Abstract
Background
High-purity RNA serves as the basic requirement for downstream molecular analysis of plant species, especially the differential expression of genes to various biotic and abiotic stimuli. But, the extraction of high-quality RNA is usually difficult from plants rich in polysaccharides and polyphenols, and their presence usually interferes with the downstream applications. The aim of the study is to optimize the extraction of high-quality RNA from diverse plant species/tissues useful for downstream molecular applications.
Results
Extraction of RNA using commercially available RNA extraction kits and routine hexadecyltrimethylammonium bromide (CTAB) methods did not yield good quality DNA-free RNA from Prosopis cineraria, Conocarpus erectus, and Phoenix dactylifera. A reliable protocol for the extraction of high-quality RNA from mature leaves of these difficult-to-extract trees was optimized after screening nine different methods. The DNase I-, and proteinase K treatment-free modified method, consisting of extraction with CTAB method followed by TRIzol, yielded high-quality DNA-free RNA with an A260/A280 and A260/A230 ratios > 2.0. Extraction of RNA from Conocarpus, the most difficult one, was successful by avoiding the heat incubation of ground tissue in a buffer at 65 oC. Pre-warming of the buffer for 5–10 min was sufficient to extract good-quality RNA. RNA integrity number of the extracted RNA samples ranged between 7 and 9.1, and the gel electrophoresis displayed intact bands of 28S and 18S RNA. A cDNA library constructed from the RNA of P. cineraria was used for the downstream applications. Real-time qPCR analysis using the cDNA from P. cineraria RNA confirmed the quality. The extraction of good quality RNA from samples of the desert-growing P. cineraria (> 20-years-old) collected in alternate months of the year 2021 (January to December covering winter, spring, autumn, and the very dry and hot summer) proved the efficacy of the protocol. The protocol’s broad applicability was further validated by extracting good-quality RNA from 36 difficult-to-extract plant species, including tissues such as roots, flowers, floral organs, fruits, and seeds.
Conclusions
The modified DNase I and Proteinase K treatment-free protocol enables to extract DNA-free, high-quality, intact RNA from a total of 39 difficult-to-extract plant species belonging to 32 angiosperm families is useful to extract good-quality RNA from dicots and monocots irrespective of tissue types and growing seasons.
Similar content being viewed by others
Background
High-quality DNA-free RNA is mandatory for a number of plant molecular biology applications such as reverse transcription polymerase chain reaction (RT-PCR), real-time fluorescent quantitation polymerase chain reaction (qPCR), cDNA library construction, northern blot, RNase protection assay, in situ hybridization, subtractive hybridization, and gene expression analysis. Also, good-quality RNA is the prime requirement for RNA-seq analysis which is highly used for gene expression analysis in different tissues and plants under stress, detection of mutations, alternative splicing, and post-transcriptional modifications.
Plants are very diverse in their biochemical profile, and the extraction of RNA at a quality desired for downstream applications is difficult with plants abundant in polysaccharides, proteins, and secondary metabolites [1, 2]. Polysaccharides co-precipitate with RNA in low ionic strength buffers, which constitute potent enzyme inhibitors that reduce yield and result in poor-quality RNA by significantly affecting the extraction procedure [1, 2]. Polyphenols bind irreversibly to RNA and proteins to form high-molecular-weight complexes [1, 2]. Both adversely affect downstream operations like gene expression [3] and RNA-seq analysis [4].
Plant growth stage (immature and mature), physiological status of the samples, age of tissue (i.e., young freshly expanding vs. mature fully expanded but non-senescing tissue), type of tissue (leaf, root, stem, flower, fruit tissues and seeds), growing season especially under abiotic conditions, biochemical profile of the plants, type of plants (herbs, shrubs, trees) and conditions of the material (dry, rigid fibrous material) are the factors affecting the extraction of high-quality RNA [1, 5, 6]. Immature tissues, preferably the seedling leaves, are mostly selected to extract high-quality RNA as they are low in polysaccharides and polyphenols [4, 5, 7, 8]. Extraction of high-quality RNA from plants growing in different environments such as deserts, mountains, seawater, or experimental stress (abiotic and biotic) conditions seems complicated due to high levels of inhibitory/RNA binding metabolites [5, 9, 10]. In the case of plants, trees, in particular having short periods of the growing season, i.e., the emergence of new foliage, mature leaves, or tissues, are the sources for extraction of RNA for downstream applications. The extraction of RNA, even from different species of the same genus, requires optimization for the protocol to yield high-quality, intact RNA. Various methods have been developed for RNA extraction from different plant tissues [5, 6, 11, 12]. RNA extraction protocols across planta use guanidinium thiocyanate, CTAB, or sodium dodecyl sulfate (SDS) based methods [5, 6, 13, 14]. However, the methods must be optimized for specific species and even for the nature of the target tissue [4]. Successful extraction of RNA using CTAB, TRIzol, and TRIzol Plus RNA Purification kits has been reported [4,5,6,7, 12, 15]. The kit formats are highly useful for extracting RNA from the model plants [16]. CTAB-based extraction and its modifications have been reported as successful for high-quality RNA from recalcitrant plants [4,5,6]. Further, special precautions are required for RNA isolation as it has a very short half-life once extracted from cells or tissues and is susceptible to degradation [13]. Our attempt to extract a fair amount of good quality DNA-free RNA for downstream applications of Prosopis cineraria, Conocarpus erectus, Phoenix dactylifera was unsuccessful using different established protocols.
Prosopis cineraria (L.) Druce, a member of the legume family (Fabaceae), is one of the prime choices for desert afforestation programs due to their high adaptability to thrive in extreme environmental conditions of deserts (https://sdgs.un.org/partnerships/give-ghaf-tree-planting-program#:~:text=With%20over%2075%2C000%20seeds%20planted,air%2 C%20for%20generations%20to%20come). Prosopis spp. are rich in tannins and polysaccharides [17], and these compounds are considered problematic to yield good quality RNA suitable for downstream applications. Total RNA extraction from seedling tissues of Prosopis spp. has been documented by CTAB methods [18, 19] and commercial kits [20].
