Abstract
Background
Inconsistencies have existed in research findings on the association between cardiovascular disease (CVD) and single nucleotide polymorphisms (SNPs) of ADIPOQ, triggering this up-to-date meta-analysis.
Methods
We searched for relevant studies in PubMed, EMBASE, Cochrane Library, CNKI, CBM, VIP, and WanFang databases up to 1st July 2017. We included 19,106 cases and 31,629 controls from 65 published articles in this meta-analysis. STATA 12.0 software was used for all statistical analyses.
Results
Our results showed that rs266729 polymorphism was associated with the increased risk of CVD in dominant model or in heterozygote model; rs2241766 polymorphism was associated with the increased risk of CVD in the genetic models (allelic, dominant, recessive, heterozygote, and homozygote). In subgroup analysis, significant associations were found in different subgroups with the three SNPs. Meta-regression and subgroup analysis showed that heterogeneity might be explained by other confounding factors. Sensitivity analysis revealed that the results of our meta-analysis were stable and robust. In addition, the results of trial sequential analysis showed that evidences of our results are sufficient to reach concrete conclusions.
Conclusions
In conclusion, our meta-analysis found significant increased CVD risk is associated with rs266729 and rs2241766, but not associated with rs1501299.
Similar content being viewed by others
Background
Cardiovascular disease (CVD) is the primary cause of death worldwide, leading to 32% of all deaths worldwide in 2013 [1]. Epidemiological and biological evidences demonstrate that multiple environmental and genetic factors are implicated in CVD, although the etiology of CVD has not been fully elucidated [2,3,4,5]. Identifying CVD-relative risk factors is critical in control of the development and progress of CVD.
Adiponectin is involved in CVD: low levels of adiponectin (hypoadipoectinemia) positively correlate with the risk of CVD, and higher levels of adiponectin protect against this disease [6,7,8,9,10,11]. Adiponectin is synthesized and secreted by adipose tissue [12], osteoblasts [13], skeletal muscle [14], and cardiomyocytes [15]. This protein, as one of the most abundant adipocytokines in blood, has anti-atherogenic, cardioprotective, anti-inflammatory, and antithrombotic properties [16,17,18,19,20].
Adiponectin is encoded by ADIPOQ which is located in chromosome 3q27 [21], and adiponectin levels are influenced by single-nucleotide polymorphisms (SNPs) in ADIPOQ [22]. SNPs in ADIPOQ have been found to be associated with CVD [23, 24], diabetes [25, 26], stroke [27, 28], myocardial infarction [29, 30], cancer [31, 32], kidney disease [33, 34], and even gynecological conditions [35, 36]. Previous studies have shown the association between SNPs in ADIPOQ (rs3774261, rs1063537, rs2082940, rs2241766, rs266729, and rs1501299) and CVD/subclinical CVD [30, 37, 38]. The three common SNPs of ADIPOQ (rs266729, rs2241766, and rs1501299) were most widely studied. However, findings from previous studies on the three SNPs in relation to CVD risk are inconsistent and inconclusive.
For rs266729 (− 11,377 C/G) in ADIPOQ, Du et al. [39] and Zhang et al. [40] found that the SNP is associated with CVD risk; Stenvinkel et al. [41] revealed that rs266729 is associated with the decreased risk of CVD; Zhang et al. [40], Cheong et al. [27], and Chiodini et al. [29] found that there is no significant association between rs266729 and CVD. For rs2241766 (+ 45 T/G), Pischon et al. [42] and Jung et al. [43] identified no association between rs2241766 and the risk of coronary artery disease (CAD) in patients with type 2 diabetic mellitus (T2DM); Du et al. [39], Oliveira et al. [44], and Mofarrah et al. [45] found that there is a significant association between rs2241766 polymorphism and CAD risk; Chang et al. [46] revealed that rs2241766 is associated with the decreased risk of CVD. Moreover, for rs1501299 (+ 276 G / T), Bacci et al. [47] and Esteghamati et al. [48] revealed that rs1501299 is associated with the decreased risk of CAD; Mohammadzadeh et al. [38], however, reported that there is an association between rs1501299 and CAD risk; Foucan et al. [49] found that there is no significant association between rs1501299 and CAD in patients with T2DM. Thus, those results are inconsistent.
Meta-analysis performed by Zhang et al. in 2012 revealed that associations between the SNPs (rs2241766, rs1501299, and rs266729) in ADIPOQ and CVD were significant but weak [50]. Since that data, several more studies have emerged to investigate the association between SNPs in ADIPOQ and susceptibility to CVD [37, 38, 45]. In this study, we further collected references and updated meta-analysis of association between SNPs (rs2241766, rs1501299, and rs266729) in ADIPOQ and CVD in order to get a more precise and reliable assessment of the association.
Methods
Search strategy
We performed an extensive literature search in PubMed, EMBASE, Cochrane Library, CNKI, CBM, VIP, and WanFang databases for published articles on the association between ADIPOQ polymorphisms and CVD risk up to July 1st, 2017. The literature search was done without any language or population restrictions imposed. During the literature search, we used various combinations of keywords, such as ‘coronary heart disease (CHD)’ or ‘coronary artery disease’ or ‘cardiovascular disease’ or ‘ischemic heart disease’ or ‘angina’ or ‘myocardial infarction (MI)’ or ‘stroke’ or ‘atherosclerosis’ or ‘arteriosclerosis’ or ‘coronary stenosis’ combined with ‘ADIPOQ’ or ‘APM1’ or ‘ACDC’ or ‘adiponectin gene’ and ‘polymorphisms’ or ‘variants’ or ‘variations’. Joseph Sam Kanu and Shuang Qiu independently performed the literature search for potential articles included in this meta-analysis. All articles retrieved were first organized in reference manager software (Endnote 6).
