Abstract
Background
Circular RNAs play an important role in tumor genesis and progression, but they have not been sufficiently studied in patients with nasopharyngeal carcinoma (NPC).
Methods
The circular RNA, circCAMSAP1, was screened in NPC cells by RNA sequencing analysis. The expression of circCAMSAP1 in NPC tissues was examined by real-time quantitative polymerase chain reaction (RT-qPCR) and in situ hybridization. Wound-healing, transwell, MTT and flow cytometry assays, and nude mouse tumor models were used to explore the effect of circCAMSAP1 on proliferation and metastasis of NPC in vitro or in vivo. The downstream proteins regulated by circCAMSAP1 were screened using mass spectrometry. The interaction between circCAMSAP1 and the SERPINH1 mRNA was identified using the circular RNA immunoprecipitation method and the luciferase reporter assay. The interaction between SERPINH1 and transcription factor c-Myc was verified through Co-immunoprecipitation (Co-IP) and immunofluorescence. The effect of c-Myc on the generation of circCAMSAP1 was examined through RT-qPCR and chromatin immunoprecipitation. Finally, the splicing factors that promote the production of circCAMSAP1 were explored by RT-qPCR and RNA immunoprecipitation (RIP).
Results
We found that circCAMSAP1 was highly expressed in NPC tissues and promoted NPC proliferation and metastasis. Additionally, circCAMSAP1 promoted SERPINH1 expression through improved SERPINH1 mRNA stability by binding to the 3′-untranslated region (3’UTR) of SERPINH1. Highly expressed SERPINH1 reduced the ubiquitination-degradation rate of c-Myc, causing increased tumorigenesis. Meanwhile, c-Myc, cooperating with splicing factor 10 (SRSF10), could also promote CAMSAP1 pre-mRNA transcription and back-splicing, forming a positive feedback of circCAMSAP1 production, resulting in the proliferation and metastasis of NPC.
Conclusions
Our findings revealed that circCAMSAP1 promotes NPC proliferation and metastasis by binding to the 3’UTR of SERPINH1, suggesting that the positive feedback of circCAMSAP1-SERPINH1-c-Myc may serve as a prognostic biomarker or therapeutic target in patients with NPC.
Similar content being viewed by others
Background
Nasopharyngeal carcinoma (NPC) is a common head and neck malignant tumor in southern China [1]. Factors contributing to the pathogenesis of NPC mainly include Epstein-Barr virus infection [2], genetic factors [3], and environmental factors [4]. The most common pathological type of NPC is non-keratinizing carcinoma with a high degree of malignancy, metastasis, and proliferative ability [5]. The onset of NPC is hard to detect, so early diagnosis is rare, and NPC has often already progressed into middle or late stage by the time it is detected [6]. While radiotherapy and chemotherapy can effectively control the growth of local tumors, local recurrence and distant metastasis contribute to the low survival rate of patients with NPC [7]. Therefore, finding biomarkers for early diagnosis of NPC and exploring the molecular mechanisms of metastasis and proliferation are essential for optimizing clinical treatment strategies.
Circular RNAs, or circRNAs, are non-coding RNAs that have recently received widespread attention. They were originally regarded as a by-product of mRNA production from the processing of precursor mRNA [8]. With the help of splicing factors, the 3′ end and 5′ end are reversely spliced to form a closed loop; this unique structure of circRNAs prevents their degradation by RNase [9]. Additionally, circRNAs are regarded as potential markers of cancer [10]. Increasingly more circRNAs are found to be involved in the growth and metastasis of a variety of malignant tumors, and are closely related to the recovery of the disease [11]. CircRNAs can function by interacting with RNA [12, 13], DNA [14], or proteins [15], or encode small peptides [16] to participate in intracellular signal transduction. Our previous studies have found that several circRNAs have played an important role in the metastasis and proliferation of NPC [17,18,19,20], but further studies searching for more such circRNAs are warranted.
In this study, we found that the circRNA circCAMSAP1 is highly expressed in patients with NPC tissues, and strongly associated with the clinical and TNM stages of NPC patients. Furthermore, we found that circCAMSAP1 affects the proliferation and metastasis of NPC in vitro and in vivo. We also revealed that circCAMSAP1 promotes SERPINH1 expression and improves SERPINH1 mRNA stability by binding to its 3’-untranslated region (3’UTR). Additionally, c-Myc was found to be involved in the transcription and formation of circCAMSAP1, cooperating with SRSF10. Therefore, circCAMSAP1-SERPINH1-c-Myc forms a positive feedback loop and may be a novel therapeutic target for treatment of NPC.
Methods
Data analysis
The RNA sequencing data of clinical NPC samples (GSE68799) was downloaded from the Gene Expression Omnibus (GEO) database. The reads per million (RPM) value was used to standardize the count value for subsequent analysis. The NPC gene expression data (GSE12452, GSE53819) were used to analyze the expression of SERPINH1 and SRSF10.
Tissue samples
NPC tissue samples from two cohorts were collected at the Xiangya Hospital of Central South University. The first cohort included 29 NPC tissues and 12 chronic nasopharyngeal inflammation tissues (Table S1) for RT-qPCR. The second cohort included 82 NPC tissues and adjacent non-NPC tissues (Table S2) for in situ hybridization. The study was approved by the Joint Ethics Committee of the Health Bureau of Central South University, and the written informed consent was provided by each participant prior to surgery.
Cell lines, plasmids, small interfering RNAs and transfection reagents
NPC cell lines (CNE2, HONE1, HNE2, HK1, CNE1, 5-8F, HNE1, 6-10B, and C666-1) and immortalized nasopharyngeal epithelial cell line NP69 were derived from the Cell Center of Central South University. All these cells were cultured on RPMI 1640 (Life Technologies, Grand Island, NY, USA) containing 10% fetal bovine serum (Gibco, Grand Island, NY, USA) at 37 °C and 5% CO2 and free of mycoplasma contamination.
The sequences of circCAMSAP1 exons 2 and 3 were inserted into the pcDNA3.1(+) circRNA Mini Vector (kindly provided to us by professor Yong Li at Baylor College of Medicine, USA) to construct an overexpression vector. The SERPINH1-Flag, c-Myc-Flag, and SRSF10-Flag overexpression plasmids were purchased from Sino Biological Company (Beijing, China). The SERPINH1 3’UTR wild-type and mutant sequences were inserted into the pMIR-REPORT Luciferase miRNA Expression Reporter Vector (Ambion, TX, USA). The CAMSAP1 promoter sequence was inserted into the PGL3 basic vector (E1751, Promega, USA).
The siRNAs that specifically targeted circCAMSAP1, SERPINH1, c-Myc, and SRSF10 were purchased from Gemma Gene Corporation (Shanghai, China), and the sequence is shown in Table S3.
Lipofectamine 3000 (Life Technologies, NY, USA) was used to transfect plasmids and Hiperfect (Qiagen, Hilden, Germany) was used to transfect siRNAs.
