglobin of 9.1 g/dL, a platelet count of 23 × 109/L, and a leukocyte count of 1.3×109/L with 30% abnormal promyelocytes. The bone marrow aspirates and biopsy showed hypercellular marrow with 95% abnormal promyelocytes, including faggot cells. Karyotypic analysis of the bone marrow revealed 46, XY, t(15;17) (q22;21) in all 20 metaphases that were analyzed. No other cytogenetic abnormalities were observed. The PML-RARA fusion gene was detected using RT-PCR and FISH analysis. A diagnosis of acute promyelocytic leukemia with t(15;17) (q22;21);PML-RARA was made based on the World Health Organization (WHO) 2008 classification of myeloid lymphoid neoplasms. The patient was treated with ATRA in combination with idarubicin. The presence of the PML-RARA rearrangement was monitored by FISH and quantitative RT-PCR. He achieved a complete remission. The patient received consolidation and maintenance chemotherapies. He was monitored at regular time intervals and no PML-RARA was detected in the bone marrow after consolidation and maintenance therapies. At a regular follow-up analysis in October 2013, peripheral blood results showed a hemoglobin of 12.3 g/dL, a platelet count of 104 × 109/L, and a leukocyte count of 19.8×109/L, with 58% neutrophils and 5% eosinophils. The bone marrow aspirates showed myeloid hyperplasia with eosinophils and eosinophil precursors (13%) without blast excess (1%). Karyotypic analysis of the bone marrow revealed 46, XY, t(5;14)(q33;q32) in 20 metaphase cells. The PDGFRB rearrangement was detected by FISH analysis, while the PML-RARA rearrangement remained undetectable by RT-PCR. Since there has been two identified PDGFRB partner genes located at 14q32, TRIP11 and KIAA1509 (http://atlasgeneticsoncology.org), RT-PCR analysis was performed to specify the partner gene. As a result, TRIP11-PDGFRB fusion transcript, not KIAA1509- PDGFRB was detected (Figures 1 and 2). A final diagnosis of t-MN with t(5;14)(q33;q32) and a TRIP11-PDGFRB rearrangement was made.
RT-PCR using primers located in TRIP11 exons 14-16 and PDGFRB exons 11-12 resulting in amplification of TRIP11-PDGFRB fusion transcripts. M: Molecular weight marker, lane 1; 5′ TRIP11 and 3′ PDGFRB specific primers (patient), lane 2; Replication of 5′ TRIP11 and 3′ PDGFRB specific primers (patient), lane 3; 5′ TRIP11 and 3′ PDGFRB specific primers (control), lane 4; 5′ KIAA1509 and 3′ PDGFRB specific primers (patient), lane 5; 5′ KIAA1509 and 3′ PDGFRB specific primers (control).
Methods
Conventional cytogenetic analysis and FISH analysis
Chromosomes were analyzed with GTG-banding and the karyotype was described according to the International System for Human Cytogenetics Nomenclature, 2013. FISH analysis was conducted using a dual color PDGFRB breakapart probe (5q33, D5S1907, centromere side, red signal/D5S2014, telomere side, green color). The manufacturer’s recommendations were followed for the hybridization and post-washing procedures (Abbott Molecular/Vysis, Des Plaines, IL, USA). The expected normal signal pattern is two red/green fusion signals. When cells have PDGFRB gene rearrangement, the fusion signal would be split into a red and a green signal.
Molecular methods
To determine whether TRIP11 or KIAA1509 were fused to PDGFRB, RNA extraction was performed with the Qiamp RNA Blood Mini Kit (Qiagen, Hilden, Germany). Reverse transcription was performed with random hexamers and MMLV reverse transcriptase (Life Technologies, NY, USA). RT-PCR was performed with Takara LA Taq DNA polymerase (Takara Bio Inc, Shiga, Japan). For the detection of fusion transcript between TRIP11 and PDGFRB, 5′ TRIP11 and 3′ PDGFRB specific primers were used as previously described [4], using 1407CF (5′-CGCTGCAGCTTTCTGTCTCTCAGGAACAAG-3′) and 2022PR (5′-GTAACGTGGCTTCTTCTGCCA-3′). RT-PCR was performed with ABI 2720 (Life Technologies, Foster City, CA, USA) and cycles were as follows: initial denaturation at 94°C for 1 min; then 35 cycles of 94°C for 30 sec, 60°C for 30 sec and 72°C for 2 min; and a final extension of 5 min at 72°C. Direct sequencing of amplified products was performed with ABI 3130 (Life Technologies). Sequences were analyzed using Sequencher software (version 4.10 Gene Codes Corporation, Ann Arbor, MI, USA). For the detection of a fusion transcript between KIAA1509 and PDGFRB, 5′ KIAA1509 and 3′ PDGFRB specific primers were used as previously described [5], using KIAA1509-RTF1 (5′-ccgggacacagataagac-3′) and PDGFRB-RTR1 (5′-catgatcttcagctccgaca-3′).
