Abstract
Background
In spite of the purification of the laying hens and broilers of avian leukosis virus (ALV) has made remarkable achievements, the infection of ALV was still serious in Chinese indigenous chickens.
Methods
In order to assess the epidemic state of avian leukosis virus in indigenous chickens in China, 10 novel strains of ALV subgroup J (ALV-J), named JS16JH01 to JS16JH10, were isolated and identified by virus isolation and immunofluorescence antibody assays from a Chinese local breed farm with a sporadic incidence of tumors. To understand their virological characteristics further, the proviral genome of ENV-LTR was sequenced and compared with the reference strains.
Results
The homology of the gp85 gene between the ten ALV-J strains and NX0101 was in the range from 89.7–94.8% at the nuclear acid level. In addition, their gp85 genes were quite varied, with identities of 92–98% with themselves at the nuclear acid level. There were several snp and indel sites in the amino acid sequence of gp85 genes after comparison with other reference strains of ALV. Interestingly, a novel insertion in the gp85 region was found in two strains, JS16JH01 and JS16JH07, compared with NX0101 and HPRS-103.
Discussion
At present, owing to the large-scale purification of ALV in China, laying hens and broiler chickens with ALV infection are rarely detected, but ALVs are still frequently detected in the local chickens, which suggests that more efforts should be applied to the purification of ALV from indigenous chickens.
Similar content being viewed by others
Background
Avian leukosis virus (ALV), belonging to the genus Alpharetrovirus of the family Retroviridae, is a notorious retrovirus that can cause various types of neoplastic diseases in birds [1]. ALV is divided into 10 different subgroups based on viral envelope interference, host range, and viral cross-neutralization patterns [2, 3]. Subgroups A, B, C, D, J, and K are exogenous viruses that infect chickens [3, 4], whereas subgroup E is an endogenous virus [5]. Avian leukosis is a neoplastic disease that primarily causes myelocytomatosis (ML) and physiological hemangioma [3, 6, 7]. ALV-J was first isolated and identified from commercial meat-type chickens in 1988 in the United Kingdom [8], and then it spread quickly worldwide causing enormous economic losses to the global poultry industry [6, 9,10,11,12,13]. In 1999, ALV-J infection was first detected in broilers in China [14] and subsequently spread to other chicken types. Chickens infected with ALV-J have been a serious issue in the China poultry industry since 2007 [15, 16].
A measure of success has been achieved in the purification, prevention, and control of ALV among white-feathered broilers and layer chickens since the purification of ALV was executed in China. But there are many breeds with excellent characteristics in China, these chickens are still susceptible to ALV-J infection [17, 18].Thus, to grasp the quasispecies evolution of ALV in a flock of chickens of local breed and accelerate the purification of ALV in China, this study isolated and identified the viruses from a local chicken farm during the process of purifying ALV and the molecular characteristics of env and LTR were determined and analyzed.
Methods
Case history
In an indigenous chicken farm, chickens suffering from hepatosplenomegaly, yellowish-white tumors on the visceral surface, and presenting a drastic decline in egg production were suspected to be infected with ALV. Necropsies were performed on the diseased birds soon after death. Part of the tissues with altered local pathology were fixed with 4% formalin for pathological section.
Virus isolation and identification
Thirty chickens were randomly selected and plasma samples were collected in sterile 2-mL tubes containing 1% sodium heparin. After centrifugation at 2000 rpm for 3 min, each plasma sample was inoculated into DF-1 cells originating from embryonic fibroblast cells of C/E type chickens resistant to subgroup E endogenous ALV. The cultures were maintained for 9 days in Dulbecco’s modified Eagle’s medium supplemented with 2% fetal bovine serum at 37 °C in a 5% CO2 incubator. Next, each supernatant sample was subjected to ELISA using an ALV antigen test kit (IDEXX, USA) to detect the ALV group-specific antigen p27. Meanwhile, the cultured cells were fixed by using an aldehyde fixative and then were detected by immunofluorescence antibody assays for ALV-J, ALV-A/B, reticuloendotheliosis (REV), and Marek’s disease virus (MDV) infection by using ALV-A/B specific polyclonal antibody, ALV-J specific monoclonal antibody JE9, REV specific monoclonal antibody 11B118, and MDV specific monoclonal antibodies BA4 and H19, respectively. Then FITC-labeled anti-mouse IgG secondary antibody was used to detect primary antibody binding, after that the cells were observed by using a fluorescence microscope.