Inclusion and exclusion criteria
A study included in this meta-analysis was based on the following criteria: 1) the study has sufficient data to allow association between CVD risk and ADIPOQ SNP to be assessed; 2) the study included original data (independence among studies); 3) evaluation of the ADIPOQ polymorphisms (rs266729, rs2241766, and rs1501299) and CVD risk; 4) the language of the study was English or Chinese; and 5) observed genotype frequencies in controls must be consistent with Hardy–Weinberg equilibrium (HWE). We excluded a study based on: 1) the study contained overlapping data; 2) the study with missing information (particularly genotype distributions), after corresponding author, who was contacted by us with email, failed to provide the required information; and 3) genome scans investigating linkages with no detailed genotype distributions between cases and controls. Where there was a disagreement on the selection of a study, the issue was resolved by discussion or consensus with the third investigator (Ri Li). For articles with missing data, we emailed the corresponding authors for the required data.
Assessment of study quality
We used the NATURE-published guidelines proposed by the NCI-NHGRI Working Group on Replication in Association Studies for assessing the quality of each study included in this meta-analysis [51]. These guidelines have a checklist of 53 conditions for authors, journal editors, and referees to interpret data and results of genome-wide or other genotype–phenotype association studies clearly and unambiguously. We used the first set of 34 conditions in assessing the quality of each study. We allocated a score of 1 point for each condition a study met, and no point (0 score) if the condition or requirement is lacking. Each study was given a total Quality Score – the sum of all points each study obtained. Study quality assessment was independently carried out by Joseph Sam Kanu and Shuang Qiu.
Data extraction
Joseph Sam Kanu and Shuang Qiu extracted data from each study independently. We summarized the information extracted from each article in Table 1. The characteristics of articles included first author, year of publication, country in which the study was done, study population (ethnicity), numbers of cases and controls, genotyping method, SNPs investigated, genotype frequency of cases and controls, and outcome (Table 1; Additional file 1: Tables S1, S2, and S3).
Statistical analysis
HWE was evaluated for each study using Goodness of fit Chi-square test in control groups, and P < 0.05 was considered as a significant deviation from HWE. The strength of association between the three ADIPOQ polymorphisms and CVD susceptibility was assessed using odds ratios (OR) and 95% confidence intervals (95% CI). The associations were measured based on five different genetic models: allelic model (rs266729: G versus C; rs2241766: G versus T; rs1501299: T versus G), dominant model (rs266729: GG + GC versus CC; rs2241766: GG + GT versus TT; rs1501299: TT + TG versus GG), recessive model (rs266729: GG versus GC + CC; rs2241766: GG versus GT + TT; rs1501299: TT versus TG + GG), heterozygote model (rs266729: GC versus CC; rs2241766: GT versus TT; rs1501299: TG versus GG), and homozygote model (rs266729: GG versus CC; rs2241766: GG versus TT; rs1501299: TT versus GG). Heterogeneity were evaluated by the Chi-square test based Q-statistic, and quantified by I2-statistic [52]. If there was no substantial statistical heterogeneity (P > 0.10, I2 ≤ 50%), data were pooled by fixed-effect model (Mantel and Haenszel methods); otherwise, the heterogeneity was evaluated by random-effect model (DerSimonian and Laird methods). Meta-regression analysis was performed to detect main sources of heterogeneity. In addition, subgroup analyses were stratified by population (European, East Asian, West Asian, and African), genotyping method (PCR-RFLP, TaqMan, and Others), sample size (< 1000 and ≥ 1000), and quality score (< 10 and ≥ 10). Sensitivity analysis was performed to examine stability of our results by omitting each study in each turn. Publication bias was measured by funnel plots [53], and quantified by the Begg’s and Egger’s tests [54] (P < 0.05 considered statistically significant publication bias). STATA 12.0 software (StataCorp. 2011. Stata Statistical Software: Release 12. College Station, TX: StataCorp LP) was used for all statistical analyses. P-value < 0.05 was considered statistically significant, except where other-wise specified. A separate analysis was performed for each SNPs included in the meta-analysis.
Trial sequential analysis (TSA)
Traditional meta-analysis may result in type I and type II errors owing to dispersed data and repeated significance testing [55, 56]. To reduce the risk of type I error, TSA was used to estimate required information size (RIS) and confirm statistical reliability with an adjusted threshold for statistical significance [57]. In present meta-analysis, we used trial sequential analysis software (TSA, version 0.9; Copenhagen Trial Unit, Copenhagen, Denmark, 2011) by setting an overall type I error of 5%, a statistical test power of 80%, and a relative risk reduction of 20% [58, 59].
If the Z-curve crosses trial sequential monitoring boundary or RIS has been reached, a sufficient level of evidence has been reached and further studies are unneeded; otherwise, additional studies are needed to reach a sufficient conclusion.