Conocarpus erectus L., commonly known as button mangrove, belongs to the Combretaceae is an associate mangrove shrub growing on shorelines in tropical and subtropical regions around the world. The plant is a rich source of polyphenols and is reported to contain more than 12 phenolic compounds, mainly flavonoids and tannins [21, 22]. There is no report of total RNA extraction from this polysaccharide and phenolic-rich plant species to date.
Phoenix dactylifera L. (Arecaceae), the Date palm, is a dioecious perennial monocot with several varieties and is the most important fruit-bearing crop in arid regions of the Middle East [23, 24]. Palms are rich in polysaccharides and polyphenolics and have leaves with a waxy cuticle and high fiber content [25], making extracting total RNA from the leaves very difficult. The date palm is reported to contain phenolic and their positional isomers, which usually interfere with the successful isolation of nucleic acids [26]. Total RNA extraction was achieved mainly from seedling tissues of Date palm [27,28,29].
To extract quality RNA from these plants for our downstream applications, we have tried different RNA extraction protocols such as Qiagen’s RNeasy Plant Mini kit, PureLink RNA Mini kit (Invitrogen, USA), Spectrum Plant Total RNA kit (Sigma, USA), Plant/Fungi DNA/RNA isolation kit (Norgen Biotek, USA), TRIzol (Ambion, USA); TRI Reagent (Sigma, USA), CTAB-based [30], and hybrid methods. We have developed a modified protocol suitable for extracting high-quality, intact RNA without DNase and proteinase K treatment, which was validated by cDNA synthesis followed by gene cloning, cDNA library preparation, and RT-qPCR analysis. Further, the applicability of the optimized RNA extraction protocol was evaluated by extracting high-quality RNA from 36 other difficult-to-extract non-model plant species consisting of different tissues such as seed, fruit (mature and immature), male and female flowers, floral parts (anther, pollen) and roots and tissues with respect to the season.
Materials and methods
Plant samples
Mature leaves of Prosopis cineraria (Fabaceae), Conocarpus erectus (Combretaceae), and Phoenix dactylifera cv. Fard (Arecaceae) were used to optimize a high-quality RNA extraction protocol. The leaf samples of P. cineraria were collected in August (42–47 oC temp.) 2020 from > 20-year-old trees growing in the desert of Sweihan area (24°17’18.3"N 55°43’36.2"E), Al Ain, UAE. The tissue samples for the extraction of the RNA with respect to the season from P. cineraria were collected in alternate months (January to December) of the year 2021. The leaf samples of C. erectus and Date palm were collected from the plants (> 20-years-old) growing nearby our Center. The plant tissues collected were frozen in liquid-Nitrogen (LN2) and stored at −80 °C to extract RNA for downstream studies.
Sample tissue preparation
Stringent measures were taken to eliminate the RNase activity from the beginning of the extraction procedure until the end and thereafter in the handling of the RNA. RNaseZap® RNase Decontamination Solution (Cat. no. AM9780; Ambion, USA) was used to eliminate the RNase activity during the extraction of the RNA. In all protocols, the tissues were ground to a fine powder in a pre-chilled mortar and pestle using liquid nitrogen and used immediately or stored at −80 °C for later use.
Kit protocols
The extraction of total RNA using RNeasy Plant Mini Kit (Cat. no. 69106; Qiagen, Germany), PureLink RNA Mini kit (Cat. no. 12183,018 A; Invitrogen, USA), Spectrum Plant Total RNA kit (Cat. no. STRN50; Sigma, USA), and Plant/Fungi DNA/RNA isolation kit (Cat. no. 26200; Norgen Biotek, USA,) was performed as per the manufacturers’ instructions. An amount of 25–50 mg of finely powdered tissue was used to extract total RNA for the kit protocols.
Trizol Protocol
In the case of TRIzol (Cat. no. 15596018; Ambion, USA) and TRI Reagent (Cat. no. T9424; Sigma, USA), ~ 50–100 mg/ml finely powdered tissue was transferred for the extraction and carried out as follows.
-
1.
Transferred 50–100 mg powdered, frozen tissue into 1 ml TRIzol/Sigma TRI Reagent in a 2 ml microcentrifuge tube. Mixed well by vigorous shaking and incubated for 5 min at room temperature (RT, 22–23 °C).
-
2.
Centrifuged the tubes at 14,000 rpm for 10 min at RT.
-
3.
The aqueous phase was transferred into a 1.5 ml microcentrifuge tube and added 0.2 ml chloroform. Mixed well by vigorous shaking, incubated for 5 min at RT, and centrifuged for 10 min at 14,000 rpm.
-
4.
Transferred the aqueous phase into a 1.5 ml microcentrifuge tube and added 0.2 ml absolute ethanol (Added 1 vol. of isopropanol for Sigma TRI Reagent). Mixed well by inversion.
-
5.
Decanted the supernatant after centrifuging at 15,000 rpm for 20 min. The pellet was washed with 500 µl of 70% (v/v) ethanol for 3–5 min. Carefully decanted or pipetted out the supernatant and repeated the step.
-
6.
The air-dried pellet was dissolved in 50 µl of RNase-free water.
TRIzol-column hybrid protocol
In this method, TRIzol (Ambion) was used as it was found superior to Sigma TRI Reagent.
Followed the TRIzol protocol up to step 4.
-
5.
Transferred the solution onto an RNeasy (pink) column of the Qiagen kit (or the column of PureLink RNA isolation kit/Spectrum RNA isolation kit).
-
6.
Centrifuged 1 min at 14,000 rpm and discarded the flow-through.
-
7.
Added 500 µl RPE buffer (Qiagen). Centrifuged 30–60 s at 12,000 rpm. Discarded the flow-through. Repeated the step.
-
8.
Centrifuged the column at 12,000 rpm for 1 min.
-
9.
Transferred the column to a new RNase-free 1.5 ml microcentrifuge tube. Added 50 µl of RNase-free water into the center of the column, incubated for 3–5 min at RT, and centrifuged for 1 min at 12,000 rpm.
