Abstract
Background
Perennial ryegrass (Lolium perenne L.) is an important temperate grass used for turf and forage purposes. With the increasing accumulation of genomic and transcriptomic data of perennial ryegrass, an efficient protoplast and transient gene expression protocol is highly desirable for in vivo gene functional studies in its homologous system.
Results
In this report, a highly efficient protoplast isolation (5.6 × 107 protoplasts per gram of leaf material) and transient expression (plasmid transformation efficiency at 55.2%) was developed and the detailed protocol presented. Using this protocol, the subcellular locations of two ryegrass proteins were visualized in chloroplasts and nuclei, respectively, and protein–protein interaction between two chlorophyll catabolic enzymes (LpNOL and LpNYC1) was recorded in its homologous system for the first time.
Conclusion
This efficient protoplast isolation and transformation protocol is sufficient for studies on protein subcellular localization and protein–protein interaction, and shall be suitable for many other molecular biology applications where the mesophyll protoplast system is desirable in perennial ryegrass.
Similar content being viewed by others
Background
Perennial ryegrass (Lolium perenne L.) is the most widely distributed and cultivated turf and forage grass in temperate zones. World-wide consorted programs are working on the molecular genetics of this grass species. The draft genome of this perennial ryegrass was recently published [1] and the assembly of the genome dataset is undergoing. The accumulation of genomic and transcriptomic datasets provided unprecedented opportunity to conduct functional genomic studies in perennial ryegrass. Stable genetic transformation systems have been established in perennial ryegrass, yet the whole transformation takes months to obtain rooted transgenic plants [2, 3].
The plant protoplast system provides a complementary or, sometimes, an alternative way to the stable genetic transformation system for gene functional analysis in many cases, such as protein subcellular localization, in vivo protein–protein and protein-DNA interactions, protein trafficking and signal transduction, etc. Currently, mesophyll protoplast-based transient expression assays are routinely used in biological studies in Arabidopsis (Arabidopsis thaliana) [4, 5], maize (Zea mays) [4], tabacco (Nicotiana tabacum) [6], rice (Oryza sativa) [7, 8], Populus [9, 10], and cucumber (Cucumis sativus) [11]. Successful ryegrass protoplast isolation from mesophyll cells has been reported previously and was used in the study on chloroplast photosynthetic and photorespiratory carbon metabolism [12]. However, its efficiency remains low and not sufficient engouth for protoplast transformation studies according to our preliminary experimental results following the previously published one (data not shown). An efficient transient expression system based on ryegrass mesophyll protoplast transformation was not reported yet.
In this study, a highly repeatable and efficient protocol for mesophyll protoplast isolation and gene transient expression was developed using ryegrass leaves as starting materials. This protocol provides a facile tool for protein subcellular localization and bimolecular fluorescence complementation (BIFC) assays as shown in this study as well as the other in vivo molecular studies where this system is applicable.
Methods
Plant material and growth conditions
Perennial ryegrass (cv. Buena vista) was grown in vermiculite: perlite: peat moss (1:3:9) in a growth chamber with temperature set at 25/20 °C (day/night), photosynthetically active radiation (PAR) of 750 µmol photons m−2 s−1 and 14 h of light per day. The potted plants were watered at about three-day intervals and fertilized with ½ MS minerals (Murashige and Skoog 1962) once a week. It is CRITICAL to water and fertilizer ryegrass plants regularly to obtain fine starting plant material.
Reagents and solutions
Recipes for the enzyme solution, Modified W5 solution, MMg solution, PEG-Ca2+ solution were listed in Table 1. Cellulase ‘Onozuka’ R-10 (Cat. No. BS197A) and macerozyme R-10 (Cat. No. YM-10-1 g) were purchased from Yakult Pharmaceutical Ind. Co., Ltd., Japan. Noting that the enzyme solution was thermally pretreated at 55 °C for 10 min to inactivate nonspecific enzymes before the addition of BSA, CaCl2 and KCl solutions.
PEG-4000 (Cat. No. 25322-68-3), D-Mannitol (Cat. No. DH190-2), and Bovine Serum Albumin (BSA) (Cat. No. 9048-46-8) were purchased from Bei Jing Ding Guo Chang Sheng Biotech Co., Ltd., China; MES (Cat. No. E169) was from Ameresco LLC, USA; CaCl2·2H2O (Cat. No. 10035-04-8) was from Xi long Chemical Co., Ltd., China; and KCl (Cat. No. 7447-40-7), NaCl (Cat. No. 7647-14-5), and MgCl2·6H2O (Cat. No. 7791-18-6) were from Sinopharm Chemical Reagent Co., Ltd., China.