Results
Overall results
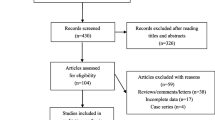
This meta-analysis included 68 studies from 65 articles after literature search and critical screening, as described in methods (Fig. 1). Meta-analysis of the rs266729 (− 11,377 C > G), rs2241766 (+ 45 T > G), and rs1501299 (+ 276 G > T) variants included 29, 40, and 44 studies, respectively. We summarize the characteristics of each primary study in Table 1. Detailed characteristics of those studies are further presented in Additional file 1: Tables S1, S2, and S3, respectively. Overall, this meta-analysis included a total of 50,735 subjects (19,106 cases and 31,629 controls).
Meta-analysis results
Association between rs266729 (− 11,377 C > G) polymorphism and CVD
The meta-analysis of the association between rs266729 (− 11,377 C > G) polymorphism and CVD included 29 studies with 29,021 subjects (10,506 cases and 18,515 controls). Significant heterogeneity among studies was observed (P h < 0.10 or I2 ≥ 50%). Thus, we selected random-effect model, and found that rs266729 polymorphism was associated with the increased risk of CVD in dominant model (GG + GC VS CC: OR = 1.129, 95% CI = 1.028–1.239, P = 0.011) and in heterozygote model (GC VS CC: OR = 1.141, 95% CI = 1.041–1.250, P = 0.005) (Table 2, Fig. 2).
Based on population, genotyping method, sample size, and quality score, we performed subgroup analyses. On the basis of population, rs266729 polymorphism was associated with the increased risk of CVD under dominant model (GG + GC VS CC: OR = 1.198, 95% CI = 1.006–1.427, P = 0.043) and under heterozygote model (GC VS CC: OR = 1.184, 95% CI = 1.002–1.398, P = 0.048) in East Asian. On the basis of genotyping methods, a significant risk association between rs266729 polymorphism and CVD was found when genotyping was performed using PCR-RFLP method under dominant model (GG + GC VS CC: OR = 1.276, 95% CI = 1.014–1.607, P = 0.038) and under heterozygote model (GC VS CC: OR = 1.282, 95% CI = 1.032–1.592, P = 0.025). On the basis of sample size or quality score, we found that rs266729 polymorphism was associated with the increased risk of CVD under allelic, dominant, and heterozygote models (all OR > 1 and P < 0.05), after pooled the ORs by the subgroups of sample size ≥ 1000 or quality score ≤ 10 (Table 2).
Association between rs2241766 (+ 45 T > G) polymorphism and CVD
The meta-analysis of the association between rs2241766 (+ 45 T > G) polymorphism and CVD included 40 studies with 25,548 subjects (10,746 cases and 14,802 controls). Using inverse-variance weighted random effect model (P h < 0.10 or I2 ≥ 50%), we found that rs2241766 polymorphism was associated with the increased risk of CVD in the five genetic models (allelic, dominant, recessive, heterozygote, and homozygote) (all OR > 1 and P < 0.05) (Table 3, Fig. 3).
Subgroup analyses were stratified by population, genotyping method, sample size, and quality score. Firstly, on the basis of population, rs2241766 polymorphism was associated with the increased risk of CVD under the five dominant models in East Asian and under allelic, recessive, and homozygote models in West Asian (all OR > 1 and P < 0.05). Secondly, on the basis of genotyping method, the results that genotyping was done by PCR-RFLP or other methods showed that rs2241766 polymorphism was associated with the increased risk of CVD under five genetic models (all OR > 1 and P < 0.05). Thirdly, on the basis of sample size, rs2241766 polymorphism was associated with the increased risk of CVD under the five genetic models in the subgroup of sample size ≤1000 (all OR > 1 and P < 0.05), but was associated with the decreased risk of CVD in the subgroup of sample size ≥1000 under recessive model (GG VS GT + TT: OR = 0.696, 95% CI = 0.539–0.885, P = 0.003) and under homozygote model (GG VS TT: OR = 0.669, 95% CI = 0.519–0.862, P = 0.002). Finally, on the basis of quality score, when we pooled the ORs by the subgroups of quality score ≤ 10, we found that rs2241766 polymorphism was associated with the increased risk of CVD under the five genetic models (all OR > 1 and P < 0.05) (Table 3).
Association between rs1501299 (+ 276 G > T) polymorphism and CVD
The meta-analysis of the association between rs1501299 (+ 276 G > T) polymorphism and CVD included 44 studies with 37,371 subjects (12,852 cases and 24,519 controls). Using the inverse-variance weighted random effect model (P h < 0.10 or I2 ≥ 50%), we found that there was no association between rs1501299 polymorphism and CVD in the five genetic models (all P > 0.05) (Table 4).
In the subgroup analysis, no significant association was found between rs1501299 polymorphism and CVD risk under the five genetic models in any subgroup (all P > 0.05) (Table 4).
Heterogeneity analysis
In this meta-analysis, meta-regression was used to investigate the source of heterogeneity by year, population, genotyping method, sample size, and quality score. We found that sample size (allelic model: P = 0.019; dominant model: P = 0.032; recessive model: P < 0.001; and homozygote model: P < 0.001) and quality score (allelic model: P = 0.035; dominant model: P = 0.032; recessive model: P < 0.001; and homozygote model: P < 0.001) contributed to the observed heterogeneity across all the studies of the association between rs2241766 polymorphisms and CVD risk. However, in the meta-analysis of the associations between rs266729/rs1501299 polymorphisms and CVD risk, we did not identify the source of heterogeneity (all P > 0.05) (Additional file 2: Table S4).