RNA extraction and RT-qPCR
RNA extraction and RT-qPCR were performed according to the manufacturer’s instructions. The RT-qPCR was conducted using the HiScript cDNA synthesis kit (R323-01, Vazyme, China) and the SYBR RT-qPCR Master Mix (Q511-02, Vazyme, China). Primers are shown in Table S3. Relative gene expression was calculated by the comparative CT method (ΔΔCT), and β-actin was used as a housekeeping gene.
Western blotting
Western blotting analysis was performed as described in previous studies [19]. Antibodies are shown in Table S4. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control to normalize the protein loading.
RNase R and actinomycin D treatment
RNase R enzyme (Thermo Fisher, Grand Island, NY, USA) digestion and actinomycin D (Sigma, Missouri, USA) treatment were used to detect the stability of RNA. For RNase R digestion, the total RNA extracted from NPC cells was added or not added to the RNase R enzyme (20 U/μL), incubated at 37 °C for 30 min and then at 70 °C for 10 min to inactivate the RNase R. This was followed by RT-qPCR detection. For the actinomycin D treatment, NPC cells were added with actinomycin D at a final concentration of 1 μg/mL at 0, 6, 12, 24 h, and RNA was collected for RT-qPCR analysis.
Cycloheximide treatment
For the cycloheximide (CHX) treatment, NPC cells were added with CHX at a final concentration of 50 μg/mL at 0, 15, 30, 60 min, and protein was collected for western blotting analysis.
In situ hybridization, fluorescence in situ hybridization, and immunohistochemistry
In situ hybridization (ISH) or fluorescence in situ hybridization (FISH) was carried out to detect circCAMSAP1 expression in NPC tissues using digoxigenin-labeled probes as previously reported [19]. The Enhanced Sensitive ISH Detection kit I (POD) (MK1030, BOSTER, China) was used for ISH in NPC tissues. The ElivisionTM + Polyer HRP (Mouse/Rabbit) immunohistochemistry kit (Kit-9902, Maxim, China) was used to evaluate immunohistochemistry.
Immunofluorescence
After 24 h of cell climbing, 4% paraformaldehyde was fixed for 30 min, 0.25% Triton X-100 was ruptured for 40 min, and 5% bovine serum albumin was blocked for 1 h. Cells were immuno-stained with antibodies overnight at 4 °C, washed and incubated with fluorescently labeled secondary antibodies (Thermo Fisher, Grand Island, NY, USA) at 37 °C for 40 min, and nuclei were stained with 6-diamidino-2-phenylindole (DAPI). A confocal laser scanning microscope (Perkin Elmer, Massachusetts, USA) was used for observation and imaging.
Hematoxylin–eosin staining (H&E)
After the paraffin sections were dewaxed and watered with xylene and gradient alcohol, the nuclei were stained with hematoxylin solution (Beyotime, Shanghai, China), the eosin solution was used for staining cytoplasm (Beyotime, Shanghai, China), and the slides were mounted with neutral resin. Pictures were taken with an Olympus BX51 fluorescence microscope (Olympus, Japan).
Wound-healing and transwell assay
Wound-healing assay was performed as previously reported [19]. An Olympus Digital Camera was used to capture the scratched area at 0, 12 and 24 h. The invasion assay was analyzed by transwell chambers with 8-μm pores (Millipore).
MTT and flow cytometry
Eight hundred transfected cells were seeded into each well of a 96-well plate. After the cells adhered to the wall, 20 μl MTT (Beyotime, Shanghai, China) was added to each well for four additional hours. The 490 nm wavelength on the enzyme-linked instrument (Molecular Devices, California, USA) was used to detect the absorbance value. Flow cytometry was used to analyze the proliferation of NPC cells using propidium iodide staining.
Liquid chromatography combined with tandem mass spectrometry (LC-MS/MS)
Protein identification by LC-MS/MS was performed using the previously reported method [19]. The peptide samples were diluted with 0.1% trifluoroacetic acid (TFA) (Sigma, Missouri, USA) and then transferred to the analysis chamber, where the Nano-LC LTQ Orbitrap ETD (Bruker Daltonic, Billerica, USA) was used for mass spectrometric analysis and identification. The Proteome Discoverer software (Thermo Scientific, Waltham, MA, USA), as well as the UniProtKB/Swiss-Prot database (release 2014_02), were used to identify the proteins.
circRNA Immunoprecipitation (circRIP) assay
The biotin-labeled circCAMSAP1 probe was used for RNA pull-down. The circCAMSAP1 overexpression plasmid and probe were transfected into NPC cells. Cells were incubated with streptavidin Dynabeads (M-280, Invitrogen) at 4 °C. Immunoprecipitated RNA was isolated with TRIzol reagent and analyzed by RT-qPCR. The primers and probe sequences used are shown in Table S3.
RNA pull-down assay and co-immunoprecipitation
RNA pull-down assay was performed using a magnetic RNA protein pull-down kit (20164, Thermo Scientific) according to the manufacturer’s instructions. Cell lysates were incubated with the streptavidin Dynabeads or antibodies, and the enriched proteins were denatured at high temperatures, then examined using western blotting.
Luciferase reporter assays and chromatin immunoprecipitation (ChIP)
The Dual-Luciferase® Reporter Assay System (E2980, Promega, USA) was used according to the manufacturer’s instructions.
ChIP analysis was performed using the Pierce™ Magnetic ChIP Kit (26157, Thermo Scientific, USA). The c-Myc antibody was used to enrich the chromatin fragments in NPC cells, and the enriched chromatin fragments were analyzed by RT-qPCR. RT-qPCR primers were designed according to the binding sites (Table S3).
RNA binding protein immunoprecipitation (RIP) assay
RIP assay was performed with the RIP kit (Millipore, Billerica, MA, USA) according to the manufacturer’s instructions. See Table S3 for detection primers.
Animal testing
First, CNE2 (2 × 106) transfected with the circCAMSAP1 overexpression vector, circCAMSAP1 siRNA1, or the empty vector and scrambled siRNA were injected through the tail vein of nude mice (5 mice per group). After 2 months, the nude mice were sacrificed by cervical dislocation. Lung tissues were collected, and pulmonary nodules were counted. ISH, H&E staining, and immunohistochemistry were performed to examine the expression of circCAMSAP1, SERPINH1 and c-Myc.
Then, cells were subcutaneously injected into the armpit of nude mice. The size of the subcutaneous tumor nodule was measured every 3 days. After 27 days, the nude mice were and the subcutaneous tumor size was measured. Tumor volume, size, and weight and the body weight of the nude mice were measured. ISH, H&E staining, or immunohistochemistry were performed to examine the expression of circCAMSAP1, SERPINH1, and c-Myc. All animal studies were approved by the Ethics Committee of the Xiangya Hospital, Central South University.
Statistical analysis
The GraphPad Prism 8 (https://www.graphpad.com/scientific-software/prism/) was used for all statistical analyses in this study. The unpaired t-test was performed on each group of data, and a p-value of < 0.05 was considered to show a significant difference. Image Pro Plus v.6.0 (Media Cybernetics, Inc. Rockville, USA) was used to analyze the images from the wound-healing and transwell assays. Image J software (https://imagej.nih.gov/ij/download.html) was used to analyze the images from western blotting.
