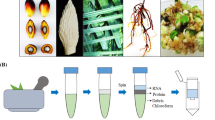
Abstract
Background
There is a growing interest in transcriptomics studies parallel to the advancement of transcriptome databases and bioinformatics, which provided the opportunity to study responses to growths, stimuli and stresses. There is an increase in demand for excellent RNA extraction techniques. General RNA extraction protocols can be used in RNA extraction, but the quality and quantity vary in different types of tissues from different organisms. Hence, a specific RNA extraction method for each organism’s tissue type is required to obtain the desired RNA quality and quantity.
Results
The improved CTAB RNA extraction method is superior to the PCI method and MRIP method for thick waxy leaves that were applied for mature sago palm (Metroxylon sagu Rottb.) leaf tissue and produce total RNA extract with good purity (OD 260/280 ≥ 1.8, OD 260/230 ≥ 2.0) and integrity (RIN ~ 7). RNA sequencing was conducted with the extracted samples and showed good assembly results (Q20 ≥ 97, Q30 ≥ 91%, assembly mean length ≥ 700 bp).
Conclusion
The improved CTAB RNA extraction method enables rapid, cost-effective, and relatively simple RNA extraction from waxy, fibrous and high-in-polyphenol sago palm (M. sagu Rottb.) leaf tissue with next-generation RNA sequencing recommended quality.
Graphical Abstract

Similar content being viewed by others
Introduction
Sago palm (Metroxylon sagu) is one of the starch-producing crops and is the best starch-producing species within the Metroxylon spp. which could be found from southern Thailand to the Solomon Islands [1, 2]. Among the Metroxylon spp. only M. sagu is both hapaxanthic (once-flowering) and soboliferous (produce suckers) [3]. M. sagu lifecycle is divided into four stages which is the rosette stage, bole forming stage, inflorescence stage and fruit ripening stage where the starch accumulation in the trunk starts at the bole formation stages and reaches its peak at the beginning of the inflorescence stage which is approximately nine years after plantation [3]. The advantages of sago palm are its abilitiy to adapt to acidic, saline soil with seasonal flooding such as peat swamps which other crops are unable to grow [4]. There is growing interest in this palm as one of the candidates to combat the world food shortage.
Advances in nucleic acid sequencing and bioinformatics technologies have revolutionized our ability to study plant growth biology and its response to stimuli and stresses [5,6,7,8]. Several methods (and their adoptees) such as Rochester et al. [9, 10], Chomczynski and Sacchi [11, 12], Chang et al. [13, 14], Schultz et al. [15,16,17], Salzman et al. [10, 18], Kiefer et al. [19, 20], Hu et al. [21, 22], Hussain [23,24,25,26,27,28,29], modified Zeng and Yang [10, 30,31,32], Wu et al. [10, 33], modified Gasic et al. [20, 22, 34,35,36,37,38,39,40], Xiao et al. [41] and several kits such as RNAqueous™ Total RNA Isolation Kit [12, 22], FavorPrep™ Plant Total RNA Mini extraction kit [22], and Qiagen RNeasy Plant Mini Kit [10] were used in sago palm leaf RNA extraction, but the actual quality and quantity were not well defined. Notably, modified Zeng and Yang [30] and modified Gasic et al. [34] methods which utilized CTAB as its extraction buffer are better in young sago palm leaf RNA extraction.
The major differences between the improved CTAB method based on Untergasser [42] with modified Zeng and Yang [30] and modified Gasic et al. [34] are the extraction buffer is not pre-heated to 55 °C–65 °C and the precipitation using ice-cooled isopropanol for 10 min instead of overnight precipitation with lithium chloride. This change minimized the damage to the RNA due to high temperature and the extraction complete more rapidly. The effect of other extraction buffers on mature sago palm leaf; which are waxier, more secondary metabolites, and less viable cells; RNA extraction are still unknown, hence a comparative study of RNA extraction on mature sago palm leaf with different extraction buffers and protocols will demonstrate the potential of the CTAB extraction method.