Cloning and sequencing of ENV
A pair of primers (ALV-F: GATGAGGCGAGCCCTCTCTTTG; ALV-R: TGTGGTGGGAGGTAAAATGGCGT) used to target the entire sequence of env was designed to specifically amplify a highly conserved region present in all ALV subgroups. PCR was performed as described by Meng et al. [19] with the DNA of p27-positive DF-1 cells. PCR products were separated by 1.0% agarose gel electrophoresis, purified by using the Omega Gel Extraction kit (Omega, USA),and were then cloned into the PMD18-T vector. The resulting construct was then used to transform Escherichia coli DH5α cells (TaKaRa, Japan). The positive clones were then sequenced by Shanghai Sangon Biotech Co., Ltd. (Shanghai, China).
Analysis of ALV-J strains and molecular evolution
The homologies of the nucleic acid and amino acid sequences were compared between gp85 from each ALV-J strains and the reference sequences. To assess the trends in molecular evolution of the ALV-J strains, gp85 from different ALV reference strains were included for phylogenetic analysis (Table 1). Phylogenetic analysis was accomplished by muscle alignment and the neighbor-joining and maximum parsimony methods with 500 bootstrap replicates using software MEGA 5.1.
Results
Clinical and histopathological analysis
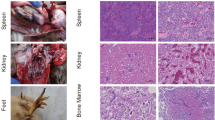
Necropsies were performed on the diseased birds soon after death. Hepatosplenomegaly and yellowish-white tumors on the visceral surface were the apparent symptoms (Fig. 1a). Histolopathological analysis of the liver revealed infiltration of eosinophilic myeloid cells around the hepatocytes (Fig. 1b).
Isolation and identification of exogenous ALV
Ten strains of ALV-J, named as JS16JH01–JS16JH10 were isolated from an indigenous chicken farm in China. The ALV p27 antigen was detected in the supernatants of the DF-1 and CEF cultures inoculated with the plasma of isolated strains. The results showed that 10 samples inoculated with plasma samples were positive for ALV p27 antigen, whereas the samples of the control group were negative, and it indicated that the chickens were infected with exogenous ALV. Immunofluorescence antibody assays revealed that the infected cells were only labeled with ALV-J specific monoclonal antibody JE9; no reaction with ALV-A/B specific polyclonal antibodies, REV specific and MDV specific monoclonal antibodies (Fig. 2) was shown.
Sequence analysis of the ENV and LTR gene
Target fragments of approximately 2200 bp were amplified from the DF1 cells infected with ALV-J using the common primers (Genbank accession no. MG700533-MG700542). Then gp85 sequences were obtained, and were compared with the sequences of reference strains of different subgroups. The results showed that the gene homology with ALV-J ranged from 89.7–94.8%, but no more than 51.8% compared with the other subgroups. What is more, their gp85 genes were quite varied, with identities of 92–98% with themselves at the nuclear acid level. Furthermore, phylogenetic analysis revealed that all the 10 strains were on the same branch as the ALV-J reference sequence (Fig. 3).
Further analysis suggested that there were multiple deletions and mutations in the gp85 gene in the isolated strains in comparison with NX0101 and HPRS-103 (Fig. 4). Compared with NX0101, 60 nucleotides were missed in rTM and DR-1. In addition, a missing asparagine and a missing arginine in the 62nd and 119th amino acid positions were observed. Furthermore, A143G, R151P, and P154S, from most of the isolated strains, changed the hydrophilicity of the amino acid in the cells and the advanced structure, as well as the function, of the protein (Table 2).
Discussion
ALV-J was first detected in China in 1999 [14], which was then followed by more reports on white-feathered broilers, layer chickens, and local breed chickens infected with ALV-J [20,21,22]. From 2007 to 2010, ALV-J caused an outbreak of hemangioma, which led to enormous economic losses to the Chinese poultry industry [15, 16]. Owing to China’s 11th Five-Year plan, high achievement was made in the purification process of avian leukosis from chicken farms; there has been fewer reports related to ALV-J in laying hens and broiler [23]. However, the strong pathogenicity and diversity of ALV still render indigenous chickens susceptible [24, 25] and seriously affect the production and promotion of excellent chicken varieties in China. In a previous national epidemiological survey, a significant proportion of local breeds (for example, 70–80%) were found to be infected with ALV-J. All these available data indicated that indigenous breeds in China were greatly infected with ALV-J.