Protoplast isolation
Fully expanded leaves (about 10–12 days after leaf emergence) were collected from healthy ryegrass plants and only the middle sectioned leaves were used as the starting material (the tips and bases of leaf blades were removed). A total of 0.4–0.6 g of leave sections were cut into 0.5 mm strips transversely with sharp razors wetted with the enzyme solution, and the leaf strips, once cut off, were immediately emerged into the 6 ml enzyme solution (Table 1). The leaf strips were then vacuumed (0.1 MPa) for 1 h under dark at room temperature. The enzymatic digestion was carried out in a thermal-stable shaker at 30 rpm, 28 °C in dark for about 6 h to the extent that the leaf strips were readily dissembled upon gentle touches with a pipette tip.
The suspended protoplasts were firstly filtered through a layer of cheese clothes and then through a nylon mesh (75 μm), and the residue was washed twice with 10 ml pre-chilled modified W5 solution (Table 1). The filtrate was centrifuged at 100×g for 2 min with low acceleration and deceleration speed (BECKMAN, Model Allegra 64R, Hamburg, Germany). The pelleted protoplasts were re-suspended in 10 ml pre-chilled modified W5 solution, and set still for about one hr on ice (or overnight at 4 °C) to allow protoplast to sediment. The precipitated protoplasts were re-suspended in 3 ml MMg solution (Table 1). According to the counted number of viable protoplasts, the suspension was centrifuged down at 100×g for 2 min and re-suspended in the MMg solution to a final concentration of ~5 × 105 cells ml−1 (depending on experimental purposes, the concentration can be readily achieved up to 7.3 × 106 cells ml−1).
Noting that the pipette tips used in transferring protoplasts should be cut with scissors to minimize mechanical damage.
Vector construction and plasmid preparation
One chloroplast-localized protein LpPPH [13] and one unnamed NAC transcription factor LpNACx (unpublished result) were used for the subcellular localization test. In brief, the LpPPH and LpNACx genes were inserted into the p2GWF7.0 vector [14] in fusion with a GFP tag at the C-terminal. For bimolecular fluorescence complementation (BiFC) test, putative LpNOL and LpNYC1 genes were amplified from perennial ryegrass (unpublished), and cloned into a pair of split citrine vectors (pN-citrine-GW and pC-citrine-GW) to generate fusion proteins with citrine N-terminal or with citrine C-terminal tags. The pair of BiFC vectors were constructed in Dr. Bingyu Zhao’s lab at Virginia Tech: the pN-citrine-GW vector was generated using the backbone of p2GW7.0 [15] with an insertion of citrine-N-terminal with a T7 tag in the C terminal of a GATEWAY ccdB cassette(B) which would result in a translational fusion of target protein and citirine-T7 after LR clonase recombination (Invitrogen, Carlsbad, CA); likewise, the pC-citrine-GW vector was constructed with the insertion of citrine-C-terminal with HA tag. The primers used in gene cloning were presented in Table 2. The constructed vectors were electro-transformed into E. coli T1 cells (Invitrogen) and the plasmids were extracted using a commercial midi-prep kit (TIANpure plasmid kit II, TianGen Co., Beijing, China) to yield quality DNAs (>300 ng ul−1).
PEG-mediated protoplast transformation
The transient gene expression system using ryegrass protoplasts was modified from a protocol reported by Yoo et al. [5]. The optimal amount of plasmid (from 1 to 10 µg) and concentration of mannitol (from 0.1 to 0.3 M) were tested. In brief, plasmids were mixed with 200 µl protoplast stock at RT, adding equal volume of PEG-Ca2+ medium (Table 1), gently mixed and setting still for 5 min at RT for plasmid transformation. Then, three ml W5 solution was added slowly and gently mixed with the protoplast suspension, followed by a centrifugation at 100×g for 2 min and the pellet was re-suspended gently in 0.5 ml of W5 solution. Finally, the protoplast suspension was transferred into 1.5 ml tubes that were pre-coated with 1% BSA to avoid protoplast attachment to the tube surface, and incubated for 10–24 h in dark at RT.
Microscopy
The density and viability of isolated protoplasts were counted by using the standard hemocytometer and FDA staining assay [16] and observed under a light microscope (OLYMPUS Model BX53, Tokyo, Japan).
Transformed protoplasts were observed with a confocal laser scanning microscope (Zeiss LSM780 Exciter) for GFP, CFP (citrine), and chloroplast auto-fluorescence with the excitation wavelengths and emission filters set at 488 nm/band-pass 505–530 nm for GFP, 458 nm/band-pass 465–530 nm for CFP, and 488 nm/band-pass 650–710 nm for chloroplast auto-fluorescence. Image processing was performed using the Volocity software (Zeiss).