In this paper, we describe a validated method for the extraction of RNA for sago palm (Metroxylon sagu Rottb) leaf tissue for RNA sequencing (RNA-seq) purpose modified from Untergasser [42] RNA Miniprep using the CTAB method [42]. These procedures demonstrated the successful extraction of RNA from sago palm leaf tissue for RNA sequencing. The procedures described here are rapid, cost-effective and relatively simple.
Materials and methods
Sampling
Sago palm leaf samples were wiped with 70% ethanol to remove debris, stored in labeled containers and snap-frozen in liquid nitrogen on-site to preserve the RNA. The samples need to be kept in liquid nitrogen before long-term storing in a − 80 °C freezer. Selection of the leaf age influences the total RNA yield and purity, in which young leaf gives better RNA yield and purity. In this study, mature sago palm leaf was selected in compliance with the motif of the main study.
Total RNA extraction
Three different extraction methods were compared in this study, i.e., improved CTAB method, PCI method [23] and MRIP method [41], to determine the most suitable method for extraction of total RNA from Metroxylon sagu leaf tissue for RNA sequencing (Table 1). The improved CTAB method is a modified method based on Untergasser [42] which focus on the simplicity, rapidity and reproducibility of RNA extraction from mature sago palm leaf sample. PCI method is an RNA extraction method practice on sago palm young leaf sample while MRIP method is a published RNA extraction method for young coconut palm leaf which shows promising results. One gram of mature sago palm leaf samples was grounded finely in prechilled mortar and pestle with liquid nitrogen for all three methods. The ribonucleic acid pellet obtained, which contains both DNA and RNA, from the three protocols were further subjected to DNase treatment. The pellet was resuspended with 0.5 ml of Tris–HCl buffer. Next DNase treatment was conducted with RQ1 RNase-Free DNase (Promega) according to protocol. Then, the mixture was centrifuged at 16,100 g, 4 °C for 15 min. After that, the mixture was transferred into a new 1.5-ml microcentrifuge tube. Then, an equal volume of ice-cooled isopropanol was added, mixed and incubated at room temperature for 10 min. After that, the mixture was centrifuged at 16,100 g, 4 °C for 15 min. Then, the supernatant was discarded and the pellet was washed with 1 ml of 70% ethanol twice. Lastly, the pellet was resuspended with 100 µl Tris–EDTA buffer (10 mM Tris–HCl, 1 mM disodium EDTA, pH 8.0) for further analysis.
Improved CTAB method
This method was modified based on RNA miniprep using the CTAB method [42] (Table 2). One gram of the powdered sago palm leaf sample was transferred into a 50-ml polypropylene centrifuge tube. Then, 15 ml of CTAB buffer with 1% (v/v) β-mercaptoethanol was added, mixed and then incubated for 5 min. After that, 15 ml of chloroform was added into the mixture, mixed and then incubated for another 5 min. The mixture was centrifuged at 20,922 g, 4 °C for 5 min. Next, the top aqueous layer from the tube was transferred into a new polypropylene centrifuge tube. Then 15 ml of chloroform was added into the aqueous layer tube, mixed and then incubated for 5 min. The mixture was centrifuged at 20,922 g, 4 °C for 5 min. Next, the top aqueous layer from tube again was transferred into new 50-ml polypropylene centrifuge tube. Then, equal volume of ice-cooled isopropanol was added, mixed and then incubated in room temperature for 10 min. After that, the mixture was centrifuged at 20,922 g, 4 °C for 15 min. Next, the supernatant from the tube was discarded and the pellet was transferred, with 1 ml of 70% ethanol, into a new 1.5-ml microcentrifuge tube. Then, the tube was centrifuged at 16,100 g, 4 °C for 2 min. The pellet was washed again with 1 ml of 70% ethanol and then discarded. Lastly, the pellet was resuspended with appropriate buffer for further analysis or storage.