CTAB protocol
The hexadecyltrimethylammonium bromide (CTAB; Cat. no. 52365; Sigma, USA) protocol of Chang et al. [30] was tried to extract RNA. The extraction using CTAB buffer was followed by SSTE buffer [30]. All the solutions were prepared with nuclease-free water. The composition of the CTAB extraction buffer was: 2% (w/v) CTAB, 2% (w/v) polyvinylpyrrolidone K-40 (PVP, Cat. no. PV40; Sigma, USA), 100 mM Tris-HCl (pH 8.0; Trizma base, Cat. no. 1503; Sigma, USA), 25 mM ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA; Cat. no. EA023, Sigma, USA), 2.0 M sodium chloride (NaCl; Cat. no. 1103414; Sigma, USA), 0.5 g/1 spermidine (free acid, S2626-1G, Sigma, USA), 5% (v/v) β-mercaptoethanol (added just before use, Cat. no. 63689; Sigma, USA). SSTE buffer consisted of 1.0 M NaCl, 0.5% (w/v) sodium dodecyl sulfate (SDS, Cat. no. L3771; Sigma, USA), and 10 mM Tris-HC1 (pH 8.0).
SDS-LiCl method
The protocol reported as universal by Vennapusa et al. [5] was attempted to extract the RNA precisely as described.
Modified protocol of the present study
The optimized protocol of the present study was a modification of Chang et al. [30]. It consisted of an extraction using the CTAB buffer [30] and was followed by TRIzol (Ambion), which is described below step by step.
-
1.
Prepared 5 ml CTAB extraction buffer by adding 5% (v/v) β-ME in a 15 ml Falcon tube and incubated at 65 °C for 5–10 min in a pre-heated heat block.
-
2.
Transferred the fine tissue powder (200–500 mg of ground tissue) into 5 ml extraction buffer, mixed well by vortexing (30 s), and shook thoroughly. Incubated for at least 5 min at RT on a Nutating mixer (or similar kind) and centrifuged at 12,000 rpm for 10 min.
-
3.
Transferred the aqueous phase to a new 15 ml tube and added an equal volume of CIA (24:1). Mixed by shaking the tube thoroughly, incubated at least 5 min at RT on a Nutating mixer and centrifuge at 12,000 rpm for 10 min.
-
4.
Repeated step 3.
-
5.
Transferred the aqueous phase to a new tube and added 1/4 vol. (for 4 ml, added 1 ml) 10 M LiCl, mixed by inversion, and precipitated overnight at 4 oC (or 4–6 h at -20 oC).
-
6.
Centrifuged at 15,000 rpm (> 20,000 xg) for 20–30 min at 4 oC, decanted and drained off all the supernatant by keeping the tube upside down on a paper towel or pipetted out.
-
7.
Dissolved the pellet in 1 ml TRIzol (Ambion), vortexed, and shook vigorously. Incubated for 3–5 min at RT.
-
8.
Centrifuged the tubes at 15,000 rpm for 10 min and transferred the aqueous phase into a 1.5 ml microcentrifuge tube.
-
9.
Added 0.2 ml of chloroform, mixed well by vortexing and vigorous shaking. Incubated for 3–5 min at RT and centrifuged for 10 min at 15,000 rpm (> 20,000 xg).
-
10.
Transferred the aqueous phase into a 1.5 ml microcentrifuge tube and added an equal volume of CIA or chloroform. Mixed well by vigorous shaking, incubated for at least 5 min on a Nutating mixer at RT, and centrifuged for 10 min at 14,000 rpm.
Optional: A repeat of step 10 with CIA was good for the complete elimination of proteins, especially in the case of challenging tissue types such as fruit, tuber, etc.
-
11.
Transferred the aqueous phase into a 1.5 ml microcentrifuge tube and added 0.5 vol. of ethanol (if isopropanol, add 1 vol. of isopropanol). Mixed well by inversion.
-
12.
Centrifuged at 15,000 rpm for 10 min. Carefully decanted or pipetted out the supernatant.
-
13.
Washed the pellet with 500 µl of 70% (v/v) ethanol for 3–5 min. Centrifuged at 15,000 rpm for 1–2 min. Carefully decanted or pipetted out the supernatant. Repeated the step and air-dried the pellet.
-
14.
Added 50 µl of RNase-free water and dissolved the pellet completely.
RNA quality and quantity assessment
The quantity and purity of the extracted RNA samples were determined by measuring 2 µl in a Nanodrop spectrophotometer (Nanodrop 2000, Thermo Scientific). The quality of the RNA samples was assessed by loading 1 µg total RNA on a Hydragreen (non-toxic nucleic acid stain, ACT gene, NJ, USA) added 1% (w/v) agarose (Cat. no. 8010.00; Conda Lab, Spain) gel, and were documented using GelDoc™ EZ Gel Documentation System (Bio-Rad, USA).
RNA integrity determination
RNA integrity Number (RIN) values were determined based on the entire electrophoretic RNA sample trace on an Agilent 2100 Bioanalyzer (Expert B.02.10.SI764). One µl of extracted total RNA was loaded to determine the integrity and concentration in the Bioanalyzer with the Plant RNA Pico chip assay following the manufacturer’s instructions (Agilent Technologies, Santa Clara, CA, USA). The Bioanalyzer uses the electrophoretic technology on a chip to separate RNA fragments by size, read by laser-induced fluorescence, is translated into electropherograms (bands and peaks), and displays the distribution and relative amounts of differently sized RNAs. The measuring of the RNA integrity was carried out as reported earlier [4, 31], based on two metrics: the ratio of the large (28S) to small (18S) ribosomal RNA subunits (28S/18S) and the RIN.
Downstream analysis of the extracted RNA.
a) cDNA synthesis, amplification of genes using PCR, and gene cloning.