Results
circCAMSAP1 is highly expressed in NPC
To evaluate the role of circRNAs in the development of NPC, the high-throughput RNA sequencing (GSE68799) of NPC including 41 NPC tissues and 4 chronically inflamed nasopharyngeal epithelium tissues was reanalyzed. A total of 568 circRNAs that are not expressed in normal nasopharyngeal tissues but are expressed in NPC tissues were obtained according to their spliced RPM values. The top 20 circRNAs expressed in NPC tissues were selected for subsequent analysis by Sanger sequencing (Table S5). Notably, hsa_circ_0001900, which is located on chromosome 9 (chr9:138773478–138774923), was not reported in NPC (Fig. S1A).
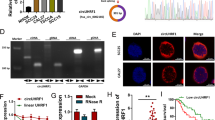
To verify the expression of circCAMSAP1 in NPC, 29 NPC tissues and 12 chronic rhinitis epithelial tissues (as normal controls) were used for RT-qPCR. The results showed that circCAMSAP1 was expressed significantly more in the 29 NPC samples (Fig. 1A) than in the 12 non-cancer nasopharyngeal epithelial (NPE) tissues. Moreover, the expression of circCAMSAP1 was significantly correlated with a high T stage (p < 0.05) in NPC patients. However, there was no significant correlation between expression of circCAMSAP1 and N stage classification (Fig. S1B).
circCAMSAP1 is highly expressed in NPC. A The expression of circCAMSAP1 was measured in 29 NPC tissues and 12 non-cancerous NPE tissue samples by RT-qPCR. β-actin was used as an internal reference. NPE, nasopharyngeal epithelium; NPC, nasopharyngeal carcinoma. ***, p < 0.001. B Representative images of circCAMSAP1 in 82 NPC tissues and 29 adjacent NPE tissues by in situ hybridization. × 200, scale bar = 50 μm; × 400, scale bar = 20 μm. The statistical data was shown in the right panel. ****, p < 0.0001. C The stability of circCAMSAP1 was detected in three RNase R-treated NPC cells by RT-qPCR. CAMSAP1 mRNA was used as a negative control. ***, p < 0.001; ****, p < 0.0001. D The stability of circCAMSAP1 was examined in NPC cells after treating with actinomycin D for 0, 6, 12 and 24 h. The relative RNA levels of circCAMSAP1 and CAMSAP1 mRNA in NPC cells were measured by RT-qPCR. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. E Intracellular localization of circCAMSAP1 (red) in three NPC cells, as determined by fluorescence in situ hybridization using a digoxigenin-labeled circCAMSAP1 probe. Nuclei were stained with DAPI (blue). Scale bar = 20 μm. F The cellular localization of circCAMSAP1 was examined in three NPC cells by nuclear and cytoplasmic separation test. GAPDH and U6 were used as positive controls for cytoplasm and nucleus, respectively
Then, the high expression of circCAMSAP1 in NPC tissue was further confirmed in 82 NPC and 29 non-cancer NPE paraffin sections using ISH. The data showed that circCAMSAP1 was expressed significantly more in NPC tissue samples (Fig. 1B) and the high expression of circCAMSAP1 was significantly correlated with high T stage (p < 0.01), N stage (p < 0.0001), and M stage (p < 0.001) classification in NPC patients (Fig. S1C). Furthermore, circCAMSAP1 was also found to be more highly expressed in NPC cell lines than in immortalized NPE cell line NP69 by RT-qPCR (Fig. S1D). Hence, circCAMSAP1 expression could serve as a prognostic biomarker for NPC.
The reverse splicing of exons 2 and 3 of the CAMSAP1 gene produces circCAMSAP1 as seen in the circBase database (Fig. S1E). Moreover, circRNAs are more stable than intracellular mRNA. As shown in Fig. 1C, resistance to digestion by RNase R exonuclease confirmed that circCAMSAP1 is indeed circular. The results of actinomycin D assays revealed that the half-life of the circCAMSAP1 transcript is longer than the half-life of the CAMSAP1 mRNA, indicating that circCAMSAP1 is more stable than the linear CAMSAP1 transcript (Fig. 1D). RNA FISH analysis and RNA nuclear and cytoplasmic isolation showed that circCAMSAP1 was distributed predominantly in the cytoplasm of NPC (Fig. 1E, F). Hence, circCAMSAP1 was selected for the evaluation of NPC tumorigenesis in the present study.
circCAMSAP1 promotes the metastasis and proliferation of NPC cells in vitro
To investigate the potential functions of circCAMSAP1 in regulating the biological behavior of NPC cells, two siRNAs for circCAMSAP1 were first designed to target the back-splice site of circCAMSAP1. The circCAMSAP1 overexpression vector was also constructed. RT-qPCR confirmed the overexpression or knockdown efficiency of the circCAMSAP1 overexpression vector or two siRNAs in CNE2, HONE1, and HNE2 cells after transfection (Fig. S2A). Wound-healing and transwell assays demonstrated that circCAMSAP1 upregulation significantly enhances migration and invasion of CNE2, HONE1, and HNE2 cells, whereas circCAMSAP1 downregulation inhibits cell migration and invasion (Fig. 2A-B, Fig. S2B). MTT assays showed that circCAMSAP1 overexpression markedly increases proliferation and viability, while circCAMSAP1 knockdown produces the opposite effect (Fig. 2C, Fig. S2C). Cell cycle analysis revealed that overexpression of circCAMSAP1 leads to increased rates of NPC cells in G2/M phases, suggesting that upregulation of circCAMSAP1 results in G2/M arrest in NPC cells (Fig. 2D, Fig. S2D). These results show that circCAMSAP1 promotes the development of NPC by promoting migration, invasion, and proliferation of NPC cells.
circCAMSAP1 promotes proliferation, migration, and invasion of NPC cells in vitro. Wound-healing assay (A), transwell invasion (B), MTT assays (C), and flow cytometry analysis of cell cycle distribution (D) in the empty vector, the circCAMSAP1 overexpression vector, two circCAMSAP1 siRNAs, and scramble siRNAs transfected NPC cells. EV, empty vector; OE, the overexpression vector; NC, scrambled negative control; Data are shown as means ± standard deviation (SD). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001
circCAMSAP1 promotes the metastasis and proliferation of NPC cells in vivo
To investigate the cancer-promoting role of circCAMSAP1 in vivo, we further evaluated the effect of circCAMSAP1 silencing and overexpression on NPC progression in xenografts. The nude mouse lung metastasis model showed that the number of intrahepatic metastatic nodules was much lower in the mice inoculated with the sicircCAMSAP1/CNE2 cells than mice transplanted with the negative control/CNE2 cells. While mice transplanted with the circCAMSAP1/CNE2 cells showed more intrahepatic metastatic nodules than that with the negative control/CNE2 cells (Fig. 3A-B).