PCI method
This method was adapted based on the RNA extraction method performed by Hussain [23]. One gram of the powdered sago palm leaf sample was transferred into a 50-ml polypropylene centrifuge tube. Then, 15 ml ice-cooled extraction buffer [150 mM lithium chloride (LiCl), 50 mM Tris–HCl (pH 9.0), 5 mM ethylene diaminetetraacetic acid (EDTA), and 5% w/v sodium dodecyl sulfate (SDS)] was added, mixed and incubated for 5 min. After that, the mixture was centrifuged at 20,922 g, 4 °C for 5 min. Next, the top aqueous layer from tube was transferred into a new 50-ml polypropylene centrifuge tube. The following PCI treatment was performed trice where 15 ml of ice-cooled PCI was added, mixed and incubated for 5 min. After that, the mixture was centrifuged at 20,922 g, 4 °C for 5 min. Then, the top aqueous layer from the tube was transferred into a new 50-ml polypropylene centrifuge tube. After the PCI treatment, the following chloroform treatment was performed thrice where 15 ml of chloroform were added and mixed for 5 min. Then, the mixture was centrifuged at 20,922 g, 4 °C for 5 min. After that, the top aqueous layer from the tube was transferred into a new 50-ml polypropylene centrifuge tube. After the chloroform treatment, 8 M of lithium chloride was added to make the final concentration of 2 M lithium chloride mixture and was incubated for 24 h at 4 °C. Then, the mixture was centrifuged at 20,922 g, 4 °C for 30 min. After that, the pellet was transferred into a new 1.5-ml microcentrifuge tube and washed with 70% ethanol. The pellet was then dissolved in 400 µl sterile dH2O. Then, 1 ml of absolute ethanol and 40 µl 3 M Sodium acetate was added into the mixture and mixed. After that, the mixture was incubated at − 80 °C for 20 min. Then, the mixture was centrifuged at 16,100 g, 4 °C for 30 min. Next, the supernatant was discarded and the pellet was washed with 70% ethanol twice. Lastly, the pellet was resuspended with appropriate buffer for further analysis or storage.
MRIP method
This method was adapted based on the RNA extraction method performed by Xiao et al. [41]. One gram of the powdered sago palm leaf sample was transferred into a 50-ml polypropylene centrifuge tube. Then, 15 ml MRIP buffer [3.05 g ammonium thiocyanate (% w/v), 9.44 g guanidine thiocyanate (% w/v), 3.33 ml 3 M sodium acetate (pH 5.2) (% v/v) and 38 ml phenol (% v/v), adjust pH to 5.0 with acetic acid] was added, mixed and incubated for 5 min. Then, 5 ml ice-cooled chloroform was added, mixed and further incubated for 5 min. After that, the top aqueous layer from tube was transferred into a new 50-ml polypropylene centrifuge tube. Next, equal volume of ice-cooled isopropanol were added, mixed and incubated for 10 min. Then, the mixture was centrifuged at 20,922 g, 4 °C for 15 min. The supernatant was discarded and the pellet was washed with 70% ethanol twice. Lastly, the pellet was resuspended with appropriate buffer for further analysis or storage.
RNA purity test
RNA purity test was performed using Eppendorf Biophotometer Plus for RNA extraction methods comparison and Nanodrop was used to test the RNA quality for RNA sequencing samples. Samples pellet were dissolved in 100 µl Tris–EDTA and transferred into Eppendorf UVette cuvette for the instrument measurement in Eppendorf Biophotometer Plus while Samples pellet were dissolved in nuclease-free water for nanodrop. The purity of the sample’s RNA extract was measured by the OD260/OD230 and OD260/OD280 ratio where the higher the value of the ratio the purer the sample’s RNA [43].
RNA quality test
RNA quality test was performed using Tris–borate–EDTA buffered Agarose Gel Electrophoresis (TBE-AGE) for RNA extraction methods comparison and Agilent 2100 Bioanalyzer was used to test the RNA quality for RNA sequencing samples. In TBE-AGE, sample pellets were dissolved in 100 µl Tris–HCl. 5 µl of the RNA samples was mixed with 2 µl loading dye. The samples mixtures and GeneRuler 1-kb DNA ladder were loaded into the gel to perform the electrophoresis. In TBE-AGE, good-quality RNA sample represented by intact 28 s and 18 s rRNA band without smearing. In Agilent 2100 Bioanalyzer, sample pellets were dissolved and analyzed. RNA Integrity Number (RIN) is use to determine the quality of the RNA sample scored from 1 (worst) to 10 (best) [44].
