Abstract
In this work, Ti3C2Tx MXene was identified as efficient nanozyme with area-dependent electrocatalytic activity in oxidation of phenolic compounds, which originated from the strong adsorption effect between the phenolic hydroxyl group and the oxygen atom on the surface of Ti3C2Tx MXene flake. On the basis of the novel electrocatalytic activity, Ti3C2Tx MXene was combined with alkaline phosphatase to construct a novel cascading catalytic amplification strategy using 1-naphthyl phosphate (1-NPP) as substrate, thereby realizing efficient electrochemical signal amplification. Taking advantage of the novel cascading catalytic amplification strategy, an electrochemical biosensor was fabricated for BCR/ABL fusion gene detection, which achieved excellent sensitivity with linear range from 0.2 fM to 20 nM and limit of detection down to 0.05 fM. This biosensor provided a promising tool for ultrasensitive fusion gene detection in early diagnosis of chronic myelogenous leukemia and acute lymphocytic leukemia. Moreover, the manageable catalytic activity of MXene broke a path for developing nanozymes, which possessed enormous application potential in not only electrochemical analysis but also the extensive fields including organic synthesis, pollutant disposal and so on.
Graphical Abstract

Similar content being viewed by others
Introduction
Electrochemical biosensor has become one of the most predominant analysis tools in clinical diagnosis due to the outstanding merits of high sensitivity and selectivity, rapid response, low cost, simple instrumentation, easy miniaturization and good quantitative ability [1,2,3,4]. Enzymatic electrocatalysis is a widely used technique in electrochemical biosensor, which improves the analytical sensitivity by promoting the electrochemical redox reactions with the help of enzymes [5,6,7]. Nevertheless, natural enzymes are general cost to manufacture and store, unstable to transfer or modify, and sensitive to harsh physiochemical conditions. Nanozymes, nanomaterials with enzyme-like catalytic activities, well overcome the shortages of natural enzymes [8, 9]. Therefore, nanozymes are attracting increasing attention in bioanalysis to substitute conventional natural enzyme [10, 11]. However, the catalytic activities of conventional nanozymes strongly rely on the defect dependent active centers, such as surface dangling bonds or heterogeneous doping domains, which is distributed unevenly on the surface [12, 13]. As a consequence, the homogeneity of particle is a very important parameter of nanozymes, which brings great challenge to prepare high-quality product and applied to precise quantitative analysis [10]. From this prospective, developing nanomaterials with enzyme-like catalytic activities independent to the morphology or crystal defect is of great significance as this kind of materials might be more easily controlled to obtain uniform catalysis activity.
MXene is termed for a series of two-dimension (2D) transition metal carbides, nitrides, and carbonitrides [14, 15]. Recent years, because typical MXene possess biocompatibility, large specific surface area, rich surface chemistry, tunable lateral size, good electrical conductivity, and mechanical robustness, allowing efficient and selective interaction with target species. MXene-based materials have been utilized as electrocatalysts for detecting small molecules, pharmaceutical drugs, environmental pollutants, and biomarkers. These characteristics render MXene an ideal platform as sensing materials for electrochemical application [16]. The electrocatalytic activity MXene have attracted enormous research interest in diverse fields, such as hydrogen evolution [17], oxygen evolution [18, 19], N2-to-NH3 conversion [20], fuel cell [21, 22], energy storage [23, 24] and carbon dioxide reduction [25, 26]. These reported works implied a probable reality that the catalysis activities of MXene could be originated from the 2D basal planes rather than the defects, which was significantly different from conventional nanozymes including metallic oxide [27,28,29], carbon nanomaterials [30, 31], and 2D metallic sulfide [32, 33]. As a result, the electrocatalytic activity of MXene was directly related to the area of flake rather than the shape or morphology. Benefiting from the unique electrocatalytic characteristics and 2D structure, MXenes were expected to provide a chance to easily control the electrocatalytic activity by simply customize the total area of flakes.
In this work, the electrocatalytic activity of Ti3C2Tx MXene for phenols oxidation was identified and applied to constructing a cascading catalytic amplification strategy for electrochemical biosensor to determine of BCR/ABL fusion gene, the key biomarker for clinical diagnosis of chronic myeloid leukemia (CML). Ti3C2Tx MXene presented efficient and area-dependent phenol adsorption on the 2D plane, thus catalyzing the electrochemical oxidation. For biosensor application, Ti3C2Tx MXene was spread on electrode and further decorated with gold nanoparticles for DNA capture probe (CP) immobilization. Besides, DNA walking machine was employed to recognize target BCR/ABL fusion gene and mediate nucleic acid amplification. As illustrated in Scheme 1, the DNA walking machine would start DNA nicking and expose DNA fragments from the magnetic beads in the presence of BCR/ABL fusion gene. The exposed DNA fragments helped the assembly of biotin labeled DNA probe (Bio-DP) on the sensing surface according to the sandwich DNA hybridization of CP-DNA fragment-Bio-DP. Finally, streptavidin modified alkaline phosphatase (SA-ALP) further was modified onto the biosensing interface via the specific biotin-streptavidin reaction. With the addition of 1-naphthyl phosphate (1-NPP) in the electrolyte solution, 1-naphthol was produced via ALP-catalytic hydrolysis of 1-NPP and generated an amplified electrochemical signal via Ti3C2Tx MXene-catalytic electrochemical oxidization. With DNA walking machine and cascading catalysis for signal amplification, the electrochemical biosensor achieved excellent sensitivity for detection of BCR/ABL fusion gene with the linear range from 0.2 fM to 20 nM and limit of detection down to 0.05 fM, which could provide a powerful bioanalysis tool for clinical diagnose of CML. Moreover, the efficient electrocatalysis activity of Ti3C2Tx MXene for phenols oxidation possessed great application potential in the more fields including sewage treatment and organic synthesis and so on.