circCAMSAP1 promotes proliferation and metastasis of NPC cells in vivo. A Representative images of nude mice lung tissues after tail vein injection of NPC cells. Mice were injected with NPC cells after overexpression or knockdown of circCAMSAP1 (Left). The number of nodules on the surface of lung tissues were counted and analyzed (Right). n = 5; ***, p < 0.001; ****, p < 0.0001. B Representative images of hematoxylin and eosin-stained metastatic lung nodules after overexpression or knockdown of circCAMSAP1. × 40, scale bar = 200 μm; × 100, scale bar = 100 μm; × 200, scale bar = 50 μm. C Growth curve of subcutaneous tumor volume in nude mice. Nude mice were injected with NPC cells subcutaneously in the armpit after overexpression or knockdown of circCAMSAP1. n = 5; *, p < 0.05; **, p < 0.01. D-F Image of subcutaneous tumor tissues in nude mice (D), tumor volume (E) and tumor weights (F) show that circCAMSAP1 promoted CNE2 xenograft growth 27 days after subcutaneous injection. n = 5; *, p < 0.05; **, p < 0.01; ***, p < 0.001
The subcutaneous tumor model of nude mice was constructed by injecting CNE2 into the armpit of nude mice. The data showed that the volume and weight of the tumors formed in the circCAMSAP1 knockdown group were significantly lower than those of the tumors in the group treated with the negative control, while circCAMSAP1 overexpression produced the opposite effect (Fig. 3C-F). Therefore, we conclude that circCAMSAP1 may act as an oncogene and promote NPC growth and metastasis.
circCAMSAP1 stabilizes the expression of SERPINH1 by binding to its 3’UTR
High-throughput mass spectrometry was carried out to explore the function of circCAMSAP1 in NPC and identify the possible signaling pathway by which circCAMSAP1 promotes proliferation, migration, and invasion in CNE2 cells after overexpression or knockdown of circCAMSAP1. A total of 2415 protein molecules were analyzed by mass spectrometry, 97 protein molecules were positively associated with circCAMSAP1 expression, while 146 protein molecules were negatively associated with circCAMSAP1 expression (Table S6, Fig. S3A). To elucidate the regulatory mechanism of the influence of circCAMSAP1 on signaling, the expression of the 97 protein molecules positively associated with circCAMSAP1 was analyzed from the NPC microarray data (GSE12452, GSE53819). Among them, SERPINH1, a molecular chaperone which has been reported to be closely related to tumor proliferation and metastasis [21] was highly expressed in NPC tissues (Fig. S3B). Our RT-qPCR results also showed that SERPINH1 mRNA was highly expressed in the 29 NPC tissues and correlated with the expression of circCAMSAP1 (Fig. S3C-D).
To depict other mechanisms that are responsible for the relationship between circCAMSAP1 and SERPINH1 in NPC, western blotting analysis, and RT-qPCR were performed in NPC cells after overexpression or knockdown of circCAMSAP1. The data showed that circCAMSAP1 can positively regulate the protein and mRNA expression of SERPINH1 (Fig. 4A-B). RNA pull-down showed that circCAMSAP1 is unable to bind directly to the SERPINH1 protein (Fig. S3E). Additionally, bioinformatics analysis using the RNA22 software showed that circCAMSAP1 has a continuous binding sequence to the 3’UTR of the SERPINH1 mRNA (Fig. S3F). In eukaryotic cells, the 3’UTR of mRNAs plays a role in maintaining mRNA stability [13]. Thus, the binding of circCAMSAP1 to the 3’UTR of SERPINH1 mRNA may delay the degradation of SERPINH1 mRNA by nucleic acid exonucleases, ultimately leading to elevated SERPINH1 expression. The dual-luciferase reporter gene assay showed that circCAMSAP1 could regulate the luciferase activity of the SERPINH1 3’UTR (Fig. 4C). The circRIP assay confirmed that circCAMSAP1 binds to the 3’UTR of SERPINH1 mRNA in NPC cells (Fig. 4D), and overexpression of circCAMSAP1 enhances the stability of SERPINH1, while knockdown of circCAMSAP1 gives the opposite result in NPC cells after actinomycin D treatment (Fig. 4E).
circCAMSAP1 stabilizes the expression of SERPINH1 by binding to its 3’UTR. A Western blotting analysis of SERPINH1 expression in three NPC cell lines after overexpression and knockdown of circCAMSAP1. B RT-qPCR analysis of SERPINH1 expression in three NPC cell lines after overexpression and knockdown of circCAMSAP1. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. C The luciferase reporter gene activity was analyzed in three NPC cell lines after overexpression or knockdown of circCAMSAP1. The wild-type (WT) or mutant (MT) of the SERPINH1 3’UTR were constructed and transfected into NPC cells. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. D The circRIP assay was used to detect the binding of circCAMSAP1 to the 3’UTR of SERPINH1 mRNA. **, p < 0.01; ****, p < 0.0001. E The stability of SERPINH1 mRNA was measured in NPC cells after overexpression or knockdown of circCAMSAP1. Cells were treated with actinomycin D for 0, 60, 120 min and RT-qPCR was used to measure the expression of SERPINH1 mRNA. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001
To illustrate whether SERPINH1 is critical for circCAMSAP1-induced growth, migration, and invasion of NPC cells, we overexpressed or knocked down circCAMSAP1 or SERPINH1 in NPC cells. The results revealed that overexpression of SERPINH1 promotes NPC cell growth, migration, and invasion, while knockdown of SERPINH1 has the opposite effect (Fig. S4). Specifically, knockdown of SERPINH1 significantly alleviates the promotive effect of circCAMSAP1 overexpression on proliferation (Fig. 5C, Fig. S5B), migration (Fig.5A, Fig. S5A) and invasion (Fig. 5B) of NPC cells. Additionally, overexpression of SERPINH1 partially restored the inhibitory effect of circCAMSAP1 knockdown on proliferation (Fig. 5C, Fig. S5B), migration (Fig.5A, Fig. S5A), and invasion (Fig. 5B) of NPC cells. Collectively, these data demonstrated that circCAMSAP1 increases the expression of SERPINH1 and promotes cell proliferation and metastasis of NPC cells by binding to the 3’UTR of SERPINH1.
circCAMSAP1 promotes proliferation, migration, and invasion of NPC through SERPINH1. Wound-healing assay (A), transwell invasion (B), and MTT assays (C) in NPC cells after knockdown or overexpression of circCAMSAP1 and SERPINH1. Data are shown as means ± SD. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001
SERPINH1 inhibits c-Myc’s ubiquitination-degradation
To identify the possible function of SERPINH1 on the proliferation, migration, and invasion of NPC cells regulated by circCAMSAP1, four different interaction databases (HitPredict, BioGRID, UniHI, and GeneMANIA) were used to identify the proteins that interact with SERPINH1. We then selected c-Myc because it was the only protein predicted to bind to SERPINH1 by all four interaction databases and it plays an important role in tumorigenesis and development [22]. Immunoprecipitation analysis (Fig. 6A) showed that SERPINH1 can bind to c-Myc in NPC cells and immunofluorescence showed the co-localization between c-Myc and SERPINH1 (Fig. 6B). Western blotting showed that SERPINH1 could positively regulate the expression of c-Myc (Fig. S6A).