Statistical analysis
IBM SPSS Statistics 28.0.1.1 (15) was used to analyse the 260/280 ratio, 260/230 ratio and RNA weight data obtained for RNA extraction method comparison. All the data were assessed using normality test to determine its parametricity. Parametric data were further subjected to multivariate analysis of variance (MANOVA) coupled with Tukey’s test while non-parametric data were subjected to Kruskal–Wallis test.
RNA sequencing
The improved CTAB method was used to extract RNA from sago palm leaf tissue and sent for sequencing. The RNA samples were pelletized in 100% absolute ethanol and shipped for sequencing service. BGISEQ-500 sequencing platform was used in this study. The RNA sequencing data were measured by Q20 and Q30 values. BGI NGS RNA sequencing guideline for good-quality data are Q20 ≥ 90%, Q30 ≥ 80 [45]. The assembly sequences median length (N50) and the mean length provides information on abundance of the sample RNA length, the closer the N50 value to the mean length the more normalized the distribution of the sample RNA length.
Results and discussion
RNA extraction methods comparison
The improved CTAB method was compared with the PCI method [23] and MRIP methods [41] and showed the best result in RNA quality and quantity (Table 3 and Fig. 1). Due to low RNA yield of MRIP method, there are no visible bands in TBE-AGE. On the other hand, there are presence of visible smear at the 5 s rRNA region, but absent at 28 s and 18 s rRNA band in TBE-AGE of PCI method indicating the considerable degradation of the RNA. CTAB method gives the best RNA quality as shown by the intact 28 s and 18 s rRNA band with slightly visible smear at the 5 s rRNA region in TBE-AGE. This outcome shows that the PCI method is not suitable for extracting RNA from mature sago palm leaf tissue. The lower sample weight used in this test improved the RNA yield and purity of the extracted RNA compared to the reported result. This outcome also shows that MRIP method is not suitable for RNA extraction from mature sago palm leaf tissue although it shows promising result when applied on RNA extraction from young coconut leaf. The purity and the RNA yield are very much lower compared to the reported result on application of MRIP method on young coconut leaf.
TBE-AGE assay of sago palm leaf tissue RNA extract between improved CTAB method, PCI method and MRIP methods. Line M = GeneRuler 1 kb DNA Ladder, lane 1 = improved CTAB method replicate 1, lane 2 = improved CTAB method replicate 2, lane 3 = PCI method replicate 1, lane 4 = PCI method replicate 2, lane 5 = MRIP method replicate 1, lane 6 = MRIP method replicate 2. Improved CTAB method shows the best results as shown by the present of intact 28 s and 18 s rRNA band. There is presence of smearing at the 5 s region for CTAB method and PCI method but more significant in the latter method. No band is observed in MRIP method
The 260/280 ratio, 260/230 ratio and the RNA weight data were subjected to normality test using IBM SPSS Statistics 28.0.1.1 (15). The null hypothesis (H0) for the normality test assumes the population is normally distributed and the standard significance level (α) is 0.05. The P-values of 260/280 ratio and 260/230 ratio data are higher than α so we accept H0, hence they are normally distributed (Table 4). On the other hand, P-value of RNA weight is lower than α so we reject H0, hence the data are not normally distributed. Since both 260/280 ratio and 260/230 ratio data are normally distributed, they were subjected to MANOVA coupled with Tukey’s test while because of the RNA weight data are not normally distributed, they were subjected to independent-samples Kruskal–Wallis test using IBM SPSS Statistics 28.0.1.1 (15). The H0 for the Tukey’s test assumes all means being compared are from the same population and the standard significance level (α) is 0.05. The P-values between all the RNA extraction methods are lower than the α (0.05) value so we reject H0, hence the 260/280 ratio and 260/230 ratio data between all the RNA extraction methods are not the same (Table 5). The H0 for the Kruskal–Wallis test assumes the distribution of the weight is the same across the RNA extraction methods and the standard significance level (α) is 0.05. The P-values of all the RNA extraction methods are higher than the α (0.05) value so we accept H0, hence distribution of the weight is the same across the RNA extraction methods (Table 6).