Experimental section
Reagents and materials
Ti3C2Tx (MXene) few layer dispersion solution (Lateral size 2–5 µm), Fe3O4 nanoparticles, TiO2 nanoparticles, bulk Ti3AlC2 and WS2 nanosheets (Diameter 2–5 µm) were obtained from Jiangsu XFNANO Materials Tech. Co., Ltd. (Nanjing, China). MoS2 nanosheets (Diameter 20–500 nm) were obtained from Nanjing JCNANO Tech. Co., Ltd. (Nanjing, China). NH2-Fe3O4 and Nafion solutions were obtained from Aladdin Biochemical Tech. Co., Ltd. (Shanghai, China). Gold chloride (HAuCl4∙4H2O), sodium citrate, 6-mercaptohexanol (MCH), 1-naphthyl phosphate (1-NPP), 1-naphthol, streptavidin–alkaline phosphatase (SA-ALP), 4-nitrophenol, β-estradiol and diethanolamine (DEA) were purchased from Sigma-Aldrich Chemical (St. Louis, USA). Tris(2-carboxyethyl) phosphine hydrochloride (TCEP) was purchased from Sangon Biotech. Co., Ltd. (Chongqing, China). Nt.BsmAI nicking endonuclease (Nt.BsmAI) and CutSmart buffer were provided by New England Biotech. Co., Ltd. (Beijing, China). All high-performance liquid chromatography (HPLC)-purified sequences (Additional file 1: Table S1) in our experiments were ordered from Sangon Biotech. Co., Ltd. (Shanghai, China). Clinical serum samples were obtained from the University-Town Hospital of Chongqing Medical University (Chongqing, China). The buffers and solutions involved in this experiment were display in Additional file 1: S1.
Modification of electrode surface
Prior to modification, the bare glassy carbon electrode (GCE) was polished with 300 nm and 50 nm alumina slurries to a mirror-like surface and then rinsed ultrasonically with ultrapure water, anhydrous ethanol and ultrapure water for 5 min in sequence. Then, the GCE was dried by nitrogen at room temperature. Simultaneously, Ti3C2Tx MXene (0.1 mg/mL) was suspended in ultrapure water containing a 0.1% Nafion solution and sonicated for 60 min. Gold nanoparticles (AuNPs) were synthesized according to a typical method, and the detailed procedure was elaborated in Additional file 1: S3. The products were stored at 4 °C protected from light for further use.
Next, 10 µL of the Ti3C2Tx MXene (0.10 mg/mL) suspension was dropped onto the GCE surface and allowed to dry slowly, followed by the addition of 10 µL AuNPs solution and drying at room temperature to obtain the modified electrodes (AuNPs/Ti3C2Tx MXene/GCE). Afterwards, 10 µL of thiolated capture probe (CP) pretreated by TCEP was dropped onto the AuNPs/Ti3C2Tx MXene/GCE surface and incubated overnight at 4 °C. After being washed with washing buffer, the modified electrode was further incubated with 1.0 mM MCH for 1 h at room temperature to block the nonspecific site, obtaining the electrochemical biosensing platform (MCH/CP/AuNPs/Ti3C2Tx MXene/GCE).
Preparation of DNA walking machine
DNA walking machine was prepared with reference to a previous report [34]. First, 1.0 µL walker probe solution (2.0 µM) and 1.0 µL protecting probe solution (2.0 µM) were mixed and heated at 95 °C for 5 min, and then naturally cooled to obtain ds-DNAs. Later, 20 µL support probe solution (2.0 µM) was sufficiently mixed with dsDNA and added to 20 µL Au@Fe3O4 (the detailed procedure was described in Additional file 1: S4), stirred overnight and magnetically separated to obtain the expected DNA walking machine (DNA-Au@Fe3O4). Finally, the well prepared DNA walking machine was further suspended in PBS and stored at 4 °C for further use.