Results and discussion
Protoplast isolation from mesophyll cells of perennial ryegrass
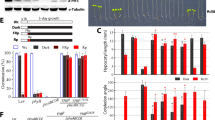
Selecting the proper starting leaf material was critical for the whole protocol. Using unselective green leaves (the 1st to the 3rd leaves from the top) for the protoplast isolation and transient gene expression assay (e.g. for the detection of subcellular localization) yielded inconsistent results. While consistent result was only achieved with the newly and fully expanded leaves (the 2nd leaf from the top which is ~10–12 days after leaf emergence) from healthy plants. The optimum concentrations of cellulose R-10 and macerozyme R-10 used in this study on ryegrass were the same as those used in rice [8], maize [4] and wheat [17]. The concentration of mannitol in the enzyme solution was critical for integrity of isolated protoplasts [18]. After testing a series of mannitol concentrations in the enzyme solution (1.50% cellulose R-10 and 0.75% macerozyme R-10), we found that mannitol at 0.6 M lead to the highly protoplast isolation efficiency (5.6 × 107 protoplasts per gram; Fig. 1) and protoplast viability (82.8%) after enzyme digestion (Fig. 1). After further removal of broken cell debris using centrifugation, the re-suspended protoplast density can be readily achieved at 7.3 × 106 protoplasts per ml (Fig. 1). Compared to the previous ryegrass protoplast isolation protocol [12], the current protocol used different digestive enzyme mix and mannitol concentration with optimized experimental procedures (see notes and details in “Methods” section), which were all indispensable factors for successful and efficient ryegrass protoplast isolation.
Effects of mannitol concentration on ryegrass protoplast isolation. A Isolated protoplast on a hemocytometer; B effect of mannitol concentration on protoplast density; C effect of mannitol concentration on protoplast viability counted using the FDA staining assay. Mannitol concentration was set at 0.3, 0.4, 0.5, 0.6, or 0.7 M, respectively. The number of intact and viable protoplasts was counted visually for round and intact protoplasts under a light microscope (OLYMPUS Model BX53, Tokyo, Japan). At least 30 protoplasts were counted in one scope, and the means were from ≥3 scopes. Different letters represent statistically significant difference at p = 0.05, and bars above columns represent standard errors
PEG-mediated protoplast transformation
The isolated protoplasts were subjected to PEG-mediated transformation. After adjusting variable parameters, we achieved up to 55.2% transformation efficiency with 10 µg plasmid DNA (plasmid size ~10 kb) prepared with a regular plasmid midi-prep kit. Higher protoplast transformation efficiency could be achieved using higher amount or purity (e.g. with CsCl purified plasmid DNA) of plasmid DNA [5]. The mannitol concentration in the PEG-Ca2+ solution was also critical for the transformation efficiency that the highest efficiency was achieved at 0.3 M mannitol (data not shown).
The developed protocol can be efficiently applied to protein subcellular localization and BiFC assays. For examples, LpPPH, a ryegrass chlorophyll catabolic enzyme, was previously shown to localize in chloroplast using Arabidopsis protoplast [13]; using the current protocol, we were able to confirm its subcellular localization in its homologous system (Fig. 2). In another experiment, a putative ryegrass NAC transcription factor was shown localized in the nucleus in ryegrass protoplast (Fig. 2).
LpNOL and LpNYC1 interact in vivo using BIFC
The current protocol is also efficient enough for protein–protein interaction assay (e.g. BIFC). We cloned two putative chlorophyll catabolic enzyme-encoding genes, LpNOL & LpNYC1, from perennial ryegrass, and fused them with split N- and C- terminal citrine fragments, respectively. Orthologs of these two genes in model plant species Arabidopsis and rice encode key chlorophyll catabolic enzymes (CCEs), which physically interact with each other and cooperatively catalyze the degradation of Chl b [19, 20]. As shown in Fig. 3, the citrine signal was only visualized in the chloroplasts when LpNOL & LpNYC1 were co-transformed, proving that these two proteins interact with each other in vivo in its homologous system.
Conclusions
In sum, a highly efficient mesophyll cell protoplast isolation and transformation protocol was developed for perennial ryegrass, which can be readily used for protein subcellular localization, and protein–protein interaction analysis. The protocol was illustrated in Fig. 4 with critical points pinpointed, and the recipes were shown in Table 1.