The RNA sample isolated from P. cineraria were reverse transcribed using Invitrogen SuperScript III Reverse Transcriptase using oligodT(20) according to the manufacturer’s instructions (Cat. no. 18080093; ThermoFisher Scientific, USA). PCR using the synthesized cDNA with gene-specific primers for HSF6a and HSF7b (Table 1) was carried out in a 50 µl reaction mixture using the Phusion High-Fidelity Taq polymerase kit (Cat. no. M0530L; NEB, USA) following the manufacturer’s protocol adding 1–2 µL cDNA. The PCR program was as follows: initial denaturation at 98 °C for 30 s followed by identical 30 cycles of 10 s denaturing at 98 °C, 20 s annealing at 55 °C, and 40 s extension and final extension of 10 min at 72 °C. The PCR products were resolved on 1% (w/v) agarose gel and were documented using the GelDoc. The amplified fragments extracted from the gel using NucleoSpin gel and PCR Clean-up (Cat. no. 740609.50 S; Macherey-Nagel, Germany) were cloned in a pCR-Blunt vector (ZeroBlunt PCR cloning kit, Cat. no. K2700-20; Invitrogen, USA) and sequenced (Macrogen, Korea). The sequences were confirmed by BLAST (NCBI) similarity search.
Real-Time-qPCR
Gene expression analysis by synthesizing the first strand cDNA from 1 µg RNA isolated from P. cineraria was carried out using QuantiTect Reverse Transcription kit (Cat. no. 205311; Qiagen, Germany) following manufacturer’s instructions in a 20 µl reaction mixture and the cDNA synthesized was stored at −20 oC until use. RT-PCR analyses were carried out in reactions containing 2 µl of diluted (1:4) cDNA, 200 nM primers, and 5 µl of 2X Fast SYBR Green PCR master mix (Cat. no. 4385614; Applied Biosystems, USA) in a 10 µl total volume using a StepOnePlus™ Real-Time PCR system (Applied Biosystems, USA). The HSF primers (Table 1) were used for the relative expression of the HSFs. The qRT-PCR reactions were performed following the fast-thermal cycles: 50 oC for 2 min, 95 oC for 20 s, followed by 40 cycles of 95 oC for 3 s and 60 oC for 30 s. The actin gene (Table 1) was used as the reference gene. The 2−∆CT method [32] was used to calculate the relative expression level.
cDNA library preparation
The cDNA library was prepared using the CloneMiner II cDNA library construction kit (A1180; Invitrogen, USA) as per the manufacturer’s instructions. The cDNA samples following the kit procedure were electroporated into ElectroMAXTM DH10BTM competent cells using Electroporator (Bio-Rad, USA), and the entry clones derived from BP recombination with pDONRTM 222 (Invitrogen, USA) were sequenced using M13(–20)-F and M13-R primers (Table 1).
Application of the optimized protocol to different plant species and tissues
The applicability of the modified protocol was evaluated by extracting RNA from different plant species and tissues of various angiosperm (monocot and dicot) families, and the details are provided in Additional file 1. The different plant species and tissues collected from various locations stored at -80 oC were powdered into fine powder using LN2. RNA was extracted from the powdered tissue using the optimized (CTAB-TRIzol) protocol described.
Statistics
All the RNA extraction procedures of P. cineraria, C. erectus and P. dactylifera were repeated five times, and three replicates of other plants/tissues. Data represent the mean ± SE and were evaluated at a 5% level of significance.
Results and discussion
Successful extraction of high-quality RNA with no degradation and traces of DNA, proteins, polyphenols, and polysaccharides accelerates fundamental and applied research on plant biology. Good quality RNA is essential for diverse downstream molecular studies such as gene expression profiling, subtractive hybridization, construction of cDNA libraries, and RNA-Seq analysis. Optimization of RNA extraction protocol specific to plant species or even to different tissues at high purity is mandatory for trustworthy end results in downstream molecular applications. Polyphenolic components during the extraction of RNA undergo oxidization readily and bind to RNA and co-precipitate, which further cause RNA degradation and become problematic in downstream studies [33].
Lysis of cells, the primary step of RNA extraction, releases large quantities of contaminants that usually hamper RNA extraction and/or result in inhibition in the downstream molecular studies. The RNA extraction buffer plays a crucial role in the rapid denaturation of nucleases and stabilization of RNA to obtain good-quality RNA [4, 5]. Commercially available kits and reagents for the extraction of RNA are simple, rapid, non-toxic, and usually yield high-quality RNA from plant tissues, especially of the model plants: Arabidopsis, Brachypodium, Setaria, and tobacco [11, 34,35,36]. Nevertheless, the use of these kits or reagents is inefficient in yielding high-quality RNA in sufficient amounts in the case of plants that are rich in polyphenols and polysaccharides [5, 33]. In the present study, the proprietary kits of Qiagen, Invitrogen, and Norgen, and the widely used RNA extraction reagent, TRIzol (Ambion)/TRI Reagent (Sigma), did not yield good quality RNA from the mature leaves of the plant species of the present study (Table 2). In all the cases, the A260/A280 and A260/A230 ratios were below 1.8 due to the presence of proteins and polysaccharides. In the present study, though TRIzol (Ambion) was better than TRI Reagent (Sigma), the quality was low (Table 2). The unsuitability of Trizol has also been accomplished in different plant tissues [4, 37]. Trizol reagent, though its success varies with respect to the manufacturers, has been widely used for rapid RNA extraction in several plant species [4, 15, 38]. However, Trizol reagents have proved unsuitable for extracting RNA from plant tissues rich in polysaccharides and polyphenolics [39]. In the TRIzol method, the use of RNA binding columns of commercial kits after ethanol precipitation enabled easy handling and early completion of the protocol, but the amount and quality were low (Table 2). The commercial kits and the TRIzol method did not efficiently separate RNA from proteins and other organic compounds of the plant species of the present study with high levels of polysaccharide and polyphenol (Table 2). This has been pointed out as one of the reasons for the degradation of RNA extracted [40]. In the case of TRIzol, though the quality of the RNA was low, the RNA samples were free of DNA; the centrifugation of the tissue in TRIzol, i.e., before the addition of chloroform, was the reason for the DNA-free status.