SERPINH1 binds to c-Myc and inhibits its ubiquitination-degradation. A Co-IP assay was shown the interaction between SERPINH1 and c-Myc in three NPC cell lines. B Co-localization of SERPINH1 and c-Myc protein in NPC cells was determined by immunofluorescence. Scale bars = 20 μm. C Western blotting analysis of c-Myc expression in CNE2 cells after overexpression of circCAMSAP1 and knockdown of SERPINH1 or circCAMSAP1 or overexpression of SERPINH1. D Western blotting analysis of the c-Myc protein stability in NPC cells after treatment with cycloheximide (CHX). E Western blotting analysis of the ubiquitination of c-Myc protein after overexpression of circCAMSAP1 in NPC cells. F The phosphorylation level of c-Myc T58 and S62 phosphorylation sites was measured by western blotting in CNE2 cells after overexpression of circCAMSAP1 and knockdown of SERPINH1, or knockdown of circCAMSAP1 and overexpression of SERPINH1. G The nuclear c-Myc expression was examined in NPC cells after overexpression of circCAMSAP1 in CNE2 cells by western blotting. H The transcript activity of c-Myc was measured in NPC cells after overexpression of circCAMSAP1 and knockdown of SERPINH1, or knockdown of circCAMSAP1 and overexpression of SERPINH1. ***, p < 0.001; ****, p < 0.0001
To identify whether circCAMSAP1 participates in the interaction of SERPINH1 with c-Myc, circCAMSAP1 or SERPINH1 was overexpressed or knocked down in NPC cells. Western blotting showed that knockdown of SERPINH1 could inhibit the upregulation of c-Myc expression caused by overexpression of circCAMSAP1, while overexpression of SERPINH1 could improve downregulation of c-Myc expression caused by circCAMSAP1 (Fig. 6C, Fig. S6B). RT-qPCR assays further showed that circCAMSAP1 and SERPINH1 had no effect on the mRNA expression level of c-Myc (Fig. S6C). This suggests that the effect of circCAMSAP1 and SERPINH1 on the expression of c-Myc may be at the post-translational level.
We then treated NPC cells with cycloheximide (CHX) to identify the effect of circCAMSAP1 and SERPINH1 on the stability of the c-Myc protein in NPC cells. The results showed that circCAMSAP1 and SERPINH1 could inhibit the degradation of the c-Myc protein (Fig. 6D, Fig. S7A).
A common degradation pathway of c-Myc is through ubiquitination. The degree of ubiquitination of the protein is estimated by the rate of binding of c-Myc to the ubiquitinated protein [23]. When NPC cells are treated with MG132, a proteasome inhibitor, overexpression of circCAMSAP1 inhibits c-Myc ubiquitination (Fig. 6E), while inhibition of SERPINH1 promotes c-Myc ubiquitination (Fig. S7B). The change of c-Myc phosphorylation level is tightly correlated to the ubiquitination-degradation of c-Myc [24]. The phosphorylation level of c-Myc at the T58 and S62 sites, and the results showed that overexpression of SERPINH1 could inhibit the phosphorylation of c-Myc, while knockdown of SERPINH1 has the opposite result (Fig. S7C). Knockdown of SERPINH1 can promote the downregulation of c-Myc phosphorylation caused by overexpression of circCAMSAP1, while overexpression of SERPINH1 can inhibit the upregulation of c-Myc phosphorylation caused by circCAMSAP1 knockdown (Fig. 6F, Fig. S7D).
As a transcription factor, c-Myc regulates the expression of many proteins, plays an important role in cell proliferation, cycle regulation, invasion, and metastasis, and affects the progression of many tumors [25]. Therefore, we tested whether circCAMSAP1 and SERPINH1 can increase the abundance of c-Myc expression in the nucleus through the nuclear-plasmin separation protein analysis. The results showed that circCAMSAP1 and SERPINH1 could increase c-Myc expression in the nucleus (Fig. 6G, Fig. S8A). The c-Myc transcription factor usually works with a promoter containing E-box (5′-CACGTG-3′), so we used a luciferase reporter plasmid containing three E-boxes to explore the effects of circCAMSAP1 and SERPINH1 on the transcriptional activity of c-Myc. The results showed that overexpression of SERPINH1 could promote the transcriptional activity of c-Myc in NPC cells, while knockdown of SERPINH1 has the opposite result (Fig. S8B). Furthermore, knockdown of SERPINH1 could inhibit the promotion of c-Myc transcriptional activity by circCAMSAP1, while overexpression of SERPINH1 could decrease c-Myc transcriptional activity caused by knockdown of circCAMSAP1 (Fig. 6H).
The expression of circCAMSAP1, SERPINH1 and c-Myc in nude mice lung metastases and subcutaneous tumors was also analyzed. ISH and immunohistochemistry showed that circCAMSAP1, SERPINH1, and c-Myc were highly expressed and positively correlated in lung metastases and subcutaneous tumors in the overexpression group, but low in the knockdown group, compared with the negative control group (Fig. S9).
c-Myc combined with SRSF10 promotes circCAMSAP1 expression
To explore the mechanism of high expression of circCAMSAP1 in NPC tissues, the expression of circCAMSAP1 was examined in NPC cells after overexpressing or knocking down c-Myc (Fig. S10A-B). It was found that c-Myc positively regulates the expression of circCAMSAP1, CAMSAP1 pre-mRNA, and CAMSAP1 mRNA (Fig. 7A-B, Fig. S10C) in NPC cells. The expression of CAMSAP1 mRNA levels in NPC tissues may be up-regulated (Fig. S10D). Bioinformatics software (PROMO and JASPER databases) showed that c-Myc could bind near the positions -1391 bp and -1211 bp of the CAMSAP1 promoter (Fig. S10E). The luciferase reporter confirmed that c-Myc binds to the CAMSAP1 promoter region (-1450 bp- -851 bp) (Fig. 7C). ChIP analysis further showed that c-Myc mainly binds to nucleotide -1391 bp and-1211 bp in the CAMSAP1 promoter (Fig. 7D).