Results and discussion
We have identified a TRIP11-PDGFRB fusion in a patient with t(5;14)(q33;q32) after treatment for APL. Cytogenetic analysis of a bone marrow aspirate revealed 46, XY, t(5;14)(q33;q32) in 20 analyzed cells. Break-apart FISH analysis demonstrated that rearrangement of PDGFRB at 5q33 was positive in 460 of 500 cells analyzed. Sequencing of RT-PCR products revealed fusion between exon 16 of TRIP11 (NM_004239.3) and exon 11 of PDGFRB (NM_002609.3), which is the same breakage and reunion point as that reported in the previous study [4]. The KIAA1509-PDGFRB fusion gene was not detected by RT-PCR (Figures 1 and 2). t-MN after treatment for APL is an infrequent but severe complication. The incidence of t-MN was 1% - 6.5% [7],[8]. t-MDS is a more common complication than t-AML when considering t-MNs [7]-[9]. The most common cytogenetic abnormalities were partial and complete deletions of chromosomes 5 and 7, and 11q23 rearrangements. Translocation of 5q31-33 and PDGFRB occur rarely in t-MN. The PDGFRB gene at 5q33 is normally expressed in erythroid and myeloid precursors in the bone marrow, and it is a receptor tyrosine kinase which plays an important role in wound healing and other processes in adults [10],[11]. PDGFRB is disrupted by other translocations, such as t(5;12), or t(5;14). The most common abnormality is t(5;12)(q31-33;p12), which forms an ETV6-PDGFRB fusion gene. The ETV-PDGFRB fusion gene was identified in patients with chronic myelomonocytic leukemia (CMML) and translocation t(5;12)(q33;p13) by Golub and Gilliland in 1994 [12]. ETV-PDGFRB is a chimeric tyrosine kinase protein which is constitutively active due to enforced homodimerization by self-association domains present on the fusion partner protein (ETV6). This in turn activates downstream signaling pathways, resulting in cell proliferation and survival [13]. Levin et al. cloned KIAA1509 as a PDGFRB fusion partner in imatinib-responsive myeloproliferative disease associated with t(5;14)(q33;q32)[5]. Expression of the CEV14-PDGFRB fusion gene has been reported in acute myeloid leukemia with t(7;11) after clonal evolution [4] in one reported case of t-MN after treatment for AML. Abe et al. hypothesized that the CEV14-PDGFRB fusion causes ectopic constitutive tyrosine kinase activation of PDGFRB, leading to transformation via the ras signal transduction pathway [4]. CEV14 is homologous to TRIP11, which was isolated as one of the proteins that interacts with thyroid hormone receptors in the presence of T3. In our case, response to therapy was monitored by FISH and RT-PCR for the PML-RARA rearrangement. At his last regular follow-up, RT-PCR for PML-PARA was negative, and eosinophilia was mild. Eosinophilia has been reported in recurrent chromosomal aberrations. 5q31-33 is often involved in myeloid disorders with eosinophilia, suggesting involvement of a PDGFRB rearrangement. t(5;14)(q33;q32) was detected with conventional cytogenetics and FISH analysis for PDGFRB was positive. In order to identify partner genes of PDGFRB, such as KIAA1509 or TRIP11, we performed RT-PCR and direct sequencing for both. We have identified a TRIP11-PDGFRB rearrangement. Detection of PDGFRB rearrangement is important because these rearrangements respond well to treatment with imatinib mesylate [6].
The majority of abnormalities involving PDGFRB are detected by conventional cytogenetics. Performing conventional cytogenetics on a regular basis in all treated APL patients for the early detection of t-MNs implied by chromosomal aberrations is important.
Conclusion
This is the first report of t-MN with a TRIP11-PDGFRB rearrangement after treatment of APL.
Consent
Written informed consent was obtained from the patient for publication of this Case report and any accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal.
References
Rund D, Krichevsky S, Bar-Cohen S, Goldschmidt N, Kedmi M, Malik E, Gural A, Shafran-Tikva S, Ben-Neriah S, Ben-Yehuda D: Therapy-related leukemia: clinical characteristics and analysis of new molecular risk factors in 96 adult patients. Leukemia 2005,19(11):1919–1928. 10.1038/sj.leu.2403947
Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW: WHO classification of tumours of haematopoietic and lymphoid tissues. 2008: 127.