The improved CTAB method gives the best RNA yield with an average of 177.5 µg RNA for 1 g mature sago palm leaf tissue while PCI method and MRIP method only gives 11.17 µg and 7.97 µg RNA, respectively, even though the difference is not significant between RNA extraction methods. The RNA purity of the CTAB method for protein contamination as shown by the 260/280 ratio was significantly higher with an average of 1.93 than the PCI method and MRIP method with an average of 1.635 and 1.18, respectively. The RNA purity of the CTAB method for other organic compounds contamination as shown by 260/230 ratio was also significantly higher with an average of 1.955 than the PCI method and MRIP method with an average of 0.86 and 0.2, respectively. Hence, the improved CTAB method is superior for RNA extraction on mature sago palm leaf samples compared with the PCI method and MRIP method.
RNA extraction of mature sago palm leaf tissue by improved CTAB method
Pre-sequencing RNA sample quality and quantity tests were performed on the samples with NanoDrop and Agilent 2100 (refer Table 7 and Figs. 2, 3, 4, 5, 6, 7). Note that the RNA yield by the sequencing service provider is underestimating the actual RNA yield because the sample pellet was not fully dissolved during pellet resuspension by the sequencing service provider (data on actual RNA yield before pelletized and shipped for RNA sequencing are provided as Additional File 1).
There is high intensity of 5 s and some consistent signals pattern at the 18 s and 5 s fast region across the samples which is not ideal compared to the RIN standard but the outcome of the sequencing results showed otherwise. RNA sequencing of the extracted sago palm leaf tissue RNA by improved CTAB method gives a good result as all the samples exceed good-quality data definition for next-generation sequencing where Q20 ≥ 90%, Q30 ≥ 80% (refer Table 8). The de novo assembly of the sequenced RNA transcripts shows mean length > 700 bps and N50 > 1300 bps (refer Table 9) can be used as a guideline for future RNA sequencing of sago palm RNA extracts. The high intensity of the 5 s and some consistent signals pattern at the 18 s and 5 s fast region could indicate the RNA profile of mature sago palm leaf tissue and do not represent the degradation of the RNA sample. The below-average sample T1 RIN and 28 s/18 s ratio do not affect the sequencing result much as the clean read ratio, clean read Q30, mean assembly length and N50 values are comparable with other samples.
Conclusion
In this study, three RNA extraction methods, i.e., improved CTAB, PCI and MRIP, were compared and evaluated based on the best RNA purity and quality extracted from sago palm leaf tissue. Overall, the best result of RNA purity, quantity and quality of RNA obtained are by the improved CTAB method. The total RNA extracts using the improved CTAB method were then sequenced and this method generates good sequencing data Q20 ≥ 97, Q30 ≥ 91. The mean length ≥ 700 bp and the N50 value ≥ 1.3 kbp of the assembly can be used as a guideline for future RNA sequencing in sago palm.
Availability of data and materials
All data are provided in this manuscript. Additional information on the sequencing data can be accessed on:
Repository name: NCBI’s Gene Expression Omnibus (GEO).
Data identification number: GSE189085.
Direct URL to data: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE189085.
Repository name: NCBI’s Sequence Read Archive (SRA).
Sample ID: GSM5694359 (ST1: Trunking Sample 1).
Data identification number: SRX13165895.
Direct URL to data: https://www.ncbi.nlm.nih.gov/sra/SRX13165895.
Sample ID: GSM5694360 (ST4: Trunking Sample 4).
Data identification number: SRX13165896.
Direct URL to data: https://www.ncbi.nlm.nih.gov/sra/SRX13165896.
Sample ID: GSM5694361 (ST5: Trunking Sample 5).
Data identification number: SRX13165897.
Direct URL to data: https://www.ncbi.nlm.nih.gov/sra/SRX13165897.
Sample ID: GSM5694362 (NT7: Trunking Sample 7).
Data identification number: SRX13165898.
Direct URL to data: https://www.ncbi.nlm.nih.gov/sra/SRX13165898.
Sample ID: GSM5694363 (NT8: Trunking Sample 8).