Procedure for BCR/ABL fusion gene detection
First, BCR/ABL fusion gene standard sample (1.0 µL) at different concentrations, 10 U Nt.BsmAI and were added in to the dispersion of DNA walking machine and kept for 2 h at 37 °C. Then, supernatant solution was collected numerous after magnetic separation, which contained the produced intermediate DNAs. After that, 10 µL of the above supernatant solution and 10 µL of 2.5 µM biotinylated detection probe were dipped onto the electrode and incubated at 37 °C for 1 h. Rinsing with washing buffer, the obtained electrode was treated in 10 µL of DEA buffer containing 1.25 µg/mL SA-ALP and 8 mg/mL BSA at 37 °C for 30 min. Finally, the electrochemical signal was measured in the DEA buffer containing 1.0 mg/mL 1-NPP by differential pulse voltammetry (DPV) after rinsing with DEA buffer to remove the unbound SA-ALP. All parameter configurations of electrochemical measurements were shown in Additional file 1: S5.
Theoretic calculation methods
The first principle calculations are performed by Vienna Ab initio Simulation Package (VASP) [35] with the projector augmented wave (PAW) method [36]. The exchange-functional is treated using the Perdew-Burke-Ernzerhof (PBE) [37] functional, in combination with the DFT-D correction [38]. The cut-off energy of the plane-wave basis is set at 500 eV. For the optimization of both geometry and lattice size, the Brillouin zone integration is performed with 2*2*1 Monkhorts-Pack k-point sampling. The self-consistent calculations apply a convergence energy threshold of 10–5 eV. The equilibrium geometries and lattice constances are optimized with maximum stress on each atom within 0.02 eV/Å.
Results and discussion
Morphological and elemental analysis of the Ti3C2Tx MXene
Transmission electron microscopy (TEM) was employed to study the morphology of the used Ti3C2Tx MXene sample, which presented a remarkably large flake and some stacked little fragments (Fig. 1A). Moreover, the high-angle annular dark-field (HAADF)-STEM image showed no observable spot on the flake, suggesting the uniform distribution of the elements (Fig. 1B). As shown in Fig. 1C–E, STEM-EDS elemental mappings of C, Ti, and O presented outlines well matched with the HAADF-STEM image, which visually displayed the elemental composition of the Ti3C2Tx MXene.
Energy dispersive X-ray spectroscopy (EDX) was also utilized to analyze the elemental composition of the Ti3C2Tx MXene (see in Additional file 1: Fig. S1), which involved of C, O, Ti, F and Al elements. Among them, F and Al were mainly from the residual impurities and their contents were notably lower than C, O, and Ti.
Atomic force microscope (AFM) was employed to further study the morphology of Ti3C2Tx MXene. As shown in Fig. 1G and 1H, AFM image presented sheets with thickness of about 4 nm, corresponding to the thickness of 3 layers.
Electrocatalytic activity of Ti3C2Tx MXene for phenolic compound oxidation
The electrocatalytic activity for phenolic compound oxidation of Ti3C2Tx MXene was confirmed by testing the electrocatalytic performances with different phenolic substrates, including 1-naphthol, 4-nitrophenol, and β-estradiol. As shown in Fig. 2 the modification of MXene significantly improved the oxidation currents for all the three phenolic compounds (DPV curves were seen in Additional file 1: Fig. S2), indicating the favourable and comprehensive electrocatalytic activity of Ti3C2Tx MXene for phenolic compound oxidation. Moreover, it's notable that the oxidation peaks presented distinct shifts to lower potential, revealing the electron transfer between Ti3C2Tx MXene and phenolic compound.
To further profile the unique electrocatalytic activity of Ti3C2Tx MXene, several nanomaterials including MoS2 nanosheets, WS2 nanosheets, Fe3O4 nanoparticles, TiO2 nanoparticles and bulk Ti3AlC2 were separately employed as constrast with 1-naphthol as substrate. It's notable that only Ti3C2Tx MXene presented observable electrocatalytic activity for 1-naphthol oxidation, indicating that the unique electrocatalytic activity was the intrinsic property of Ti3C2Tx MXene.
To quantitatively present the electrocatalytic activity of Ti3C2Tx MXene for 1-naphthol oxidation, DPV curves were measured with the addition of 1-naphthol at different concentrations. As shown in Fig. 3A, the peak current increased with the increasing concentration of 1-naphthol at low concentrations until reaching about 90 μA. According to Faraday's laws of electrolysis and Michaelis–Menten equation, the fitting curve was achieved with hyperbola function, where the Michaelis constant was calculated to be 0.22 mM, indicating that Ti3C2Tx MXene possessed strong affinity to 1-naphthol [39]. Moreover, the catalytic activity of Ti3C2Tx MXene for 1-naphthol oxidation in homogeneous phase solution was explored using hydrogen peroxide as oxidant. Figure 3B displayed the real-time absorbance of the aqueous solutions at 387.5 nm, including 1-naphthol (curve a), mixture of 1-naphthol and hydrogen peroxide (curve b), mixture of 1-naphthol, hydrogen peroxide and Ti3C2Tx MXene (curve c), respectively. It could be seen that the reaction ratio was significantly improved with the addition of Ti3C2Tx MXene, indicating that Ti3C2Tx MXene could efficiently catalyze the 1-naphthol oxidation by hydrogen peroxide as well [40].