Publication bias and sensitivity analysis
Publication bias was measured by funnel plots and quantified by Begg’s and Egger’s tests. No publication bias was found among the studies regarding the association between rs266729 polymorphisms and CVD risk (all P > 0.05). Publication biases were found in analyses of the associations between rs2241766 polymorphisms and CVD risk (allelic model: PEgger = 0.001, PBegg = 0.031; dominant model: PEgger = 0.001, PBegg = 0.003; and heterozygote mode: PEgger = 0.003, PBegg = 0.003), and between rs1501299 polymorphisms and CVD risk (recessive model: PEgger = 0.031, PBegg = 0.035) (Table 5 and Additional file 3: Figures S1, S2, and S3). Sensitivity analyses showed that this meta-analysis was relatively stable and credible (Figs. 4, 5, and 6).
TSA
In the TSA of rs266729 and CVD, the Z-curve crossed trial sequential monitoring boundary and the sample size reached RIS in dominant and heterozygote models (Fig. 7). In allelic, recessive, and homozygote models, the sample size also reached RIS, although the Z-curve did not cross trial sequential monitoring boundary (Fig. 7). In the TSA of rs2241766/rs1501299 and CVD, the sample size reached RIS in the five genetic models (Figs. 8 and 9). Thus, concrete conclusions were reached and further studies were not required.
Discussion
In this meta-analysis, we collected up-to-date information (July 1st, 2017) to investigate the association between ADIPOQ SNPs and the risk of CVD. Our results demonstrate that rs266729 and rs2241766 variants of ADIPOQ are associated with the increased risk of CVD, but rs1501299 is not associated with CVD risk.
In view of the association between rs266729 and CVD risk, Yang et al. (2012) [60], Zhou et al. (2012) [61], and Zhang et al. (2012) [50] performed meta-analyses. Yang et al. reported that rs266729 is associated with the increased risk of CAD in allelic and dominant models [60]. Zhou et al. found the same association in overall population, Europeans, and East Asian in allelic, dominant and heterozygote models [61]. Zhang et al. also revealed that rs266729 is associated with the increased risk of CAD in overall population and East Asian in allelic model [50]. Our results further identified that rs266729 is associated with the increased risk of CVD in overall population and East Asian in dominant and heterozygote models. In addition, our results revealed that the significant association in studies on the basis of PCR-RFLP method, indicating that different genotyping method may result in different statistical results.
The association between rs2241766 and CVD risk also has been the subject of meta-analysis [60,61,62,63,64]. These studies are inconsistent. Yang et al. found no significant association between rs2241766 and CAD risk [60]. Zhang et al. found no overall significant risk association between CHD and rs2241766 in Han Chinese population [62]. Zhou et al. reported that rs2241766 is associated with the decreased risk of CVD in recessive and homozygote models, and the decreased risk of CVD in East Asian in allelic, dominant, recessive, and homozygote models [61]. Zhou et al. performed a meta-analysis of the association between rs2241766 and CVD risk in allelic model, and they found that rs2241766 is associated with the increased risk of CVD [63]. In our meta-analysis, we found that rs2241766 is associated with the increased risk of CVD in overall population and East Asian in all the five genetic models, and in West Asian in allelic, recessive, and homozygote models. Our findings is in agreement with the results of Zhou et al., but is in disagreement with the results of Yang et al., Zhang et al., and Zhou et al.
With regard to the association between 1,501,299 and CVD, the results are also conflicting [50, 60, 61]. Zhou et al. revealed no significant association between rs1501299 polymorphism with CAD susceptibility [61]. Qi et al. reported the extremely large decrease in CVD risk associated with rs1501299 polymorphism in diabetic patients [24]. Zhang et al. reported only the weak protective effect of the rs1501299 variant against CVD in general study subjects [50]. The meta-analysis by Zhao et al. revealed that rs1501299 polymorphism may play a protective role for CAD among patients with T2DM [22]. In comparison, our results revealed no significant association.
Different genetic admixture and environmental factors among human populations, which tend to explain ethnic background, strongly modulate the effects of ADIPOQ polymorphisms on adiponectin levels [65, 66]. Studies have reported that low levels of adiponectin (hypoadipoectinemia) correlate with the risk of CVD, and high levels of adiponectin protect against this disease [6,7,8,9,10,11]. These conflicting results of associations between the ADIPOQ polymorphism and CVD risk may be due to differences in publication bias, sample size, or insufficient statistical power. In addition, evidences have showed that studies which deviate from HWE in controls may reflect the presence of genotyping errors, population stratification, and selection bias in the controls (or without representation of studied sample). Thus, including those studies may decrease the quality of a meta-analysis or generate inconsistent results [67].
Heterogeneity across all the studies of the associations should be noted because it may potentially affect the strengths of the present meta-analysis. We, thus, used random effect model. Our results showed that sample size and quality score are the factors of heterogeneity across all studies of association between rs2241766 polymorphisms and CVD, but no factors contribute the heterogeneity across all studies of association between rs266729/rs1501299 polymorphisms and CVD. However, heterogeneity was still high in the subgroup analysis of the two factors. For these reasons, heterogeneity might be explained by other confounding factors, such as gene-gene interaction and gene-environment interaction.