c-Myc combines with SRSF10 to promote the expression of circCAMSAP1. A The expression of CAMSAP1 pre-mRNA was examined by RT-qPCR in NPC cells after overexpression or knockdown of c-Myc. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. B The expression of circCAMSAP1 was examined by RT-qPCR in NPC cells after overexpression or knockdown of c-Myc. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. C The luciferase reporter gene activity of the CAMSAP1 promoter was analyzed in NPC cells after overexpression of c-Myc. ***, p < 0.001; ****, p < 0.0001. D The binding of transcription factor c-Myc to the CAMSAP1 promoter detected by ChIP assay. Two sites on the CAMSAP1 promoter were selected according to the c-Myc binding sites (site 1 and site 2) using the Jasper and PROMO software. A non-binding region (site 3) was used as a negative control. **, p < 0.01; ***, p < 0.001. E The expression of circCAMSAP1 was examined by RT-qPCR in NPC cells after overexpression or knockdown of SRSF10. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. F SRSF10 bound to the flanking intron sequence of circCAMSAP1 detected by RIP assay. Intron 1 and intron 3 were selected for RIP experiments using the SRSF10 antibody and referred to the SRSF10 binding site of CAMSAP1 pre-mRNA. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. G The expression of SRSF10 was examined by RT-qPCR in NPC cells after knockdown of c-Myc. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. H The SRSF10 expression in NPC cells after knockdown of c-Myc examined by western blotting
CircRNA is produced by alternative splicing of pre-mRNA [9]. Splicing factors act on the RNA surrounding the circRNA sequence on the pre-mRNA strand to promote the occurrence of splicing and increase the expression of circRNA [26]. We used the RBPmap database to predict splicing factors that may bind to the introns flanking exons 2 and 3 of CAMSAP1 pre-mRNA. SRSF10 had been predicted binding to the introns flanking exons 2 and 3 of CAMSAP1 pre-mRNA and was also highly expressed in NPC cells (Fig. S10F). When SRSF10 is overexpression or knocked down in NPC cells (Fig. S10G), the expression of circCAMSAP1 is increased or decreased (Fig. 7E). RIP assay confirmed the binding of SRSF10 to the flanking introns of circCAMSAP1 (Fig. 7F). Moreover, c-Myc could also promote the expression of SRSF10 (Fig. 7G-H). Taken together, these results indicate that c-Myc is involved in the transcription and formation of circCAMSAP1, cooperating with SRSF10. The positive feedback of the circCAMSAP1-SERPINH1-c-Myc axis may play crucial roles in NPC progression.
Discussion
The local recurrence and distant metastasis of NPC are major obstacles in the clinical treatment of NPC. Future research should be aimed to explore the molecular mechanism of NPC metastasis and proliferation and find the molecules active in the mechanism [27, 28]. Recent studies have shown that non-coding RNA, including microRNA [29], long non-coding RNA [30], and circRNA [31] play an important role in the occurrence and development of NPC. Among them, circCRIM1 [32], circTGFBR2, circHIPK3 [33] and circ_0046263 [34] play an important role in the metastasis and proliferation of NPC. Our research group found that circSETD3 acts as a molecular sponge to absorb miR-615-5p and miR-1538, which promotes the expression of MAREP1 and the metastasis of NPC [20]. circARHGAP12 promotes the metastasis of NPC by regulating the Ezrin complex [17]. Furthermore, circRNF13 inhibits the metastasis and proliferation of NPC via regulating the cell metabolic reprogramming [19]. However, research on the mechanism of circRNAs in the development of NPC is still insufficient.
In this study, we identified a large number of circRNAs through RNA sequencing of NPC clinical samples. We focused on circCAMSAP1 because of its high expression in NPC and found it was significantly correlated with high TNM stages in NPC. To further explore the relationship between the expression of circCAMSAP1 and the prognosis of NPC patients, we will conduct follow-up analyses of the cases, especially multicenter analyses with large samples. More samples should also be collected to verify whether circCAMSAP1 can be used as a new marker for liquid biopsy of patients with nasopharyngeal cancer.
It has been reported that circCAMSAP1 promotes tumor proliferation or metastasis in colorectal cancer [35], osteosarcoma [36], and liver cancer [37] through the competitive endogenous RNA (ceRNA) mechanism. In this study, we found that circCAMSAP1 does not directly bind with the SERPINH1 or c-Myc proteins, but circCAMSAP1 can directly bind to the SERPINH1 3’UTR, and circCAMSAP1 stabilized SERPINH1 expression by directly binding to its 3’UTR, ultimately promoting the expression of the SERPINH1 protein. Moreover, circCAMSAP1 may be involved in various mechanisms in tumors.
SERPINH1, a collagen synthesis protein, is required for the correct folding and secretion of collagen. Although previous studies have shown that SERPINH1 is a potential molecular marker and therapeutic target for many tumors including gastric cancer [38], hepatocellular carcinoma [21], cervical squamous cell carcinoma [39], renal cell carcinoma [40], and head and neck squamous cell carcinoma [41], the mechanism by which SERPINH1 affects tumor progression through the production of collagen is unclear. In this study, we found that SERPINH1 could inhibit c-Myc ubiquitination-degradation. Furthermore, c-Myc, as a transcription factor, can regulate the expression of many proteins, play an important role in cell proliferation, cycle regulation, invasion, and metastasis, and affect the development of many tumors including NPC [25].
c-Myc directly promotes the expression of EMT signaling pathway-related proteins [42] and cell cycle-related proteins [43]. Meanwhile, c-Myc upregulates the expression of NPC-related proteins, such as BCAT1 [44], BMI1 [45] and BRD7 [46], to indirectly aggravate NPC development. The c-Myc-induced overexpression of non-coding RNAs contributes to the regulation of downstream genes, further enhancing the metastasis and proliferation of NPC [46,47,48,49,50]. c-Myc also increases the levels of metabolic-related proteins for the execution of metabolic reprogramming, which in turn leads to metastasis and proliferation [51, 52]. Furthermore, c-Myc interacts with signaling networks associated with NPC metastasis and proliferation, including the PI3K/Akt [53] and JNK/c-JUN [54, 55] pathways, exacerbating the malignant progression of NPC.
Since c-Myc has many target genes, we continued to analyze the differentially expressed proteins in NPC cells obtained from the proteomics platform after overexpression or knockdown of circCAMSAP1. Among them, circCAMSAP1 up-regulated 97 proteins and down-regulated 146 proteins. The functional and signaling pathway enrichment were conducted through the Metascape database and DAVID database (Table S7). The results show that some proteins functionally enriched to cell-cell adherens junctions [56], the cell cycle, and G2/M checkpoints [57], whereas signaling pathway enrichment included the Rho GTPase signaling pathway [58], VEGFA/VEGFR signaling pathway [59], noncanonical NF-κB signaling [60], WNT/β-catenin signaling pathway [61], and PI3K/Akt signaling pathway [53], all of which are closely related to tumor metastasis and proliferation. In addition, the function of differential proteins is closely related to glycolysis/gluconeogenesis [19] and amino acid metabolism [62], promoting the malignant progression of tumors through metabolic reprogramming. Therefore, circCAMSAP1 may either affect the expression of metastatic proliferation-related proteins and pathways through SERPINH1-c-Myc, or regulate metabolic reprogramming, which in turn affects the malignant progression of NPC. Further research concerning the specific mechanisms and targets of circCAMSAP1/SERPINH1/c-Myc axis should be addressed in the future.
In this study, we found that c-Myc promoted the transcription of CAMSAP1 to form pre-mRNA and the expression of SRSF10. The high expression of SRSF10 could promote pre-mRNA splicing to form circCAMSAP1. The splicing process is usually completed by a complicated splicing complex [63]; we have identified that SRSF10 participates in the splicing of circCAMSAP1. Whether SRSF10 combines with other splicing factors or RBPs to form a splicing complex warrants further exploration.