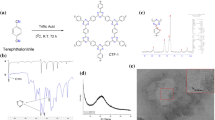
A Oxidation currents of 1-naphthol at different concentrations measured with Ti3C2Tx MXene modified electrode; B Absorbancy at 387.5 nm of 1-naphthol solution (curve a), mixture of 1-naphthol and hydrogen peroxide (curve b), mixture of 1-naphthol, hydrogen peroxide and Ti3C2Tx MXene (curve c), respectively. C The Faradic currents measured by electrode modified with Ti3C2Tx MXene at different concentrations in solution containing 7.0 mM 1-naphthol. D Charge density difference of the "lying-down" adsorption mode. Yellow and blue represent charge accumulation and depletion (isovalue: 0.005 au)
To survey the quantitative relation between electrocatalytic activity and amount of Ti3C2Tx MXene, Ti3C2Tx MXene solutions at different concentrations were employed to modify GCE to test the electrocatalytic activities for 1-naphthol oxidation, respectively. As shown in Fig. 3C, the Faradic current increased linearly with the increasing concentration of Ti3C2Tx MXene in low concentration range and reached a constant in high concentration range, indicating that the electrocatalytic activity was in direct proportion to the total modified area of Ti3C2Tx MXene flakes but not further improved by the stacking of the flakes.
Mechanism study for the catalytic activity of Ti3C2Tx MXene
The first principle calculations are performed to explain the catalytic activity of Ti3C2Tx MXene for 1-naphthol oxidation. The free energies of 1-naphthol adsorption were first calculated to determine whether 1-naphthol could be adsorbed by Ti3C2Tx MXene. The optimized geometries of 1-naphthol on and Ti3C2Tx MXene were given in Fig. S3 (see in Additional file 1), which showed 1-naphthol molecule adsorbed on the plane surface of Ti3C2Tx MXene in a "lying-down" or "standing-up" manner, with adsorption energies of − 1.18476 and − 0.75625 eV, respectively, indicating relative strong physical adsorption. As a result, the "lying-down" adsorption of 1-naphthol on Ti3C2Tx MXene was critical in the electrocatalytic oxidation. To further investigate the origin of 1-naphthol adsorption, the differential charge density of 1-naphthol adsorbed on Ti3C2Tx MXene was calculated. As shown in Fig. 3D, the changes in charge density caused by 1-naphthol adsorption mainly came from hydroxyl group and oxygen atoms. Moreover, changes in charge density were also found on the aromatic rings, indicating that the aromatic structure played a role in leading to the "lying-down" adsorption mode of 1-naphthol on Ti3C2Tx MXene.
Characterization of the DNA walking machine
To quantify the efficiency of DNA Walking machine, fluorescence dynamics experiments were carried out to verify it. Firstly, AuNPs and ssDNA-functionalized AuNPs were prepared according to previous literature with little adjustment [41]. In short, sodium citrate (3 mL, 1%) was added rapidly to the boiling solution of HAuCl4 (100 mL, 1%). After the color changed from pale yellow to wine-red, the mixture was stopped heating and cooled to room temperature (RT) with continued stirring. The AuNPs had an average particle size of 13 nm and were stored at 4 °C for further use. The preparation method of ssDNA (support probe and walker-protect dsDNA) -functionalized AuNPs is as follows. Firstly, the 1428 μL denature-supporting probe (2 μM) and 72 μL denature-walker-protect dsDNA (2 μM) were mixed with 1 μL acetic acid (500 mM, pH 5.2) and 0.5 μL TCEP (100 mM) at RT for 1 h, respectively. Then the mixture was added into 1 mL AuNP solution, and the resultant solution was stored in a drawer at RT for at least 16 h. After 25 μL Tris–acetate (500 mM, pH 8.2) was added to the mixture, 250 μL NaCl (1 M) was dropwise added to the mixture every three hours (30, 40, 50, 60, 70 μL were added respectively). Subsequently, the resulting mixture was stored in a drawer overnight. Lastly, the mixture was centrifuged (10 000 rpm, 10 min) to remove the excess reagents, and the red precipitate was washed and dispersed in DNA preparation solution for further use.