Our meta-analysis has some limitations. Firstly, significant publication bias was found in the analysis of rs2241766 (under allelic, dominant, and heterozygote models) and rs1501299 (under recessive model). Secondly, our meta-analysis mainly included Europeans and Asians with only few other races, thus limiting our power to generalize our findings in other races. Finally, our results might be affected by the potential weaknesses of genetic association studies, such as phenotype misclassifications, genotyping error, population stratification, gene-environment or gene-gene interactions, and selective reporting biases [68, 69].
Despite the limitations highlighted above, our meta-analysis also had some strength. Firstly, we searched extensively and investigated more studies and more participants than any other meta-analyses performed on the association between ADIPOQ variant and CVD, which give our study more statistical power to draw valid conclusion on this issue. Secondly, sensitivity analysis showed that the results of our meta-analysis are stable and robust. Thirdly, the evidence of our results are sufficient to reach concrete conclusions, which were proved by TSA for the first time. We strongly believe our findings will help settle some of the controversies surrounding the ADIPOQ-CVD association research.
Conclusions
Our meta-analysis found significant increased CVD risk is associated with rs266729 and rs2241766, but not associated with rs1501299. Investigating gene–gene and gene–environment interactions is needed to give more insight into the genetic association between ADIPOQ variants and CVD.
Abbreviations
- CAD:
-
Coronary artery disease
- CHD:
-
Coronary heart disease
- CVD:
-
Cardiovascular disease
- HWE:
-
Hardy–Weinberg equilibrium
- MI:
-
Myocardial infarction
- RIS:
-
Required information size
- SNPs:
-
Single-nucleotide polymorphisms
- T2DM:
-
Type 2 diabetic mellitus
- TSA:
-
Trial sequential analysis
References
Roth GA, Huffman MD, Moran AE, Feigin V, Mensah GA, Naghavi M, Murray CJ. Global and regional patterns in cardiovascular mortality from 1990 to 2013. Circulation. 2015;132:1667–78.
Bhatnagar A. Environmental determinants of cardiovascular disease. Circ Res. 2017;121:162–80.
Roberts R, Stewart AF. Genes and coronary artery disease: where are we? J Am Coll Cardiol. 2012;60:1715–21.
Ji F, Ning F, Duan H, Kaprio J, Zhang D, Zhang D, Wang S, Qiao Q, Sun J, Liang J, et al. Genetic and environmental influences on cardiovascular disease risk factors: a study of Chinese twin children and adolescents. Twin Res Hum Genet. 2014;17:72–9.
Sun LY, Lee EW, Zahra A, Park JH. Risk factors of cardiovascular disease and their related socio-economical, environmental and health behavioral factors: focused on low-middle income countries- a narrative review article. Iran J Public Health. 2015;44:435–44.
Stojanovic S, Ilic MD, Ilic S, Petrovic D, Djukic S. The significance of adiponectin as a biomarker in metabolic syndrome and/or coronary artery disease. Vojnosanit Pregl. 2015;72:779–84.
Zhang HL, Jin X. Relationship between serum adiponectin and osteoprotegerin levels and coronary heart disease severity. Genet Mol Res. 2015;14:11023–9.
Md Sayed AS, Zhao Z, Guo L, Li F, Deng X, Deng H, Xia K, Yang T. Serum lectin-like oxidized-low density lipoprotein receptor-1 and adiponectin levels are associated with coronary artery disease accompanied with metabolic syndrome. Iran Red Crescent Med J. 2014;16:e12106.
Di Chiara T, Argano C, Scaglione A, Duro G, Corrao S, Scaglione R, Licata G. Hypoadiponectinemia, cardiometabolic comorbidities and left ventricular hypertrophy. Intern Emerg Med. 2015;10:33–40.
Obata Y, Yamada Y, Kyo M, Takahi Y, Saisho K, Tamba S, Yamamoto K, Katsuragi K, Matsuzawa Y. Serum adiponectin levels predict the risk of coronary heart disease in Japanese patients with type 2 diabetes. J Diabetes Investig. 2013;4:475–82.
Pischon T, Girman CJ, Hotamisligil GS, Rifai N, Hu FB, Rimm EB. Plasma adiponectin levels and risk of myocardial infarction in men. JAMA. 2004;291:1730–7.
Scherer PE, Williams S, Fogliano M, Baldini G, Lodish HF. A novel serum protein similar to C1q, produced exclusively in adipocytes. J Biol Chem. 1995;270:26746–9.
Shinoda Y, Yamaguchi M, Ogata N, Akune T, Kubota N, Yamauchi T, Terauchi Y, Kadowaki T, Takeuchi Y, Fukumoto S, et al. Regulation of bone formation by adiponectin through autocrine/paracrine and endocrine pathways. J Cell Biochem. 2006;99:196–208.
Krause MP, Liu Y, Vu V, Chan L, Xu A, Riddell MC, Sweeney G, Hawke TJ. Adiponectin is expressed by skeletal muscle fibers and influences muscle phenotype and function. Am J Physiol Cell Physiol. 2008;295:C203–12.