Conclusions
We reported the identification of a new circRNA, circCAMSAP1 that has potent oncogenic activities in NPC (Fig. 8). Further, we demonstrated that circCAMSAP1 promotes the growth and aggressiveness of NPC cells by stabilizing SERPINH1 expression through binding to its SERPINH1 3’UTR. Additionally, the binding of SERPINH1 to c-Myc inhibited the ubiquitination-degradation of c-Myc. Meanwhile, c-Myc along with SRSF10 could promote CAMSAP1 pre-mRNA transcript and back-splicing, and form a positive feedback on circCAMSAP1 production, resulting in proliferation and metastasis of NPC. This study expands the understanding of circRNA function in NPC pathogenesis and suggests a novel circRNA as a potential biomarker and therapeutic target for NPC.
Schematic illustration of circCAMSAP1 promoting proliferation and metastasis of NPC through a positive SERPINH1/c-Myc feedback. circCAMSAP1 stabilizes SERPINH1 expression by binding to the SERPINH1 3’UTR. The binding of SERPINH1 to c-Myc inhibits the ubiquitination and degradation of c-Myc, leading to CAMSAP1 pre-mRNA transcription and back-splicing under the action of SRSF10, forming a positive feedback on circCAMSAP1 production, resulting in proliferation, migration, and invasion of NPC
Availability of data and materials
All data that support the findings of this study are available from the corresponding authors upon reasonable request.
Abbreviations
- FISH:
-
Fluorescence in situ hybridization
- GEO:
-
Gene Expression Omnibus
- ISH:
-
In situ hybridization
- NPC:
-
Nasopharyngeal carcinoma
- NPE:
-
Nasopharyngeal epithelial
- RPM:
-
Reads per million
- WT:
-
Wild type
- ChIP:
-
chromatin immunoprecipitation
- CHX:
-
cycloheximide
- CircRNA:
-
Circular RNA
- DAPI:
-
6-diamidino-2-phenylindole
- EV:
-
empty vector
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- MTT:
-
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide
- NC:
-
scrambled negative control
- OE:
-
overexpression vector
- RBP:
-
RNA-binding proteins
- RIP:
-
RNA immunoprecipitation
- TFA:
-
trifluoroacetic acid
- TNM:
-
tumor (T), nodes (N), and metastases (M)
References
Zeng Z, Huang H, Zhang W, Xiang B, Zhou M, Zhou Y, et al. Nasopharyngeal carcinoma: advances in genomics and molecular genetics. Sci China Life Sci. 2011;54(10):966-75.
Wei J, Ye J, Luo Y, Weng J, He Q, Liu F, et al. EB virus promotes metastatic potential by boosting STIM1-dependent Ca(2+) signaling in nasopharyngeal carcinoma cells. Cancer Lett. 2020;478:122-32.
Xiong W, Zeng ZY, Xia JH, Xia K, Shen SR, Li XL, et al. A susceptibility locus at chromosome 3p21 linked to familial nasopharyngeal carcinoma. Cancer Res. 2004;64(6):1972–4.
Hildesheim A, Wang CP. Genetic predisposition factors and nasopharyngeal carcinoma risk: a review of epidemiological association studies, 2000-2011: Rosetta stone for NPC: genetics, viral infection, and other environmental factors. Semin Cancer Biol. 2012;22(2):107-16.
Paiar F, Di Cataldo V, Zei G, Pasquetti EM, Cecchini S, Meattini I, et al. Role of chemotherapy in nasopharyngeal carcinoma. Oncol Rev. 2012;6(1):e1.
Liew K, Yu GQS, Wei Pua LJ, Wong LZ, Tham SY, Hii LW, et al. Parallel genome-wide RNAi screens identify lymphocyte-specific protein tyrosine kinase (LCK) as a targetable vulnerability of cell proliferation and chemoresistance in nasopharyngeal carcinoma. Cancer Lett. 2021;504:81-90.
Siak PY, Khoo AS, Leong CO, Hoh BP, Cheah SC. Current status and future perspectives about molecular biomarkers of nasopharyngeal carcinoma. Cancers (Basel). 2021;13(14):3490.
Wang Y, Mo Y, Gong Z, Yang X, Yang M, Zhang S, et al. Circular RNAs in human cancer. Mol Cancer. 2017;16(1):25.
Xu S, Zhou L, Ponnusamy M, Zhang L, Dong Y, Zhang Y, et al. A comprehensive review of circRNA: from purification and identification to disease marker potential. PeerJ. 2018;6:e5503.
Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H, et al. Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer. 2021;20(1):13.
Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in Cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6(4):319-36.
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79.
Zhu P, Zhu X, Wu J, He L, Lu T, Wang Y, et al. IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3. Nat Immunol. 2019;20(2):183-94.
Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19(1):218.
Li B, Zhu L, Lu C, Wang C, Wang H, Jin H, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun. 2021;12(1):295.
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 circular RNA in repressing Glioma tumorigenesis. J Natl Cancer Inst. 2018;110(3):304–15.
Fan C, Qu H, Xiong F, Tang Y, Tang T, Zhang L, et al. CircARHGAP12 promotes nasopharyngeal carcinoma migration and invasion via ezrin-mediated cytoskeletal remodeling. Cancer Lett. 2021;496:41-56.
Ge J, Wang J, Xiong F, Jiang X, Zhu K, Wang Y, et al. Epstein-Barr virus-encoded circular RNA circBART2.2 promotes immune escape of nasopharyngeal carcinoma by regulating PD-L1. Cancer Res. 2021;81(19):5074–88.
Mo Y, Wang Y, Zhang S, Xiong F, Yan Q, Jiang X, et al. Circular RNA circRNF13 inhibits proliferation and metastasis of nasopharyngeal carcinoma via SUMO2. Mol Cancer. 2021;20(1):112.
Tang L, Xiong W, Zhang L, Wang D, Wang Y, Wu Y, et al. circSETD3 regulates MAPRE1 through miR-615-5p and miR-1538 sponges to promote migration and invasion in nasopharyngeal carcinoma. Oncogene. 2021;40(2):307-21.
Wu G, Ju X, Wang Y, Li Z, Gan X. Up-regulation of SNHG6 activates SERPINH1 expression by competitive binding to miR-139-5p to promote hepatocellular carcinoma progression. Cell Cycle. 2019;18(16):1849-67.
Rong Z, Shi S, Tan Z, Xu J, Meng Q, Hua J, et al. Circular RNA CircEYA3 induces energy production to promote pancreatic ductal adenocarcinoma progression through the miR-1294/c-Myc axis. Mol Cancer. 2021;20(1):106.
Geng C, Kaochar S, Li M, Rajapakshe K, Fiskus W, Dong J, et al. SPOP regulates prostate epithelial cell proliferation and promotes ubiquitination and turnover of c-MYC oncoprotein. Oncogene. 2017;36(33):4767-77.