References
Byrne SL, Nagy I, Pfeifer M, Armstead I, Swain S, Studer B, Mayer K, Campbell JD, Czaban A, Hentrup S. A synteny-based draft genome sequence of the forage grass Lolium perenne. Plant J. 2015;84:816–26.
Patel M, Dewey RE, Qu R. Enhancing Agrobacterium tumefaciens-mediated transformation efficiency of perennial ryegrass and rice using heat and high maltose treatments during bacterial infection. Plant Cell Tiss Org. 2013;114:19–29.
Zhang WJ, Dewey RE, Boss W, Phillippy BQ, Qu R. Enhanced Agrobacterium-mediated transformation efficiencies in monocot cells is associated with attenuated defense responses. Plant Mol Biol. 2013;81:273.
Sheen J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 2001;127:1466–75.
Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–72.
Locatelli F, Vannini C, Magnani E, Coraggio I, Bracale M. Efficiency of transient transformation in tobacco protoplasts is independent of plasmid amount. Plant Cell Rep. 2003;21:865–71.
Chen SB, Tao LZ, Zeng LR, Vega-Sanchez ME, Umemura K, Wang GL. A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol. 2006;7:417–27.
Yu CC, Wang LL, Chen C, He CL, Hu J, Zhu YG, Huang WC. Protoplast: a more efficient system to study nucleo-cytoplasmic interactions. Biochem Biophys Res Commun. 2014;450:1575–80.
Guo JJ, Morrellfalvey JL, Labbé JL, Muchero W, Kalluri UC, Tuskan GA, Chen JG. Highly eficient isolation of populus mesophyll protoplasts and its application in transient expression assays. PLoS ONE. 2012;7:e44908.
Tan BY, Xu M, Chen Y, Huang MR. Transient expression for functional gene analysis using Populus protoplasts. Plant Cell Tiss Organ Cult. 2013;114:1–8.
Huang HY, Wang ZY, Cheng JT, Zhao WC, Li X, Wang HY, Zhang ZX, Sui XL. An efficient cucumber (Cucumis sativus L.) protoplast isolation and transient expression system. Sci Hortic-Amst. 2013;50:206–12.
Rathnam CKM, Chollet R. Photosynthetic and photorespiratory carbon metabolism in mesophyll protoplasts and chloroplasts isolated from isogenic diploid and tetraploid cultivars of ryegrass (Lolium perenne L.). Plant Physiol. 1980;65:489–94.
Zhang J, Yu GH, Wen WW, Ma XQ, Xu B, Huang BR. Functional characterization and hormonal regulation of the PHEOPHYTINASE gene LpPPH controlling leaf senescence in perennial ryegrass. J Exp Bot. 2016;67:935–45.
Karimi M, De MB, Hilson P. Modular cloning in plant cells. Trends Plant Sci. 2005;10:103–5.
Karimi M, Inzé D, Depicker A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–5.
Larkin PJ. Purification and viability determinations of plant protoplasts. Planta. 1976;128:213–6.
Shan QW, Wang YP, Li J, Zhang Y, Chen KL, Liang Z, Zhang K, Liu JX, Xi JJ, Qiu JL. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31:686–8.
Iguti S. Effects of mannitol on protoplasts of Saccharomyces cerevisiae. Plant Cell Physiol. 1968;9:573–6.
Sakuraba Y, Schelbert S, Park SY, Han SH, Lee BD, Andrès CB, Kessler F, Hörtensteiner S, Paek NC. STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell. 2012;24:507–18.
Sato Y, Morita R, Katsuma S, Nishimura M, Tanaka A, Kusaba M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009;57:120–31.
Authors’ contributions
GY, QC and ZX performed vector construction, ryegrass protoplast isolation and transformation experiments, and participated in manuscript preparation; QC and BZ designed and constructed the pair of BiFC vectors; BX and BH conceived and designed the experiments, analyzed data and prepared the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets supporting the conclusions and description of a complete protocol are included within the article.
Consent for publication
All authors agreed to publish this manuscript.
Ethics approval and consent to participate
All authors read and approved the manuscript.
Funding
This project was supported by a Grant from 31572455 from the National Science Foundation of China, by grant BK20140693 from the Natural Science Foundation of Jiangsu Province, China, and by Grant KYZ201552 from the Fundamental Research Funds for the Central Universities.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yu, G., Cheng, Q., Xie, Z. et al. An efficient protocol for perennial ryegrass mesophyll protoplast isolation and transformation, and its application on interaction study between LpNOL and LpNYC1. Plant Methods 13, 46 (2017). https://doi.org/10.1186/s13007-017-0196-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13007-017-0196-0