Pineiro R, Iglesias MJ, Gallego R, Raghay K, Eiras S, Rubio J, Dieguez C, Gualillo O, Gonzalez-Juanatey JR, Lago F. Adiponectin is synthesized and secreted by human and murine cardiomyocytes. FEBS Lett. 2005;579:5163–9.
Fujita K, Maeda N, Sonoda M, Ohashi K, Hibuse T, Nishizawa H, Nishida M, Hiuge A, Kurata A, Kihara S, et al. Adiponectin protects against angiotensin II-induced cardiac fibrosis through activation of PPAR-alpha. Arterioscler Thromb Vasc Biol. 2008;28:863–70.
Ouchi N, Kihara S, Arita Y, Maeda K, Kuriyama H, Okamoto Y, Hotta K, Nishida M, Takahashi M, Nakamura T, et al. Novel modulator for endothelial adhesion molecules: adipocyte-derived plasma protein adiponectin. Circulation. 1999;100:2473–6.
Shibata R, Sato K, Pimentel DR, Takemura Y, Kihara S, Ohashi K, Funahashi T, Ouchi N, Walsh K. Adiponectin protects against myocardial ischemia-reperfusion injury through AMPK- and COX-2-dependent mechanisms. Nat Med. 2005;11:1096–103.
Kato H, Kashiwagi H, Shiraga M, Tadokoro S, Kamae T, Ujiie H, Honda S, Miyata S, Ijiri Y, Yamamoto J, et al. Adiponectin acts as an endogenous antithrombotic factor. Arterioscler Thromb Vasc Biol. 2006;26:224–30.
Ouchi N, Kihara S, Arita Y, Nishida M, Matsuyama A, Okamoto Y, Ishigami M, Kuriyama H, Kishida K, Nishizawa H, et al. Adipocyte-derived plasma protein, adiponectin, suppresses lipid accumulation and class a scavenger receptor expression in human monocyte-derived macrophages. Circulation. 2001;103:1057–63.
Takahashi M, Arita Y, Yamagata K, Matsukawa Y, Okutomi K, Horie M, Shimomura I, Hotta K, Kuriyama H, Kihara S, et al. Genomic structure and mutations in adipose-specific gene, adiponectin. Int J Obes Relat Metab Disord. 2000;24:861–8.
Zhao N, Li N, Zhang S, Ma Q, Ma C, Yang X, Yin J, Zhang R, Li J, Yang X, Cui T. Associations between two common single nucleotide polymorphisms (rs2241766 and rs1501299) of ADIPOQ gene and coronary artery disease in type 2 diabetic patients: a systematic review and meta-analysis. Oncotarget. 2017;8:51994.
Katakami N, Kaneto H, Matsuoka TA, Takahara M, Maeda N, Shimizu I, Ohno K, Osonoi T, Kawai K, Ishibashi F, et al. Adiponectin G276T gene polymorphism is associated with cardiovascular disease in Japanese patients with type 2 diabetes. Atherosclerosis. 2012;220:437–42.
Qi L, Doria A, Manson JE, Meigs JB, Hunter D, Mantzoros CS, Hu FB. Adiponectin genetic variability, plasma adiponectin, and cardiovascular risk in patients with type 2 diabetes. Diabetes. 2006;55:1512–6.
Kaftan AN, Hussain MK. Association of adiponectin gene polymorphism rs266729 with type two diabetes mellitus in Iraqi population. A pilot study. Gene. 2015;570:95–9.
Tsai MK, Wang HM, Shiang JC, Chen IH, Wang CC, Shiao YF, Liu WS, Lin TJ, Chen TM, Chen YH. Sequence variants of ADIPOQ and association with type 2 diabetes mellitus in Taiwan Chinese Han population. ScientificWorldJournal. 2014;2014:650393.
Cheong MY, Bang OS, Cha MH, Park YK, Kim SH, Kim YJ. Association of the adiponectin gene variations with risk of ischemic stroke in a Korean population. Yonsei Med J. 2011;52:20–5.
Liu F, He Z, Deng S, Zhang H, Li N, Xu J. Association of adiponectin gene polymorphisms with the risk of ischemic stroke in a Chinese Han population. Mol Biol Rep. 2011;38:1983–8.
Chiodini BD, Specchia C, Gori F, Barlera S, D'Orazio A, Pietri S, Crociati L, Nicolucci A, Franciosi M, Signorini S, et al. Adiponectin gene polymorphisms and their effect on the risk of myocardial infarction and type 2 diabetes: an association study in an Italian population. Ther Adv Cardiovasc Dis. 2010;4:223–30.
Shaker OG, Ismail MF. Association of genetic variants of MTHFR, ENPP1, and ADIPOQ with myocardial infarction in Egyptian patients. Cell Biochem Biophys. 2014;69:265–74.
Gu CY, Li QX, Zhu Y, Wang MY, Shi TY, Yang YY, Wang JC, Jin L, Wei QY, Ye DW. Genetic variations of the ADIPOQgene and risk of prostate cancer in Chinese Han men. Asian J Androl. 2014;16:878–83.
Zhang G, Gu C, Zhu Y, Luo L, Dong D, Wan F, Zhang H, Shi G, Sun L, Ye D. ADIPOQ polymorphism rs182052 is associated with clear cell renal cell carcinoma. Cancer Sci. 2015;106:687–91.