CTAB-based protocols were widely used to extract RNA from plant tissues, and success has been accomplished in several plant species, especially where the kit formats were unsuccessful [4, 5, 14]. However, the yield and quality varied with respect to the plant tissues [4]. In oil palm, the total RNA extracted from suspension cells and embryogenic callus using the CTAB protocol was useful for subsequent RT-PCR and real-time RT-PCR [41]. But the method was reported as unsuitable for isolating RNA from the leaves of coconut palm [37]. In the present study, the A260/230 ratios of the extracted RNA using the modified CTAB method of Chang et al. [30], which was reported as efficient for several recalcitrant plant species was inferior and always displayed DNA contamination (Table 2), which required additional procedures to have DNA-free RNA. Nevertheless, the ratio of the CTAB method was superior to that of the kit formats and TRIzol (Table 2). Several documented results emphasize that CTAB-based protocols as the most efficient technique to extract RNA from forest trees [42] and various species [4]. The protocol of Vennapusa et al. [5], claimed as universal, showed clear DNA bands in their gel images and the RNA extracted with their protocol in the present study. DNase I treatment necessitates additional procedures, which usually result in insufficient RNA for downstream applications [8, 43, 44]. The presence of impurities, such as salts or proteins, can affect the movement of the RNA in agarose gel. During electrophoresis, the RNA extracted from protocols other than the optimized one, RNA loaded wells of the gel upon documentation displayed fluorescence, which is usually due to the presence of impurities (polysaccharides/proteins).
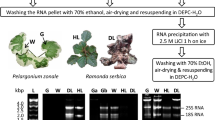
CTAB protocol of Chang et al. [30] followed by TRIzol extraction of the present study, facilitated the isolation of high quantities of DNA-free good quality RNA from the selected woody recalcitrant species (Table 2). This combined method improved the quality of the RNA significantly compared to that of the CTAB and TRIzol methods individually (Table 2). The protocol yielded a high amount of DNA-free RNA with A260/A280 and A260/A230 ratios > 2.0 (Table 2; Fig. 1A-E). The ratios indicated high purity and the polyphenol and polysaccharide contamination-free status of the RNA samples. In the present study, the extraction of the precipitated RNA of the CTAB method followed by TRIzol efficiently eliminated the DNA and protein contamination without the treatment of DNase I and Proteinase K (Fig. 1A-E). Unlike the other methods described above, no fluorescence was observed in the wells of the gels loaded with RNA extracted through the optimized protocol (Fig. 2).
Gel electrophoresis of isolated RNA from different plant species. (1) Mature leaves of Prosopis cineraria; (2) Mature leaves of Phoenix dactylifera; (3) Mature leaves of Conocarpus erectus; (4) Pollen of P. dactylifera; (5) Roots of P. dactylifera; (6) Male flower of P. dactylifera; (7) Mature fruits of Prunus salicina; (8) Mature leaves of Litchi sinensis; (9) Immature stem of Leptadenia pyrotechnica; (10) Mature leaves of Memecylon umbellatum; 11. Endosperm of Cocos nucifera. (M − 1 kb plus DNA ladder)
The centrifugation of the LiCl-precipitated RNA dissolved in TRIzol was a crucial step in removing the DNA. The second chloroform, or CIA extraction after the extraction with TRIzol, was necessary to remove the contaminants completely. Proteinase K often separates proteins from nucleic acids and inhibits ribonucleases [44]. DNase and Proteinase K use usually demand extended treatment with phenol:chloroform: isoamyl alcohol, as has been reported in several plants [8, 43,44,45].
Most protocols, including the kit formats, suggested heating the samples in the extraction buffer at 65 oC for up to 30 min. In our study, heat incubation for 30 min did not yield good-quality RNA from Conocarpus erectus. In C. erectus, the pre-warming of the buffer at 65 oC for 5–10 min yielded good-quality of RNA (Table 2). In the case of other plant species, the pre-warming of the buffer was sufficient, and the heat incubation did not result significant change in the quality and quantity of the RNA extracted (Data not shown). C. erectus is reported as a source of polyphenols and reported to contain more than 12 phenolic compounds, mainly flavonoids and tannins [21, 22]. During extraction with other methods, in the case of C. erectus, the pellet became brown, possibly due to the biochemical changes by heat activation of some of the compounds in tissues and their binding to the nucleic acids.
During the RNA extraction, the RNase activity should be inhibited; if not, it will result in the smearing of RNA. The present protocol used 5% β-ME and 0.5 g/l spermidine in the CTAB extraction buffer. β-ME, a potent reducing agent, plays a significant role in the RNA extraction protocols that irreversibly denature RNases by reducing disulfide bonds and destroying the native conformation required for enzyme functionality and also other contaminating proteins released during tissue disruption and homogenization [40, 46, 47]. The use of 1–2% β-ME was reported as effective in most protocols [4] but relied on even the tissue types [37]. Ouyang et al. [40] documented the smearing of RNA from the flower, fruit, leaf, and bud tissues of Neolamarckia cadamba, even at 5% β-ME. Siles et al. [43] reported the efficacy of β-ME in avoiding the proteinase K treatment. However, in the present study, 5% β-ME was efficient in the case of C. erectus, and we used 5% in all other species. Spermidine plays a significant role as an RNase inhibitor [40, 46, 48]. RNA degradation depends not only on the presence of RNase but also on poly(A) length and spermidine concentration [47]. The protocol of CTAB-based RNA extraction buffer for pine needles was the first extraction buffer containing 0.5 g/l of spermidine [30]. In this case, C. erectus 1 g/l spermidine reduced the smearing of RNA compared to that with 0.5 g/l. Inhibition of smearing using 1 g/i of spermidine has also been reported in Jatropha curcas [49]. The efficacy of a higher concentration of spermidine has been reported in Maqui berry [50] and N. cadamba [40].
In the present study, the integrity of the extracted RNA was assessed by the visualization of ribosomal RNA bands on 1% agarose gel and Agilent 2100 Bioanalyzer microfluidic electrophoresis chip. A sharp and distinct cytosolic and plastid ribosomal bands, i.e., DNA-free, intact 28S (4.5 kb) and 18S (1.9 kb), with a double intensity of 28S rRNA band to 18S rRNA band on the agarose gel were considered as good quality RNA (Fig. 2). The extracted RNA using the optimized protocol of the present study was of good quality with the above standards (Fig. 2). In the cases of kits and TRIzol reagents extracted RNAs, the 28S and 18S rRNA bands were not intact and exhibited degradation, usually due to polysaccharides and polyphenols.