Indigenous chickens infected with myeloma leukemia caused by ALV-J were first detected in the Shandong province in 2005; a death rate of about 10% occurred on the farm [26]. Lin [27] isolated 12 strains of avian leukosis virus subgroup J from chickens in south China during 2013–2014 and detected several amino acid variations and potential glycosylation sites in gp85 by sequencing. In addition, they found that many genetic variations of the gp85 exist in an indigenous chicken flock [17]. The monitoring of ALV in local chickens in our country has been continuous; simultaneous, our laboratory has executed the purification of ALV-J from indigenous chickens.
In this study, 10 ALV strains were isolated from golden silky fowl by a quality gene bank analysis during the process of purification. By virus isolation and identification, sequencing, and phylogenetic analysis, it was found they all belonged to subgroup J. On the phylogenetic evolutional tree, all the 10 strains were placed on the same branch as the ALV-J referential sequences. Gp85 nucleic acid homology ranged from 89.6–92.0% compared with NX0101 whereas the homology was between 84.3% and 87.3% at the amino acid level. However, gp85 sequences of the strains were quite disparate, with identities of only 50% compared with those of other subgroups. Owing to the high error and high recombination rates of polymerase, RNA viruses are highly susceptible to genetic variation and ALV-J also exhibits extremely high mutation levels. Gp85 homology between the strains was 86.1–98.4% at the nucleic acid level and 86.1%–98.4% at the amino acid level. These data indicate that an enormous diversity of ALV exists among indigenous chickens.
The envelope protein gp85 plays an essential role in the viral neutralization response and specificity of virus identifying host cells. Gp85 genes of the strains isolated in the present study showed several amino acid deletions and mutation sites compared with the reference strains. Asparagine and arginine residues were missing at the 62nd and 119th positions, respectively. Most of the mutations (e.g., A143G, R151P, and P154S) in the isolated strains, when compared with the reference strains, changed the hydrophilicity of the amino acid in the cells, and the higher structure or the function of protein. All changes affected the identities of virus and caused the virus to evolve and form a new branch.
Conclusions
The 10 strains of ALV-J isolated from the indigenous chickens in this study deviated from the original Chinese strains, suggesting that chickens in China may be a gene pool for ALV and the presence of large numbers of variants in chickens of local breeds. Moreover, our results indicated the rapid evolution of ALV-J during the process of purification. This study focused on the characterization of ALV-J and has helped us to further understand the diversity and complexity of ALV in the local chickens; importantly, it has informed us that more efforts should be put towards the purification of indigenous chickens.
References
Payne LN, Brown SR, Bumstead N, et al. A novel subgroup of exogenous avian leukosis virus in chickens. J Gen Virol. 1991;72(4):801–7.
McNally MM, Wahlin KJ, Canto-Soler MV. Endogenous expression of ASLV viral proteins in specific pathogen free chicken embryos: relevance for the developmental biology research field. BMC Dev Biol. 2010;10(1):106.
Payne LN, Gillespie AM, Howes K. Myeloid leukaemogenicity and transmission of the HPRS-103 strain of avian leukosis virus. Leukemia. 1992;6(11):1167–76.
Cui N, Su S, Chen Z, et al. Genomic sequence analysis and biological characteristics of a rescued clone of avian leukosis virus strain JS11C1, isolated from indigenous chickens. J Gen Virol. 2014;95(11):2512–22.
Fadly AM, Nair V. Leukosis/sarcoma group. Dis Poult. 2008;11:465–516.
Xu B, Dong W, Yu C, et al. Occurrence of avian leukosis virus subgroup J in commercial layer flocks in China. Avian Pathol. 2004;33(1):13–7.
Lai H, Zhang H, Ning Z, et al. Isolation and characterization of emerging subgroup J avian leukosis virus associated with hemangioma in egg-type chickens. Vet Microbiol. 2011;151(3):275–83.
Payne LN, Nair V. The long view: 40 years of avian leukosis research. Avian Pathol. 2012;41(1):11–9.
Cui Z, Du Y, Zhang Z, et al. Comparison of Chinese field strains of avian leukosis subgroup J viruses with prototype strain HPRS-103 and United States strains. Avian Dis. 2003;47(4):1321–30.
Fadly AM, Smith EJ. Isolation and some characteristics of a subgroup J-like avian leukosis virus associated with myeloid leukosis in meat-type chickens in the United States. Avian Dis. 1999:391–400.