Secondly, fluorescence kinetics curve was shown in Fig. 4A to prove the cutting efficiency of Nt.BsmAI nicking endonuclease (Nt.BsmAI). The curve a, b and c showed corresponding changes when different concentrations of target BCR/ABL fusion gene and 10 U Nt.BsmAI were added into the ssDNA (support probe and walker-protect dsDNA) -functionalized AuNPs solution, respectively. Curve d was the blank control. The slope of the curve reflects the reaction rate of enzyme shearing, which is correlated with the concentration of target gene. As can be seen from the figure, the reaction rate is fast. At the same time, when the reaction reached 2 h, the shearing enzyme still did not reach the maximum shearing value, which means that the shearing enzyme has not been completely reacted. Thus, the cutting efficiency of the Nt.BsmAI is excellent. In addition, the time of releasing hairpin structure DNA can also be known from the curve. The sharply rising stage in the curve mainly the enzymatic cleaving on the prehybridized Support DNA-Walker. It can be seen from the figure that it takes about 6–8 min to cleave and release the hairpin. The slope of tangent at t = 0 s was calculated with the fitting curves indicating the cleaving rate was highly related to the concentration of substrate in Fig. 4B. The slope of tangent at t = 0 s and logarithmic value of target BCR/ABL fusion gene concentrations presents well linear dependence range from 2 pM to 2 μΜ with pearson correlation coefficient of 0.99192, which is corresponding to the kinetic characteristic of first-order reaction.
A Fluorescence kinetics curves of the nanomachine with addition of 10 U Nt.BsmAI and target BCR/ABL fusion gene at different concentrations (from a to d: 2 μM, 2 nM, 2 pM and blank, separately). B The relationship between the slope of tangent at t = 0 s and logarithmic value of target BCR/ABL fusion gene concentrations (range of concentration: 2 pM, 2 nM, and 2 uM separately). C Time optimization of intermediate DNAs hybridizing with the capture DNA
Thirdly, as for the time of hairpin structure intermediate DNAs hybridized with the thiolated capture DNA on the electrode surface to form the sensing interface, we also conducted relevant optimization experiments. As shown in Fig. 4C, the time needed was only 1 h.
Lastly, we have consulted relevant literature, which shows that the amount of DNA loading on 15 nm gold nanoparticles is 20–30 when the concentration of NaCl is 140 mM [42]. According to the concentration and dosage of AuNPs, the amount of ssDNA fabricated on the interface of this sensor was calculated to be about 1 × 1010.
Optimization of experimental conditions
To achieved optimal analytical performance of the biosensor, some critical experimental conditions were optimized, including the ratio of walker probe to support probe, the cleaving time of Nt.BsmAI nicking endonuclease, the pH of DEA buffer and the cleaving temperature of Nt.BsmAI nicking endonuclease. As shown in Fig. 5, optimal ratio of walker probe to support probe, cleaving time, pH of DEA buffer and temperature were achieved to be 1:20, 120 min, 9.6 and 37 °C, respectively.
Analytical performance of the proposed electrochemical biosensor
To estimate analytical performance of the biosensor, the current responses toward BCR/ABL fusion gene at different concentrations were recorded under the optimal conditions through DPV measurements. As shown in Fig. 6A, the detection signal increased with the increasing concentration of target BCR/ABL fusion gene. The corresponding calibration plots of the peak currents showed a strong linear relationship to the logarithm value of target BCR/ABL fusion gene concentrations range from 0.2 fM to 20 nΜ with pearson correlation coefficient of 0.99836 (Fig. 6B). The linear regression equation was I = 1.00012 × lg (c/pM) + 11.23074 (c and I stood for the concentration of target BCR/ABL fusion gene and corresponding peak current value, respectively). The limit of detection was obtained based on three times the average standard deviation corresponding to blank sample detection, which was calculated to be 0.05 fM. Comparisons of this biosensor with some reported works for BCR/ABL fusion gene detection are shown in Additional file 1: Table S2, which highlighted the excellent sensitivity of this method in BCR/ABL fusion gene detection due to the cascading catalytic strategy and DNA walking machine for signal amplification.
Evaluation of the sensitivity and specificity of the biosensor: A DPV curves response of the electrochemical biosensor upon the increase in target BCR/ABL fusion gene concentration (from bottom to top: 0 fM, 0.2 fM, 2 fM, 20 fM, 200 fM, 2 pM, 20 pM, 200 pM, and 2 nM, 20 nM, separately) and B the corresponding linear relationship between DPV signal and logarithmic value of target BCR/ABL fusion gene concentrations (range of concentration: 20 aM, 0.2 fM, 2 fM, 20 fM, 200 fM, 2 pM, 20 pM, 200 pM, and 2 nM, 20 nM, 200 nM, 2 uM separately). C DPV responses of the electrochemical biosensor to different oligonucleotides (20 fM): (a) BCR/ABL fusion gene (target), (b) single-base-mismatched strand (B1), (c) two-base-mismatched strand (B2), (d) noncomplementary strand (B3), and (e) blank. D Stability of the proposed biosensor. The error bars represent the standard deviation of three parallel measurements
Moreover, the specificity of the biosensor was evaluated by using 3 different DNA oligonucleotides as references, including a single-base-mismatched strand (B1), a two-base-mismatched strand (B2) and a noncomplementary strand (B3), all at concentrations of 20 fM. As depicted in Fig. 6C, the response signals of the single-base-mismatched strand and two-base-mismatched strand were much lower than the response signal of the target, revealing the good capacity of the biosensor to distinguish base-mismatch. The response signal of noncomplementary sequences were approximate to the blank solution, indicating that the biosensor presented good selectivity for DNA detection.