The RIN values of the RNA samples of P. cineraria and P. dactylifera using the modified protocol ranged from 7.6 to 9.1, while that of C. erectus ranged from 7.1 to 8.6. In the present study, the results of the Nanodrop values and gel electrophoresis were corroborated with the RIN analysis of the RNA samples. RNA isolated with the optimized protocol displayed the highest peak corresponding to the 28S rRNA and 28S:18S ratio of 1.8. RIN values of 7 to 9.1 indicated the integrity of RNAs (Fig. 3C-F). RNA with a RIN value above 7.0 is required to produce good results in next-generation sequencing analysis. The RNA samples of the other protocols showed additional peaks between the 18S and 5S bands, which may be due to the degradation of 28S or 18S rRNA (Fig. 3A,B). The gel electrophoresis and the RIN analysis confirmed the high quality of RNA from all the species, which authenticated the modified protocol.
Extraction of RNA samples with respect to the seasons from the desert-growing tree
The metabolic profile of the trees varies in the growing season, and it is one of the determinant factors of the quality of the RNA. We checked the efficacy of the optimized protocol to extract good-quality RNA from the desert tree, P. cineraria covering all seasons (Table 3). In the case of trees growing under stress conditions further enhance the difficulty to obtain good quality RNA due to the high metabolic profile to negate or minimize the adverse impact on cellular functions. The protocol resulted in good quality RNA from leaves > 20-year-old desert tree P. cineraria collected in alternate months starting from January of the year 2021, which covered the different seasons, especially the very hot and dry summer with a temperature around 50 oC. The extraction of the RNA from leaf samples collected in August and October necessitated two CIA extractions following the TRIzol extraction of the optimized method.
Application of the protocol to different plant species and tissues
The RNA yield and quality rely on the type and age of tissues. The young leaves, especially the seedlings, are the prime choice for extracting intact RNA free from the impurities inhibitory to molecular applications [4]. The RNA of the present studies was extracted from mature leaves. The mature tissues, usually with high polyphenols and polysaccharides, hinder the extraction of pure and intact RNA, and it is difficult to obtain the sequence quality [4]. According to Torales et al. [19], tissue age (i.e., young freshly expanding vs. mature fully expanded but non-senescing tissue) had only a weak effect on RNA quality and no apparent effect on sequencing results in Prosopis alba. In their study, the younger leaves gave higher yields of RNA, and the impact of tissue age on RNA quality was observed in RIN values.
Extraction of RNA from leaves of various plant species, fruit (mature plum), tuber (Curculigo orchioides), rice seeds, the endosperm of coconut, and different tissues of Date palm such as root, flowers, floral parts (anther and pollen), fruits (immature and mature), and seeds were attempted with the optimized protocol (Additional file 1). The quality of the RNA using the optimized protocol was good and is useful to proceed with the downstream applications (Additional file 1). The extraction of high quality RNA from different plants and tissues, which are considered as difficult-to-extract and also from plants exposed to different seasons authenticate the protocol’s extendibility to several plants, rich in polyphenols and polysaccharides.
Validation by downstream applications
The outcome of purified RNA on downstream applications such as real-time RT-PCR, microarray analysis, next-generation RNA sequencing (RNA-Seq), northern blotting, and cloning authenticate a protocol. The RNA extracted from stress-tolerant P. cineraria was used for downstream applications such as cDNA synthesis, gene cloning, cDNA library construction, and RT-qPCR and transcriptome profiling validated the modified RNA extraction protocol.
The quality of the RNA was evaluated by synthesizing cDNA from the extracted RNA of P. cineraria. Heat shock factors (HSF6a and HSF7b) using gene-specific primers (Table 1) were amplified from the cDNA synthesized (Fig. 4). The gel-purified amplicons were cloned, and the sequences were confirmed by sequencing (Additional File 2) followed by a BLAST (NCBI) similarity search. The cDNA library was constructed using the P. cineraria RNA, and the clones confirmed by PCR using the M13F and M13R (Fig. 5) were sequenced, and several genes imparting abiotic tolerance were identified and cloned for crop modification (unpublished).
High-quality, intact RNA in sufficient amounts is the key for gene expression studies to understand the biological processes, especially the upregulation and downregulation of genes with respect to specific tissues and also under environmental stimuli. The cDNA synthesized using the QuantiTect Transcription kit followed by Real-Time qPCR used for the expression analysis of the heat shock factors (HSF6a and HSF7b) with Actin as reference gene showed a significant difference in expression level (Fig. 6), and this validated the modified RNA extraction protocol of the present study.
The samples of RNA extracted through the modified protocol from P. cineraria, a total of 54 samples of three desert trees (> 20-years-old) collected in every alternate month (triplicates) from January through December of the year 2021 (Table 3) qualified the sequence quality parameters (Novogene) for different NGS platforms, and the analysis is in progress (Data not shown).
Conclusion
No universal protocol is available for isolating total RNA as the plants vary significantly in their structural and biochemical profile. However, the modified protocol yielded high-quality DNA-free intact RNA from various plant species, tissue types, and with respect to season, which was authenticated by bioanalyzer results and successful downstream use. The protocol is useful for extracting DNA-free intact RNA from plants with high phenolics, starch, and polysaccharides.
Data Availability
Available on request to the corresponding authors.
References
Gudenschwager O, González-Agüero M, Defilippi BG. A general method for high-quality RNA isolation from metabolite-rich fruits. South Afr J Bot. 2012;83:186–92.
Huang HH, Xu LL, Tong ZK, Lin EP, et al. De novo characterization of the chinese fir (Cunninghamia lanceolata) transcriptome and analysis of candidate genes involved in cellulose and lignin biosynthesis. BMC Genomics. 2012;13:648. https://doi.org/10.1186/1471-2164-13-648.