Thu WL, Wang CH. Phylogenetic analysis of subgroup J avian leucosis virus from broiler and native chickens in Taiwan during 2000-2002. J Vet Med Sci. 2003;65(3):325–8.
Malkinson M, Banet-Noach C, Davidson I, et al. Comparison of serological and virological findings from subgroup J avian leukosis virus-infected neoplastic and non-neoplastic flocks in Israel. Avian Pathol. 2004;33(3):281–7.
Thapa BR, Omar AR, Arshad SS, et al. Detection of avian leukosis virus subgroup J in chicken flocks from Malaysia and their molecular characterization. Avian Pathol. 2004;33(3):359–63.
Du Y, Cui Z, Qin A. Subgroup J avian leukosis viruses in China. China Poult Sci. 1999;3:1–4.
Cheng Z, Liu J, Cui Z, et al. Tumors associated with avian leukosis virus subgroup J in layer hens during 2007 to 2009 in China. J Vet Med Sci. 2010;72(8):1027–33.
Gao YL, Qin LT, Pan W, et al. Avian leukosis virus subgroup J in layer chickens, China. Emerg Infect Dis. 2010;16(10):1637.
Li Y, Fu J, Cui S, et al. Gp85 genetic diversity of avian leukosis virus subgroup J among different individual chickens from a native flock. Poult Sci. 2016;96(5):1100–7.
Li X, Lin W, Chang S, et al. Isolation, identification and evolution analysis of a novel subgroup of avian leukosis virus isolated from a local Chinese yellow broiler in South China. Arch Virol. 2016;161(10):2717–25.
Meng F, Li X, Fang J, et al. Genomic diversity of the avian leukosis virus subgroup J gp85 gene in different organs of an infected chicken. J Vet Sci. 2016;17(4):497–503.
Cui Z, Sun S, Zhang Z, et al. Simultaneous endemic infections with subgroup J avian leukosis virus and reticuloendotheliosis virus in commercial and local breeds of chickens. Avian Pathol. 2009;38(6):443–8.
Mao YQ, Li WH, Dong X, et al. Different quasispecies with great mutations hide in the same subgroup J field strain of avian leukosis virus. Sci China Life Sci. 2013;56(5):414.
Sun S, Cui Z. Epidemiological and pathological studies of subgroup J avian leukosis virus infections in Chinese local “yellow” chickens. Avian Pathol. 2007;36(3):221–6.
Reinišová M, Plachý J, Kučerová D, et al. Genetic diversity of NHE1, receptor for subgroup J avian leukosis virus, in domestic chicken and wild Anseriform species. PLoS One. 2016;11(3):e0150589.
Zhang Q, Zhao D, Guo H, et al. Isolation and identification of a subgroup a avian leukosis virus from imported meat-type grand-parent chickens. Virol Sin. 2010;25(2):130–6.
Zhao DM, Zhang QC, Cui ZZ. Isolation and Identification of a subgroup B avian leukosis virus from chickens of Chinese native breed Luhua. Chin J Virol. 2010;26(1):53–7.
Cheng ZQ, Zhang L, Liu SD, et al. Emerging of avian leukosis virus subgroup J in a flock of Chinese local breed. Acta Microbiol Sin. 2005;45(4):584–7.
Lin W, Li X, Dai Z, et al. Molecular epidemiology of J-subgroup avian leukosis virus isolated from meat-type chickens in southern China between 2013 and 2014. Arch Virol. 2016;161(11):3039–46.
Acknowledgements
We thank the funding from the National Key Research and Development Program of China (2016YFD0501606) and Funds of Shandong “Double Tops” Program (SYL2017YSTD11).
Funding
This work was supported by the National Key Research and Development Program of China (2016YFD0501606) and Funds of Shandong “Double Tops” Program (SYL2017YSTD11).
Availability of data and materials
The sequences generated during the current study are available in the NCBI GenBank repository.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: MFF. Performed the experiments: LQC. Analyzed the data: CS. Wrote the paper: MFF, LQC. Checked and revised the manuscript: ZP, CZZ. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was reviewed and approved by the Shandong Agricultural University Animal Care and Use Committee and performed in accordance with the “Guidelines for Experimental Animals” of the Ministry of Science and Technology (Beijing, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Meng, F., Li, Q., Zhang, Y. et al. Characterization of subgroup J avian Leukosis virus isolated from Chinese indigenous chickens. Virol J 15, 33 (2018). https://doi.org/10.1186/s12985-018-0947-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-018-0947-1