Data identification number: SRX13165899.
Direct URL to data: https://www.ncbi.nlm.nih.gov/sra/SRX13165899.
Sample ID: GSM5694364 (NT9: Trunking Sample 9).
Data identification number: SRX13165900.
Direct URL to data: https://www.ncbi.nlm.nih.gov/sra/SRX13165900.
Abbreviations
- RNA:
-
Ribonucleic acid
- CTAB:
-
Cetyltrimethylammonium bromide
- spp.:
-
Species (plural)
- PCI:
-
Phenol–chloroform–isoamyl alcohol
- MRIP:
-
Methods for RNA isolation from palms
- Tris:
-
(Hydroxymethyl)aminomethane
- HCl:
-
Hydrochloric acid
- EDTA:
-
Ethylenediaminetetraacetic acid
- BGI:
-
Beijing Genome Institute
- NGS:
-
Next-Generation Sequencing
References
Ehara H. Genetic variation and agronomic features of Metroxylon palms in Asia and Pacific. In: Ehara H, Toyoda Y, Johnson DV, editors. Sago Palm : multiple contributions to food security and sustainable livelihoods. Singapore: Springer Nature; 2018. p. 45–59.
Ellen R. Local Knowledge and Management of Sago Palm (Metroxylon sagu Rottboell) Diversity in South Central Seram, Maluku, Eastern Indonesia. J Ethnobiol. 2006;26(2):258–98.
Flach M. Sago palm: Metroxylon sagu Rottb. Rome: Institute of Plant Genetics and Crop Plant Research, Gatersleben/International Plant Genetic Resources Institute; 1997.
Mochamad HB, Muhammad IN, Agief JP, Fendri A, Liska A. Growing area of sago palm and its environment. In: Ehara H, Toyoda Y, Johnson DV, editors. Sago palm: multiple contributions to food security and sustainable livelihoods. Singapore: Springer Nature; 2018. p. 17–29.
Abdurakhmonov IY. Bioinformatics: basics, development, and future, bioinformatics—updated features and applications. London: IntechOpen; 2016. https://doi.org/10.5772/63817.
Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. https://doi.org/10.1038/nrg2484.
Firtzlaff V, Oberländer J, Geiselhardt S, et al. Pre-exposure of Arabidopsis to the abiotic or biotic environmental stimuli “chilling” or “insect eggs” exhibits different transcriptomic responses to herbivory. Sci Rep. 2016;6:28544. https://doi.org/10.1038/srep28544.
Liu H, Wang J, Sun H, Han X, Peng Y, Liu J, Liu K, Ding Y, Wang C, Du B. Transcriptome profiles reveal the growth-promoting mechanisms of Paenibacillus polymyxa YC0136 on tobacco (Nicotiana tabacum L.). Front Microbiol. 2020;11:584174. https://doi.org/10.3389/fmicb.2020.584174.
Rochester DE, Winer JA, Shah DM. The structure and expression of maize genes encoding the major heat shock protein, hsp70. Eur Mol Biol Org. 1986;5(3):451–8.
Chong IP. Isolation, characterisation of partial small and large subunits of the ADP-glucose pyrophosphorylase gene from sago palm (Metroxylon sagu). (Master dissertation, Universiti Malaysia Sarawak). UNIMAS Institutional Repository. 2008.
Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem. 1987;162:156–9.
Lau JS. Isolation and characterisation of near complete cDNA and genomic sequences coding for granule-bound starch synthase in sago palm (Metroxylon sagu). (Master dissertation, Universiti Malaysia Sarawak). UNIMAS Institutional Repository. 2001.
Shu-Jun C, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–6.
Budiani A, Putranto RA, Minarsih H, Riyadi I, Abbas B. Ekspresi dan kloning gen penyandi ADP-Glucose Phyrophosphorylase dari tanaman sagu (Metroxylon sagu Rottb.). Menara Perkeb. 2015;83(2):76–85.
Schultz DJ, Craig R, Cox-Foster DL, Mumma RO, Medford JI. RNA isolation from recalcitrant plant tissue. Plant Mol Biol Rep. 1994;12(4):310–6.