Birtic S, Kranner I. Isolation of high-quality RNA from polyphenol-, polysaccharide- and lipid-rich seeds. Phytochem Anal. 2006;17(3):144–8. https://doi.org/10.1002/Pca.903.
Johnson MTJ, Carpenter EJ, Tian Z, Bruskiewich R, et al. Evaluating methods for isolating total RNA and predicting the success of sequencing phylogenetically diverse plant transcriptomes. PLoS ONE. 2012;7(11):e50226. https://doi.org/10.1371/journal.pone.0050226.
Vennapusa AR, Somayanda1 CJ, Jagadish SVK. A universal method for highquality RNA extraction from plant tissues rich in starch, proteins and fiber. Scientific Rep. 2020;10:16887. https://doi.org/10.1038/s41598-020-73958-5.
Ahmed M, Sarwar MB, Ashfaq R, Ahmed A, et al. Low-cost and highly efficient: a method for high-quality nucleic acid isolation from cotton fibres. BioRxiv preprint. 2022. https://doi.org/10.1101/2022.10.07.511236.
Nath O, Fletcher SJ, Hayward A, Shaw LM, et al. A comprehensive high-quality DNA and RNA extraction protocol for a range of cultivars and tissue types of the woody crop Avocado. Plants. 2022;11(3). https://doi.org/10.3390/plants11030242.
Hadi M, Stacy EA. An optimized RNA extraction method for diverse leaves of hawaiian Metrosideros, a hypervariable tree species complex. Appl Plant Sci. 2023;11(3):e11518. https://doi.org/10.1002/aps3.11518.
Yang G, Zhou R, Tang T, Shi S. Simple and efficient isolation of high-quality total RNA from Hibiscus tiliaceus, a mangrove associate and its relatives. Prep Biochem Biotechnol. 2008;38:257–64. https://doi.org/10.1080/10826060802164991.
Jensen T, Saleh L, Bents D, et al. Optimised protocols for RNA extraction from a broad taxonomic range of algae. J Appl Phycol. 2023. https://doi.org/10.1007/s10811-023-02980-7.
Zhu C, Yang J, Box MS, Kellogg EA, Eveland AL. A dynamic co-expression map of early inflorescence development in Setaria viridis provides a resource for gene discovery and comparative genomics. Front Plant Sci. 2018;9. https://doi.org/10.3389/fpls.2018.01309.
Yan WJ, Pendi FH, Hussain H. Improved CTAB method for RNA extraction of thick waxy leaf tissues from sago palm (Metroxylon sagu Rottb). Chem Biol Technol Agric. 2022;9:63. https://doi.org/10.1186/s40538-022-00329-9.
Tan SC, Yiap BC. DNA, RNA, and protein extraction: the past and the present. J Biomed Biotechnol. 2009;574398. https://doi.org/10.1155/2009/574398.
Rajakani R, Narnoliya L, Sangwan NS, Sangwa RS, Gupta V. Activated charcoal-mediated RNA extraction method for Azadirachta indica and plants highly rich in polyphenolics, polysaccharides and other complex secondary compounds. BMC Res Notes. 2013;6:125. http://www.biomedcentral.com/1756-0500/6/125.
Carpinetti PdA, Fioresi VS, da Cruz IT, de Almeida FAN, et al. Efficient method for isolation of high-quality RNA from Psidium guajava L. tissues. PLoS ONE. 2021;16(7):e0255245. https://doi.org/10.1371/journal.pone.0255245.
Lee S, Moon JS, Ko T, Petros D, Goldsbrough PB, Korban SS. Overexpression of Arabidopsis phytochelatin synthase paradoxically leads to hypersensitivity to cadmium stress. Plant Physiol. 2003;131(2):656–63.
Nakano H, Nakajima E, Hiradate S, Fujii Y, Yamada K, Shigemori H, Hasegawa K. Growth inhibitory alkaloids from mesquite (Prosopis juliflora (Sw.) DC.) Leaves. Phytochem. 2004;65(5):587–91.
George S, Venkataraman G, Parida A. Identification of stress-induced genes from the drought-tolerant plant Prosopis juliflora (Swartz) DC. Through analysis of expressed sequence tags. Genome. 2007;50(5):470–8.
Torales SL, Rivarola M, Pomponio MF, et al. De novo assembly and characterization of leaf transcriptome for the development of functional molecular markers of the extremophile multipurpose tree species Prosopis alba. BMC Genomics. 2013;14:705. https://doi.org/10.1186/1471-2164-14-705.
Padaria JC, Tarafdar A, Yadav R. Development of a heat-responsive cDNA library from Prosopis cineraria and molecular characterisation of the Pchsp17.9 gene. J Horticult Sci Biotechnol. 2015;90(3):318–24.
Ayoub NA. A trimethoxyellagic acid glucuronide from Conocarpus erectus leaves: isolation, characterization and assay of antioxidant capacity. Pharm Biol. 2010;48(3):328–32. https://doi.org/10.3109/13880200903131567.
Hameed ESA, Bazaid SA, Shohayeb MM, El-Sayed MM, El-Wakil EA. Phytochemical studies and evaluation of antioxidant, anticancer and antimicrobial properties of Conocarpus erectus L. growing in Taif, Saudi Arabia. Eur J Med Plants. 2012;2:93–112.
Chao CT, Krueger RR. The date palm (Phoenix dactylifera L.): overview of biology, uses, and cultivation. HortScience. 2007; 421077–1082.
Johnson DV, Introduction. Date palm biotechnology from theory to practice. Date Palm Biotechnology. 2011.1–1.
Angeles JGC, Laurena AC, Tecson-Mendoza EM. Extraction of genomic DNA from the lipid-, polysaccharide-, and polyphenol-rich coconut (Cocos nucifera L). Plant Mol Biol Rep. 2005;23:297–8. https://doi.org/10.1007/BF02772760.
Ziouti A, El-Modafar C, Fleuriet A, El-Boustani S, Macheix JJ. Phenolic compounds in date palm cultivars sensitive and resistant to Fusarium oxysporum. Biol Plant. 1996;38:451–7.