To evaluate the stability of the proposed biosensor, the modified electrodes were stored at 4 °C before use. As presented in Fig. 6D, there were no obvious differences during the first 5 days of storage, and the current changes were less than 1.58%. After 20 days of storage, the designed biosensor retained 89.40% of its initial current response, indicating that the proposed biosensor offers satisfactory stability for target BCR/ABL detection.
Detection of BCR/ABL fusion gene in human serum samples
To further validate the applicability of the biosensor to complex biological matrix in clinical application, different concentrations of target BCR/ABL fusion gene were added to tenfold-diluted clinical serum samples and tested with the proposed biosensor. The detection results of BCR/ABL fusion gene in human serum samples are summarized in Additional file 1: Table S3. Satisfactory recovery values were obtained ranging from 93.60% to 110.42% with relative standard deviations (RSD) between 0.27% and 0.64%. In addition, we extracted RNA from clinical serum of BCR/ABL positive patients using spin columns CB3 according to the manufacture's protocol and tested with the proposed biosensor. The detection concentrations of BCR/ABL were compared with clinical results (by reverse transcription PCR), which were summarized in Additional file 1: Table S4. It could be seen that the proposed biosensor achieved results well matched with the clinical assay, manifesting the application potential of the proposed biosensor in clinical diagnosis.
Conclusions
In summary, this work reported the unique catalytic activity of Ti3C2Tx MXene nanozyme for phenols oxidation and the application to electrochemical biosensor for BCR/ABL fusion gene detection. The catalytic activity originated from the adsorption capacity of the Ti3C2Tx MXene plane surface towards 1-naphthol. Theoretic calculation revealed the mechanism that relatively strong interaction was existed between the surface oxygen atoms of Ti3C2Tx MXene and the phenolic hydroxyl groups. Moreover, the catalytic activity was strictly proportional to the cover area of the MXene fakes but independent to the number of stacking layers, which was quite different from traditional nanozymes and significant benefited the quality control of biosensor. Ultrasensitive detection was achieved with the proposed biosensor, which provided a promising analytical tool for the clinical diagnosis of CML.
Availability of data and materials
All data generated or analyzed during this study are included in this article and the Additional Information.
Change history
26 March 2022
A Correction to this paper has been published: https://doi.org/10.1186/s12951-022-01365-1
References
Kimmel DW, LeBlanc G, Meschievitz ME, Cliffel DE. Electrochemical sensors and biosensors. Anal Chem. 2012;84:685–707.
Maduraiveeran G, Sasidharan M, Ganesan V. Electrochemical sensor and biosensor platforms based on advanced nanomaterials for biological and biomedical applications. Biosens Bioelectron. 2018;103:113–29.
Rackus DG, Shamsi MH, Wheeler AR. Electrochemistry, biosensors and microfluidics: a convergence of fields. Chem Soc Rev. 2015;44:5320–40.
Wongkaew N, Simsek M, Griesche C, Baeumner AJ. Functional nanomaterials and nanostructures enhancing electrochemical biosensors and lab-on-a-chip performances: recent progress, applications, and future perspective. Chem Rev. 2019;119:120–94.
Bocanegra-Rodriguez S, Molins-Legua C, Campins-Falco P, Giroud F, Gross AJ, Cosnier S. Monofunctional pyrenes at carbon nanotube electrodes for direct electron transfer H2O2 reduction with HRP and HRP-bacterial nanocellulose. Biosens Bioelectron. 2021;187:113304.
Manesh KM, Santhosh P, Uthayakumar S, Gopalan AI, Lee KP. One-pot construction of mediatorless bi-enzymatic glucose biosensor based on organic–inorganic hybrid. Biosens Bioelectron. 2010;25:1579–86.
Wang D, Chai Y, Yuan Y, Yuan R. Simple and regulable DNA dimer nanodevice to arrange cascade enzymes for sensitive electrochemical biosensing. Anal Chem. 2020;92:14197–202.
Liu M, Chen G, Qin Y, Li J, Hu L, Gu W, Zhu C. Proton-regulated catalytic activity of nanozymes for dual-modal bioassay of urease activity. Anal Chem. 2021;93:9897–903.
Sun Y, Xu H, Zhao X, Hui Z, Yu C, Wang L, Xue J, Zhao Y, Zhou R, Dai H, Miao C, Chen Q, Zhou J, Sun G, Huang W. Identifying the active site of ultrathin NiCo LDH as an efficient peroxidase mimic with superior substrate affinity for sensitive detection of hydrogen peroxide. J Mater Chem B. 2019;7:6232–7.
Jiang D, Ni D, Rosenkrans ZT, Huang P, Yan X, Cai W. Nanozyme: new horizons for responsive biomedical applications. Chem Soc Rev. 2019;48:3683–704.