Bong SK. Screening for Flower-Specific cDNA Sequences in Sago Palm (Metroxylon sagu) via a Differential Display Technique. (Master dissertation, Universiti Malaysia Sarawak). UNIMAS Institutional Repository. 2004.
Jamel B, Hussain MH, Salleh MA, Busri N. Total RNA isolation from sago palm. CRAUN Sago Res J. 2006;2:165–9.
Salzman RA, Fujita T, Zhu-Salzman K, Hasegawa PM, Bressan RA. An Improved RNA Isolation Method for Plant Tissues Containing High Levels of Phenolic Compounds or Carbohydrates. Plant Mol Biol Report. 1997;17:11–7.
Kiefer E, Heller W, Ernst D. A Simple and Efficient Protocol for Isolation of Functional RNA from Plant Tissues Rich in Secondary Metabolites. Plant Mol Biol Report. 2012;18:33–9.
Siti Izyan-Liyana Kamarol (2015). Representational Difference Analysis (RDA) for Identification of Molecular Factors Contributing to Trunking and Non Trunking Sago Palm (Metroxylon sagu). [Master dissertation, Universiti Malaysia Sarawak]. UNIMAS Institutional Repository
Chun-Gen Hu, Honda C, Kita M, Zi-Lian Z, Tsuda T, Moriguchi T. A Simple Protocol for RNA Isolation from Fruit Trees Containing High Levels of Polysaccharides and Polyphenol Compounds. Plant Mol Biol Report. 2002;20:69.
Anastasia-Shera Edward Atit (2013). Differential Expression Gene Profiling Studies on Trunking and Non-Trunking Sago Palm (Metroxylon sagu Rottb.). [Master dissertation, Universiti Malaysia Sarawak]. UNIMAS Institutional Repository
Mohd Hasnain Hussain (2002). Analysis of Debranching Enzymes from Pea and Potato. [Doctor of Philosophy dissertation, University of East Anglia].
Baharina Wati Barozah (2005). Synthesis of First Strand cDNA from Total RNA of Metroxylon Sagu Leaf. [Bachelor dissertation, Universiti Malaysia Sarawak]. UNIMAS Institutional Repository
Farizan Bin Nan (2006). Isolation of cDNA Fragment Encoding Starch Synthase Gene from Metroxylon sagu by RT-PCR Method. [Bachelor dissertation, Universiti Malaysia Sarawak]. UNIMAS Institutional Repository
Tajuddin Sidek Hairaddin (2006). Isolation of cDNA Fragment Encoding Starch Branching Enzyme (Isoform I) Gene from Metroxylon sagu by RT-PCR method. [Bachelor dissertation, Universiti Malaysia Sarawak]. UNIMAS Institutional Repository
Rohana Musa (2006). Isolation of cDNA Fragment Encoding Isoamylase Gene from Metroxylon Sagu by RT-PCR Method. [Bachelor dissertation, Universiti Malaysia Sarawak]. UNIMAS Institutional Repository
Mohd-Azinuddin Ahmad Mokhtar (2006). Isolation of cDNA fragment encoding starch branching enzyme (isoform II) gene from Metroxylon sagu by RT-PCR method. [Bachelor dissertation, Universiti Malaysia Sarawak]. UNIMAS Institutional Repository
Zulaikha Pol Ong (2019). Rapid Amplification of cDNA Ends of Differentially Expressed Genes from Trunking and Non-Trunking Sago Palm (Metroxylon Sagu Rottb.). [Master dissertation, Universiti Malaysia Sarawak]. UNIMAS Institutional Repository
Zhen Ying and Yang Tao. RNA Isolation from Highly Viscous Samples Rich in Polyphenols and Polysaccharides. Plant Mol Biol Report. 2002;20:417a–417e.
Jerry, anak Gerunsin (2014) Isolation, Characterisation and Expression of Fructose-1,6-bisphophate Aldolase from Sago Palm (Metroxylon sagu) Leaves. [Master dissertation, Universiti Malaysia Sarawak]. UNIMAS Institutional Repository
Roslan HA, Md.-Anowar Hossain, Jerry Gerunsin,. Molecular and 3D-Structural Characterisation of Fructose-1,6-Bisphosphate Aldolase Derived from Metroxylon Sagu. Braz Arch Biol Technol. 2017;60: e17160108. https://doi.org/10.1590/1678-4324-2017160108.