Fang Y, Wu H, Zhang T, Yang M, et al. A complete sequence and transcriptomic analyses of date palm (Phoenix dactylifera L.) mitochondrial genome. PLoS ONE. 2012;7(5):e37164. https://doi.org/10.1371/journal.pone.0037164.
Radwan O, Arro J, Keller C, et al. RNA-Seq transcriptome analysis in date palm suggests multi-dimensional responses to salinity stress. Trop Plant Biol. 2015;8:74–86. https://doi.org/10.1007/s12042-015-9155-y.
Yaish MW, Patankar HV, Assaha DVM, et al. Genome-wide expression profiling in leaves and roots of date palm (Phoenix dactylifera L.) exposed to salinity. BMC Genomics. 2017;18:246. https://doi.org/10.1186/s12864-017-3633-6.
Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–6. https://doi.org/10.1007/BF02670468.
Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3.
Livak KJ, Schmittgen TD. Analysis of the relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. https://doi.org/10.1006/meth.2001.1262.
Ma Z, Huang B, Xu S, Chen Y, Li S, Lin S. Isolation of high- quality total RNA from chinese fir (Cunninghamia lanceolata (Lamb.) Hook). PLoS ONE. 2015;10(6):e0130234. https://doi.org/10.1371/journal.pone.0130234.
Hong SY, Seo PJ, Yang MS, Xiang F, Park CM. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 2008;7(8):112. https://doi.org/10.1186/1471-2229-8-112.
Thielen PM, Pendleton AL, Player RA, Bowden KV, Lawton TJ, Wisecaver JH. Reference genome for the highly transformable Setaria viridis ME034V. G3. Genes Genomes Genet. 2020;10(10):3467–78. https://doi.org/10.1534/g3.120.401345.
Zhang Q, Zhao F, Wu Z, et al. A simple and robust method for isolating and analyzing chromatin-bound RNAs in Arabidopsis. Plant Methods. 2022;18:135. https://doi.org/10.1186/s13007-022-00967-y.
Xiao Y, Yang Y, Cao H, Fan H, et al. Efficient isolation of high quality RNA from tropical palms for RNA-seq analysis. POJ. 2012;5(6):584–9.
Wang C, Hou X, Qi N, Li C, et al. An optimized method to obtain high-quality RNA from different tissues in Lilium davidii var. unicolor. Sci Rep. 2022;12. https://doi.org/10.1038/s41598-022-06810-7.
Tattersall EAR, Ergul A, AlKayal F, DeLuc L, Cushman JC, Cramer GR. Comparison of methods for isolating high-quality of RNA from leaves of grapevine. Am J Enol Vitic. 2005;56:400–06.
Ouyang K, Lia J, Huang H, Quea Q, Lic P, Chena X. A simple method for RNA isolation from various tissues of the tree Neolamarckia cadamba. Biotechnol Biotechnol Equip. 2014;28(6):10081013. https://doi.org/10.1080/13102818.2014.981086.
Tranbarger TJ, Kluabmongkol W, Sangsrakru D, Morcillo F, et al. SSR markers in transcript of genes linked to post-transcriptional land transcriptional regulatory functions during vegetative and reproductive development of Elaeis guineensis. BMC Plant Biol. 2012;12:1.
le Provost G, Herrera, Paiva JAP, Chaumeil P, Salin F, Plomion F. A micromethod for high throughput RNA extraction in forest trees. Biol Res. 2007;40:291–7.
Siles L, Eastmond P, Kurup S. Big data from small tissues: extraction of high-quality RNA for RNA-sequencing from different oilseed Brassica seed tissues during seed development. Plant Methods. 2020;16:80. https://doi.org/10.1186/s13007-020-00626-0.
Biskup E, Schejbel L, Oliveira DN, Høgdall E. Test of an improved DNA and RNA purification protocol–importance of proteinase K and co-purified small RNAs. Separations. 2022;9(11):324. https://doi.org/10.3390/separations9110324.
Box MS, Coustham V, Dean C, Mylne JS, Protocol. A simple phenol-based method for 96-well extraction of high quality RNA from Arabidopsis. Plant Methods. 2011;13(7):7. https://doi.org/10.1186/1746-4811-7-7.
Perevoshchikova KA, Prokoph H, Hering B, Koen I, Zbarskii IB. Effect of heparin, spermidine and Be2C ions on the phosphatase and RNase activity of rat liver cell nuclei. Biull Eksp Biol Med. 1979;87:542544.
Karpetsky TP, Hieter PA, Frank JJ, Levy CC. Polyamines, ribonucleases, and the stability of RNA. Mol Cell Biochem. 1977;17:8999.
Gasic K, Hernandez A, Korban SS. RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Rep. 2004;22:437438.
Sangha JS, Gu K, Kaur J, Yin Z. An improved method for RNA isolation and cDNA library construction from immature seeds of Jatropha curcas L. BMC Res Notes. 2010;3:126.
Sánchez C, Villacreses J, Blanc N, et al. High quality RNA extraction from Maqui berry for its application in next-generation sequencing. SpringerPlus. 2016;5:1243. https://doi.org/10.1186/s40064-016-2906-x.
Acknowledgements
Acknowledge the financial support of The Presidential Court, United Arab Emirates. We gratefully acknowledge Mr. Mubarak Jebbar for his help in sample collections.
Funding
The project was funded by The Presidential Court, United Arab Emirates.
Author information
Authors and Affiliations
Contributions
MK designed the experiment. SS and SK optimized the protocol. PK, AAS, MA, and GL validated the protocol with different plant species. SS, SK, MK, PK, AAS, and MA performed the downstream applications. MK, SS, and SK prepared the first draft of the manuscript. KA supervised the project. All the authors reviewed and corrected the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Sasi, S., Krishnan, S., Kodackattumannil, P. et al. DNA-free high-quality RNA extraction from 39 difficult-to-extract plant species (representing seasonal tissues and tissue types) of 32 families, and its validation for downstream molecular applications. Plant Methods 19, 84 (2023). https://doi.org/10.1186/s13007-023-01063-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13007-023-01063-5