Sun Y, Xu H, Wang L, Yu C, Zhou J, Chen Q, Sun G, Huang W. Ultrathin NiMn layered double hydroxide nanosheets with a superior peroxidase mimicking performance to natural HRP for disposable paper-based bioassays. J Mater Chem B. 2021;9:983–91.
Fan K, Wang H, Xi J, Liu Q, Meng X, Duan D, Gao L, Yan X. Optimization of Fe3O4 nanozyme activity via single amino acid modification mimicking an enzyme active site. Chem Commun. 2017;53:424–7.
Liang M, Yan X. Nanozymes: from new concepts, mechanisms, and standards to applications. Acc Chem Res. 2019;52:2190–200.
Huang K, Li Z, Lin J, Han G, Huang P. Two-dimensional transition metal carbides and nitrides (MXenes) for biomedical applications. Chem Soc Rev. 2018;47:5109–24.
Novoselov KS, Geim AK, Morozov SV, Jiang D, Zhang Y, Dubonos SV, Grigorieva IV, Firsov AA. Electric field effect in atomically thin carbon films. Science. 2004;306:666.
Pei Y, Zhang X, Hui Z, Zhou J, Huang X, Sun G, Huang W. Ti3C2TX MXene for sensing applications: recent progress, design principles, and future perspectives. ACS Nano. 2021;15:3996–4017.
Jiang Y, Sun T, Xie X, Jiang W, Li J, Tian B, Su C. Oxygen-functionalized ultrathin Ti3C2Tx MXene for enhanced electrocatalytic hydrogen evolution. Chemsuschem. 2019;12:1368–73.
Ma T, Cao J, Jaroniec M, Qiao S. Interacting carbon nitride and titanium carbide nanosheets for high-performance oxygen evolution. Angew Chem, Int Ed. 2016;55:1138–42.
Yu M, Zhou S, Wang Z, Zhao J, Qiu J. Boosting electrocatalytic oxygen evolution by synergistically coupling layered double hydroxide with MXene. Nano Energy. 2017;44:181–90.
Zhao J, Zhang L, Xie X, Li X, Ma Y, Liu Q, Fang W, Shi X, Cui G, Sun X. Ti3C2Tx (T = F, OH) MXene nanosheets: conductive 2D catalysts for ambient electrohydrogenation of N2 to NH3. J Mater Chem A. 2018;6:24031–5.
Handoko AD, Khoo KH, Tan TL, Jin H, Seh ZW. Establishing new scaling relations on two-dimensional MXenes for CO2 electroreduction. J Mater Chem A. 2018;6:21885–90.
Jiang L, Duan J, Zhu J, Chen S, Antonietti M. Iron clusters-directed synthesis of 2D/2D Fe-N-C/MXene superlattice-like heterostructure with enhanced oxygen reduction electrocatalysis. ACS Nano. 2020;14:2436–44.
Yu C, Gong Y, Chen R, Zhang M, Zhou J, An J, Lv F, Guo S, Sun G. A solid-state fibriform supercapacitor boosted by host-guest hybridization between the carbon nanotube scaffold and MXene nanosheets. Small. 2018;14:e1801203.
Zhao X, Xu H, Hui Z, Sun Y, Yu C, Xue J, Zhou R, Wang L, Dai H, Zhao Y, Yang J, Zhou J, Chen Q, Sun G, Huang W. Electrostatically assembling 2D nanosheets of MXene and MOF-derivatives into 3D hollow frameworks for enhanced lithium storage. Small. 2019;15:e1904255.
Lang Z, Zhuang Z, Li S, Xia L, Zhao Y, Zhao Y, Han C, Zhou L. MXene surface terminations enable strong metal-support interactions for efficient methanol oxidation on palladium. ACS Appl Mater Interfaces. 2020;12:2400–6.
Wang H, Lee J. Recent advances in structural engineering of MXene electrocatalysts. J MaterChem A. 2020;8:10604–24.
Gao L, Fan K, Yan X. Iron oxide nanozyme: a multifunctional enzyme mimetic for biomedical applications. Theranostics. 2017;7:3207–27.
Pratsinis A, Kelesidis GA, Zuercher S, Krumeich F, Bolisetty S, Mezzenga R, Leroux J-C, Sotiriou GA. Enzyme-mimetic antioxidant luminescent nanoparticles for highly sensitive hydrogen peroxide biosensing. ACS Nano. 2017;11:12210–8.
Zhu Y, Zhang Z, Song X, Bu Y. A facile strategy for synthesis of porous Cu2O nanospheres and application as nanozymes in colorimetric biosensing. J Mater Chem B. 2021;9:3533–43.
Fan K, Xi J, Fan L, Wang P, Zhu C, Tang Y, Xu X, Liang M, Jiang B, Yan X, Gao L. In vivo guiding nitrogen-doped carbon nanozyme for tumor catalytic therapy. Nat Commun. 2018;9:1440.
Zhang P, Sun D, Cho A, Weon S, Lee S, Lee J, Han JW, Kim D-P, Choi W. Modified carbon nitride nanozyme as bifunctional glucose oxidase-peroxidase for metal-free bioinspired cascade photocatalysis. Nat Commun. 2019;10:940.