Wu Y, Llewellyn DJ, Dennis ES. A quick and easy method for isolating good-quality RNA from cotton (Gossypium hirsutum L.) tissues. Plant Mol Biol Rep. 2002;20:213–8.
Gasic K, Hernandez A, Korban SS. RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Rep. 2004;22:437a–437g.
Holling N. Screening of leafy-like gene in sago meristem and various tissues. (Bachelor dissertation, Universiti Malaysia Sarawak). UNIMAS Institutional Repository. 2008.
Julai N. Rapid amplification of cDNA ends and cDNA screening of alcohol dehyrdrogenase genes from Metroxylon sagu. (Bachelor dissertation, Universiti Malaysia Sarawak). UNIMAS Institutional Repository. 2008.
Wee CC. Generation of expressed sequence tags (Ests) database and isolation of alcohol dehydrogenase gene from young leaf samples of Metroxylon sagu. (Master dissertation, Universiti Malaysia Sarawak). UNIMAS Institutional Repository. 2008.
Roslan HA, Anji SB. Characterisation of inflorescence-predominant chitinase gene in Metroxylon sagu via differential display. 3 Biotech. 2011;1:27–33. https://doi.org/10.1007/s13205-011-0004-x.
Wee CC, Roslan HA. Isolation of alcohol dehydrogenase cDNA and basal regulatory region from Metroxylon sagu. Int Sch Res Netw. 2012;2012:839427. https://doi.org/10.5402/2012/839427.
Rosli SS. Isolation, heterologous expression and functional characterisation of phenyalanine ammonia lyase gene (msPAL) from sago palm (Metroxylon sagu Rottbl.). (Master dissertation, Universiti Malaysia Sarawak). UNIMAS Institutional Repository. 2017.
Xiao Y, Yang Y, Cao H, Fan H, Ma Z, Lei X, Mason AS, Xia Z, Huang X. Efficient isolation of high quality RNA from tropical palms for RNA-seq analysis. Plant Omics. 2012;5(6):584–9.
Untergasser A. “RNA Miniprep using CTAB” Untergasser’s Lab. 2008. http://www.untergasser.de/lab/protocols/miniprep_rna_ctab_v1_0.htm. Accessed 1 Aug 2018.
DeNovix Inc., Purity ratios explained. Technical note 130. DeNovix incorporated. 2022. https://www.denovix.com/tn-130-purity-ratios-explained/. Accessed 5 Aug 2022.
Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006. https://doi.org/10.1186/1471-2199-7-3.
BGI. DNBseqTM service overview: RNA-Seq (Transcriptome) sequencing. PN: BGIASO4_061319. BGI Genomics. 2019. https://www.bgi.com/api/assets/01b07d24-e382-4c07-b657-3ee75c9f7c0d. Accessed 5 Aug 2022.
Acknowledgements
This research is supported by Sarawak Research Development Council Grant: (RDCRG/CAT/2019/23). The authors thank Land Custody and Development Authority (LCDA) Sdn. Bhd. for the permission to obtain samples from their plantation and for logistical supports.
Funding
Open Access funding provided by Universiti Malaysia Sarawak. This study was supported and funded by Sarawak Research Development Council Grant: (RDCRG/CAT/2019/23).
Author information
Authors and Affiliations
Contributions
WJY planned and performed the experiments, analysis, and is the main author of the manuscript. HH designed the overall study, is the major contributor in data analysis and manuscript preparation. FHP performed the methods verification experiments and contributed to the manuscript drafting. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
The RNA purity profile prior to shipment for RNA sequencing.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yan, WJ., Pendi, F.H. & Hussain, H. Improved CTAB method for RNA extraction of thick waxy leaf tissues from sago palm (Metroxylon sagu Rottb.). Chem. Biol. Technol. Agric. 9, 63 (2022). https://doi.org/10.1186/s40538-022-00329-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40538-022-00329-9