Chung HF, Long KZ, Hsu CC, Mamun AA, Chiu YF, Tu HP, Chen PS, Jhang HR, Hwang SJ, Huang MC. Adiponectin gene (ADIPOQ) polymorphisms correlate with the progression of nephropathy in Taiwanese male patients with type 2 diabetes. Diabetes Res Clin Pract. 2014;105:261–70.
Elshamaa MF, Sabry SM, El-Sonbaty MM, Elghoroury EA, Emara N, Raafat M, Kandil D, Elsaaid G. Adiponectin: an adipocyte-derived hormone, and its gene encoding in children with chronic kidney disease. BMC Res Notes. 2012;5:174.
Yoshihara K, Yahata T, Kashima K, Mikami T, Tanaka K. Association of single nucleotide polymorphisms in adiponectin and its receptor genes with polycystic ovary syndrome. J Reprod Med. 2009;54:669–74.
Machado JS, Palei AC, Amaral LM, Bueno AC, Antonini SR, Duarte G, Tanus-Santos JE, Sandrim VC, Cavalli RC. Polymorphisms of the adiponectin gene in gestational hypertension and pre-eclampsia. J Hum Hypertens. 2014;28:128–32.
Kanu JS, Gu Y, Zhi S, Yu M, Lu Y, Cong Y, Liu Y, Li Y, Yu Y, Cheng Y, Liu Y. Single nucleotide polymorphism rs3774261 in the AdipoQ gene is associated with the risk of coronary heart disease (CHD) in northeast Han Chinese population: a case-control study. Lipids Health Dis. 2016;15:6.
Mohammadzadeh G, Ghaffari MA, Heibar H, Bazyar M. Association of two common single nucleotide polymorphisms (+45T/G and +276G/T) of ADIPOQ gene with coronary artery disease in type 2 diabetic patients. Iran Biomed J. 2016;20:152–60.
Du SX, Lu LL, Liu Y, Dong QJ, Xuan SY, Xin YN. Association of Adiponectin Gene Polymorphisms with the risk of coronary artery disease in patients with nonalcoholic fatty liver disease in a Chinese Han population. Hepat Mon. 2016;16:e37388.
Zhang M, Peng Y, Lyu S. Association between genetic variants in the adiponectin gene and premature myocardial infarction. Chinese Journal of Cardiology. 2016;44:577–82.
Stenvinkel P, Marchlewska A, Pecoits-Filho R, Heimburger O, Zhang Z, Hoff C, Holmes C, Axelsson J, Arvidsson S, Schalling M, et al. Adiponectin in renal disease: relationship to phenotype and genetic variation in the gene encoding adiponectin. Kidney Int. 2004;65:274–81.
Pischon T, Pai JK, Manson JE, Hu FB, Rexrode KM, Hunter D, Rimm EB. Single nucleotide polymorphisms at the adiponectin locus and risk of coronary heart disease in men and women. Obesity (Silver Spring). 2007;15:2051–60.
Jung CH, Rhee EJ, Kim SY, Shin HS, Kim BJ, Sung KC, Kim BS, Lee WY, Kang JH, Oh KW, et al. Associations between two single nucleotide polymorphisms of adiponectin gene and coronary artery diseases. Endocr J. 2006;53:671–7.
Oliveira CS, Saddi-Rosa P, Crispim F, Canani LH, Gerchman F, Giuffrida FM, Vieira JG, Velho G, Reis AF. Association of ADIPOQ variants, total and high molecular weight adiponectin levels with coronary artery disease in diabetic and non-diabetic Brazilian subjects. J Diabetes Complicat. 2012;26:94–8.
Mofarrah M, Ziaee S, Pilehvar-Soltanahmadi Y, Zarghami F, Boroumand M, Zarghami N. Association of KALRN, ADIPOQ, and FTO gene polymorphism in type 2 diabetic patients with coronary artery disease: possible predisposing markers. Coron Artery Dis. 2016;27:490–6.
Chang YC, Jiang JY, Jiang YD, Chiang FT, Hwang JJ, Lien WP, Chuang LM. Interaction of ADIPOQ genetic polymorphism with blood pressure and plasma cholesterol level on the risk of coronary artery disease. Circ J. 2009;73:1934–8.
Bacci S, Menzaghi C, Ercolino T, Ma X, Rauseo A, Salvemini L, Vigna C, Fanelli R, Di Mario U, Doria A, Trischitta V. The +276 G/T single nucleotide polymorphism of the adiponectin gene is associated with coronary artery disease in type 2 diabetic patients. Diabetes Care. 2004;27:2015–20.
Esteghamati A, Mansournia N, Nakhjavani M, Mansournia MA, Nikzamir A, Abbasi M. Association of +45(T/G) and +276(G/T) polymorphisms in the adiponectin gene with coronary artery disease in a population of Iranian patients with type 2 diabetes. Mol Biol Rep. 2012;39:3791–7.
Foucan L, Maimaitiming S, Larifla L, Hedreville S, Deloumeaux J, Joannes MO, Blanchet-Deverly A, Velayoudom-Céphise FL, Aubert R, Salamon R, et al. Adiponectin gene variants, adiponectin isoforms and cardiometabolic risk in type 2 diabetic patients. Journal of Diabetes Investigation. 2014;5:192–8.