Valent P, Gleich GJ, Reiter A, Roufosse F, Weller PF, Hellmann A, Metzgeroth G, Leiferman KM, Arock M, Sotlar K, Butterfield JH, Cerny-Reiterer S, Mayerhofer M, Vandenberghe P, Haferlach T, Bochner BS, Gotlib J, Horny HP, Simon HU, Klion AD: Pathogenesis and classification of eosinophil disorders: a review of recent developments in the field. Expert Rev Hematol 2012,5(2):157–176. 10.1586/ehm.11.81
Abe A, Emi N, Tanimoto M, Terasaki H, Marunouchi T, Saito H: Fusion of the platelet-derived growth factor receptor beta to a novel gene CEV14 in acute myelogenous leukemia after clonal evolution. Blood 1997,90(11):4271–4277.
Levine RL, Wadleigh M, Sternberg DW, Wlodarska I, Galinsky I, Stone RM, DeAngelo DJ, Gilliland DG, Cools J: KIAA1509 is a novel PDGFRB fusion partner in imatinib-responsive myeloproliferative disease associated with a t(5;14)(q33;q32). Leukemia 2005,19(1):27–30.
Malfuson JV, Konopacki J, Fagot T, Desangles F, Bories D, Souleau B, de Revel T: Therapy-related myeloproliferative neoplasm with ETV6-PDGFRB rearrangement following treatment of acute promyelocytic leukemia. Ann Hematol 2011,90(12):1477–1479. 10.1007/s00277-011-1188-1
Lobe I, Rigal-Huguet F, Vekhoff A, Desablens B, Bordessoule D, Mounier C, Ferrant A, Sanz M, Fey M, Chomienne C, Chevret S, Degos L, Fenaux P: European APL group experience: Myelodysplastic syndrome after acute promyelocytic leukemia: the European APL group experience. Leukemia 2003,17(8):1600–1604. 10.1038/sj.leu.2403034
Latagliata R, Petti MC, Fenu S, Mancini M, Spiriti MA, Breccia M, Brunetti GA, Avvisati G, Lo Coco F, Mandelli F: Therapy-related myelodysplastic syndrome-acute myelogenous leukemia in patients treated for acute promyelocytic leukemia: an emerging problem. Blood 2002,99(3):822–824. 10.1182/blood.V99.3.822
Montesinos P, Gonzalez JD, Gonzalez J, Rayon C, de Lisa E, Amigo ML, Ossenkoppele GJ, Penarrubia MJ, Perez-Encinas M, Bergua J, Deben G, Sayas MJ, de la Serna J, Ribera JM, Bueno J, Milone G, Rivas C, Brunet S, Lowenberg B, Sanz M: Therapy-related myeloid neoplasms in patients with acute promyelocytic leukemia treated with all-trans-retinoic acid and anthracycline-based chemotherapy. J Clin Oncol 2010,28(24):3872–3879. 10.1200/JCO.2010.29.2268
Steer EJ, Cross NC: Myeloproliferative disorders with translocations of chromosome 5q31–35: role of the platelet-derived growth factor receptor Beta. Acta Haematol 2002,107(2):113–122. 10.1159/000046641
Yoon SY, Tefferi A, Li CY: Cellular distribution of platelet-derived growth factor, transforming growth factor-beta, basic fibroblast growth factor, and their receptors in normal bone marrow. Acta Haematol 2000,104(4):151–157. 10.1159/000046507
Golub TR, Barker GF, Lovett M, Gilliland DG: Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5;12) chromosomal translocation. Cell 1994,77(2):307–316. 10.1016/0092-8674(94)90322-0
Montano-Almendras CP, Essaghir A, Schoemans H, Varis I, Noel LA, Velghe AI, Latinne D, Knoops L, Demoulin JB: ETV6-PDGFRB and FIP1L1-PDGFRA stimulate human hematopoietic progenitor cell proliferation and differentiation into eosinophils: the role of nuclear factor-kappaB. Haematologica 2012,97(7):1064–1072. 10.3324/haematol.2011.047530
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
J-HJ carried out chromosomal analysis, FISH analysis, RT-PCR and sequencing; H-GK wrote the manuscript; E-HK supervised the study and drafted the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Kim, HG., Jang, JH. & Koh, EH. TRIP11-PDGFRB fusion in a patient with a therapy-related myeloid neoplasm with t(5;14)(q33;q32) after treatment for acute promyelocytic leukemia. Mol Cytogenet 7, 103 (2014). https://doi.org/10.1186/s13039-014-0103-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-014-0103-6