Chen T, Zou H, Wu X, Liu C, Situ B, Zheng L, Yang G. Nanozymatic antioxidant system based on MoS2 nanosheets. ACS Appl Mater Interfaces. 2018;10:12453–62.
Zhu J, Peng X, Nie W, Wang Y, Gao J, Wen W, Selvaraj JN, Zhang X, Wang S. Hollow copper sulfide nanocubes as multifunctional nanozymes for colorimetric detection of dopamine and electrochemical detection of glucose. Biosens Bioelectron. 2019;141:111450.
Xu Z, Liao L, Chai Y, Wang H, Yuan R. Ultrasensitive electrochemiluminescence biosensor for microRNA detection by 3D DNA walking machine based target conversion and distance-controllable signal quenching and enhancing. Anal Chem. 2017;89:8282–7.
Kresse G, Furthmüller J. Efficiency of ab-initio total energy calculations for metals and semiconductors using a plane-wave basis set. Comput Mater Sci. 1996;6:15–50.
Joubert D, Kresse G. From ultrasoft pseudopotentials to the projector augmented-wave method. Phys Rev B. 1999;59:1758–75.
Burke K, Ernzerhof M, Perdew JP. Generalized gradient approximation made simple [Phys. Rev. Lett. 77, 3865 (1996)]. Phys Rev Lett 1997; 78: 1396-.
Grimme S, Antony J, Ehrlich S, Krieg H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J Chem Phys. 2010;132:154104.
Wang J, Huang R, Qi W, Su R, Binks BP, He Z. Construction of a bioinspired laccase-mimicking nanozyme for the degradation and detection of phenolic pollutants. Appl Catal, B. 2019;254:452–62.
Jiang B, Duan D, Gao L, Zhou M, Fan K, Tang Y, Xi J, Bi Y, Tong Z, Gao GF, Xie N, Tang A, Nie G, Liang M, Yan X. Standardized assays for determining the catalytic activity and kinetics of peroxidase-like nanozymes. Nat Protoc. 2018;13:1506–20.
Juewen L, Yi L. Preparation of aptamer-linked gold nanoparticle purple aggregates for colorimetric sensing of analytes. Nat Protoc. 2006;1:246–52.
Hurst SJ, Lytton-Jean AK, Mirkin CA. Maximizing DNA loading on a range of gold nanoparticle sizes. Anal Chem. 2006;78:8313–8.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (Grant Nos. 31071093, 31170129, and 31200064), China Postdoctoral Science Foundation (2020M683259) and the Science and Technology Planning Project of Yuzhong District of Chongqing city, China (No. 20140119).
Funding
This work was supported by National Natural Science Foundation of China (Grant Nos. 31071093, 31170129, 31200064), China Postdoctoral Science Foundation (Grand No. 2020M683259), Science and Technology Planning Project of Yuzhong District of Chongqing city, China (Grant No. 20140119)
Author information
Authors and Affiliations
Contributions
RY: Conceptualization, Methodology, Software, Investigation, Data Curation, Writing-Original Draft. JX: Software, Investigation, Project Administration. YW: Formal Analysis, Software. JQ: Supervision, Methodology, Data Curation, Administration. XH: Methodology, Software, Investigation, Project Administration. AC: Writing-Review & Editing. JX: Funding Acquisition, Supervision, Administration, Data Curation. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Consent for publication
All authors of this study agreed to publish.
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: the author group is updated with three corresponding authors
Supplementary Information
Additional file 1: S1
. Buffers. S2. Apparatus. S3. Synthesis of AuNPs. S4. Synthesis of magnetically responsive AuNPs-coated Fe3O4 (Au@Fe3O4). S5. Electrochemical measurement. Fig. S1. Energy dispersive X-ray spectroscopy (EDX) patterns of the Ti3C2Tx MXene. Fig. S2. DPV curves measured with bare GCE and Ti3C2Tx modified GCE with different substrates. Fig. S3. Optimized geometries of 1-naphthol on Ti3C2Tx MXene. Fig. S4. Gel electrophoresis characterization for the DNA walking strategy. Fig. S5. Reproducibility of the designed biosensor. Fig. S6. Characterization of electrode assembly process. Table S1. Sequences of oligonucleotides used in this work. Table S2. Comparison of available methods for the detection of BCR/ABL. Table S3. Determination of BCR/ABL in human serum samples (n=3) with the developed electrochemical biosensor. Table S4. Comparison of the concentration of clinical samples (n=3) by proposed method (DPV) and current widely used methods (reverse transcription PCR).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yu, R., Xue, J., Wang, Y. et al. Novel Ti3C2Tx MXene nanozyme with manageable catalytic activity and application to electrochemical biosensor. J Nanobiotechnol 20, 119 (2022). https://doi.org/10.1186/s12951-022-01317-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12951-022-01317-9