Zhang H, Mo X, Hao Y, Gu D. Association between polymorphisms in the adiponectin gene and cardiovascular disease: a meta-analysis. BMC Med Genet. 2012;13:40.
Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–60.
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60.
Stuck AE, Rubenstein LZ, Wieland D. Bias in meta-analysis detected by a simple, graphical test. Asymmetry detected in funnel plot was probably due to true heterogeneity. BMJ. 1998;316:469. author reply 470-461
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34.
Turner RM, Bird SM, Higgins JP. The impact of study size on meta-analyses: examination of underpowered studies in Cochrane reviews. PLoS One. 2013;8:e59202.
Brok J, Thorlund K, Wetterslev J, Gluud C. Apparently conclusive meta-analyses may be inconclusive--trial sequential analysis adjustment of random error risk due to repetitive testing of accumulating data in apparently conclusive neonatal meta-analyses. Int J Epidemiol. 2009;38:287–98.
Higgins JP, Whitehead A, Simmonds M. Sequential methods for random-effects meta-analysis. Stat Med. 2011;30:903–21.
Wang F, Qin Z, Si S, Tang J, Xu L, Xu H, Li R, Han P, Yang H: Lack of association between NAT2 polymorphism and prostate cancer risk: a meta-analysis and trial sequential analysis. Oncotarget 2017.
Zhu M, Wen X, Liu X, Wang Y, Liang C, Tu J: Association between 8q24 rs6983267 polymorphism and cancer susceptibility: a meta-analysis involving 170,737 subjects. Oncotarget 2017.
Yang Y, Zhang F, Ding R, Wang Y, Lei H, Hu D. Association of ADIPOQ gene polymorphisms and coronary artery disease risk: a meta-analysis based on 12 465 subjects. Thromb Res. 2012;130:58–64.
Zhou L, Xi B, Wei Y, Pan H, Yang W, Shen W, Li Y, Cai J, Tang H. Association between adiponectin gene polymorphisms and coronary artery disease across different populations. Thromb Res. 2012;130:52–7.
Zhang BC, Li WM, Xu YW. A meta-analysis of the association of adiponectin gene polymorphisms with coronary heart disease in Chinese Han population. Clin Endocrinol. 2012;76:358–64.
Zhou D, Jin Y, Yao F, Duan Z, Wang Q, Liu J. Association between the adiponectin +45T>G genotype and risk of cardiovascular disease: a meta-analysis. Heart Lung Circ. 2014;23:159–65.
Sun K, Li Y, Wei C, Tong Y, Zheng H, Guo Y. Recessive protective effect of ADIPOQ rs1501299 on cardiovascular diseases with type 2 diabetes: a meta-analysis. Mol Cell Endocrinol. 2012;349:162–9.
Gu HF. Biomarkers of adiponectin: plasma protein variation and genomic DNA polymorphisms. Biomark Insights. 2009;4:123–33.
Enns JE, Taylor CG, Zahradka P. Variations in Adipokine genes AdipoQ, Lep, and LepR are associated with risk for obesity-related metabolic disease: the modulatory role of gene-nutrient interactions. J Obes. 2011;2011:168659.
Boccia S, De Feo E, Galli P, Gianfagna F, Amore R, Ricciardi G. A systematic review evaluating the methodological aspects of meta-analyses of genetic association studies in cancer research. Eur J Epidemiol. 2010;25:765–75.
Ioannidis JP, Trikalinos TA. Early extreme contradictory estimates may appear in published research: the Proteus phenomenon in molecular genetics research and randomized trials. J Clin Epidemiol. 2005;58:543–9.
Daly AK, Day CP. Candidate gene case-control association studies: advantages and potential pitfalls. Br J Clin Pharmacol. 2001;52:489–99.
Acknowledgements
The authors would like to thank staff and students of the Meta-analysis Group of the Department of Epidemiology and Biostatistics, School of Public Health of Jilin University for their contributions to this work.
Funding
This work was supported by The National Natural Science Foundation of China (Grant 81573230), the Ministry of Science and Technology of the People’s Republic of China (grant number: 2015DFA31580), and the Science and Technology Commission of Jilin Province (grant number: 20150101130JC).
Availability of data and materials
Please contact author for data requests.
Author information
Authors and Affiliations
Contributions
Conception and design: JSK, SQ, YC, and YL. Provision of study materials: JSK, SQ, RL, and CK; Collection and assembly of data: JSK, SQ, and RL. Data analysis and interpretation: JSK, SQ, and RL. Manuscript writing: JSK and SQ. Revised the language/article: All authors. Final approval of manuscript: All authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declared that they have no competing interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Summary of three SNPs characteristics. (DOCX 48 kb)
Additional file 2:
Meta-regression results of the association between the SNPs and CVD risk. (DOCX 59 kb)
Additional file 3:
Funnel plots of three SNPs for publication bias. (DOCX 1250 kb)
Additional file 4:
Additional references. (DOCX 22 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kanu, J.S., Qiu, S., Cheng, Y. et al. Associations between three common single nucleotide polymorphisms (rs266729, rs2241766, and rs1501299) of ADIPOQ and cardiovascular disease: a meta-analysis. Lipids Health Dis 17, 126 (2018). https://doi.org/10.1186/s12944-018-0767-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12944-018-0767-8