Zhao R, Liu Y, Wu C, Li M, Wei Y, Niu W, et al. BRD7 promotes cell proliferation and tumor growth through stabilization of c-Myc in colorectal Cancer. Front Cell Dev Biol. 2021;9:659392.
Xu Y, Zhang G, Zou C, Qi W, Gong Z, Zhang G, et al. Long non-coding RNA LINC01225 promotes proliferation, invasion and migration of gastric cancer via Wnt/beta-catenin signalling pathway. J Cell Mol Med. 2019;23(11):7581-91.
Zheng X, Huang M, Xing L, Yang R, Wang X, Jiang R, et al. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer. 2020;19(1):73.
Zhou L, Liu R, Liang X, Zhang S, Bi W, Yang M, et al. lncRNA RP11-624L4.1 is associated with unfavorable prognosis and promotes proliferation via the CDK4/6-Cyclin D1-Rb-E2F1 pathway in NPC. Mol Ther Nucleic Acids. 2020;22:1025-39.
Zhao CX, Zhu W, Ba ZQ, Xu HJ, Liu WD, Zhu B, et al. The regulatory network of nasopharyngeal carcinoma metastasis with a focus on EBV, lncRNAs and miRNAs. Am J Cancer Res. 2018;8(11):2185-209.
Lee KT, Tan JK, Lam AK, Gan SY. MicroRNAs serving as potential biomarkers and therapeutic targets in nasopharyngeal carcinoma: a critical review. Crit Rev Oncol Hematol. 2016;103:1-9.
Tang Y, He X. Long non-coding RNAs in nasopharyngeal carcinoma: biological functions and clinical applications. Mol Cell Biochem. 2021;476(9):3537-50.
Yang M, Huang W. Circular RNAs in nasopharyngeal carcinoma. Clin Chim Acta. 2020;508:240-8.
Hong X, Liu N, Liang Y, He Q, Yang X, Lei Y, et al. Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol Cancer. 2020;19(1):33.
Ke Z, Xie F, Zheng C, Chen D. CircHIPK3 promotes proliferation and invasion in nasopharyngeal carcinoma by abrogating miR-4288-induced ELF3 inhibition. J Cell Physiol. 2019;234(2):1699-706.
Yin L, Chen J, Ma C, Pei S, Du M, Zhang Y, et al. Hsa_circ_0046263 functions as a ceRNA to promote nasopharyngeal carcinoma progression by upregulating IGFBP3. Cell Death Dis. 2020;11(7):562.
Zhou C, Liu HS, Wang FW, Hu T, Liang ZX, Lan N, et al. circCAMSAP1 promotes tumor growth in colorectal Cancer via the miR-328-5p/E2F1 Axis. Mol Ther. 2020;28(3):914-28.
Chen Z, Xu W, Zhang D, Chu J, Shen S, Ma Y, et al. circCAMSAP1 promotes osteosarcoma progression and metastasis by sponging miR-145-5p and regulating FLI1 expression. Mol Ther Nucleic Acids. 2021;23:1120-35.
Luo Z, Lu L, Tang Q, Wei W, Chen P, Chen Y, et al. CircCAMSAP1 promotes hepatocellular carcinoma progression through miR-1294/GRAMD1A pathway. J Cell Mol Med. 2021;25(8):3793-802.
Tian S, Peng P, Li J, Deng H, Zhan N, Zeng Z, et al. SERPINH1 regulates EMT and gastric cancer metastasis via the Wnt/β-catenin signaling pathway. Aging (Albany NY). 2020;12(4):3574–93.
Yamamoto N, Kinoshita T, Nohata N, Yoshino H, Itesako T, Fujimura L, et al. Tumor-suppressive microRNA-29a inhibits cancer cell migration and invasion via targeting HSP47 in cervical squamous cell carcinoma. Int J Oncol. 2013;43(6):1855-63.
Qi Y, Zhang Y, Peng Z, Wang L, Wang K, Feng D, et al. SERPINH1 overexpression in clear cell renal cell carcinoma: association with poor clinical outcome and its potential as a novel prognostic marker. J Cell Mol Med. 2018;22(2):1224-35.
Fan G, Tu Y, Wu N, Xiao H. The expression profiles and prognostic values of HSPs family members in head and neck cancer. Cancer Cell Int. 2020;20:220.
Su B, Xu T, Bruce JP, Yip KW, Zhang N, Huang Z, et al. Hsa-miR-24 suppresses metastasis in nasopharyngeal carcinoma by regulating the c-Myc/epithelial-mesenchymal transition axis. Oncol Rep. 2018;40(5):2536-46.
Niu Z, Liu H, Zhou M, Wang H, Liu Y, Li X, et al. Knockdown of c-Myc inhibits cell proliferation by negatively regulating the Cdk/Rb/E2F pathway in nasopharyngeal carcinoma cells. Acta Biochim Biophys Sin Shanghai. 2015;47(3):183-91.
Zhou W, Feng X, Ren C, Jiang X, Liu W, Huang W, et al. Over-expression of BCAT1, a c-Myc target gene, induces cell proliferation, migration and invasion in nasopharyngeal carcinoma. Mol Cancer. 2013;12:53.
Wang HB, Liu GH, Zhang H, Xing S, Hu LJ, Zhao WF, et al. Sp1 and c-Myc regulate transcription of BMI1 in nasopharyngeal carcinoma. FEBS J. 2013;280(12):2929-44.
Liu Y, Zhao R, Wei Y, Li M, Wang H, Niu W, et al. BRD7 expression and c-Myc activation forms a double-negative feedback loop that controls the cell proliferation and tumor growth of nasopharyngeal carcinoma by targeting oncogenic miR-141. J Exp Clin Cancer Res. 2018;37(1):64.
Huang W, Song W, Jiang Y, Chen L, Lu H. c-Myc-induced circ-NOTCH1 promotes aggressive phenotypes of nasopharyngeal carcinoma cells by regulating the miR-34c-5p/c-Myc axis. Cell Biol Int. 2021;45(7):1436-47.
Xing H, Sun H, Du W. LINC01116 accelerates nasopharyngeal carcinoma progression based on its enhancement on MYC transcription activity. Cancer Med. 2020;9(1):269-77.
Li M, Liu Y, Wei Y, Wu C, Meng H, Niu W, et al. Zinc-finger protein YY1 suppresses tumor growth of human nasopharyngeal carcinoma by inactivating c-Myc-mediated microRNA-141 transcription. J Biol Chem. 2019;294(15):6172-87.
Yang J, Wu SP, Wang WJ, Jin ZR, Miao XB, Wu Y, et al. A novel miR-200c/c-myc negative regulatory feedback loop is essential to the EMT process, CSC biology and drug sensitivity in nasopharyngeal cancer. Exp Cell Res. 2020;391(2):111817.
Xiao L, Hu ZY, Dong X, Tan Z, Li W, Tang M, et al. Targeting Epstein-Barr virus oncoprotein LMP1-mediated glycolysis sensitizes nasopharyngeal carcinoma to radiation therapy. Oncogene. 2014;33(37):4568-78.
Shi F, He Y, Li J, Tang M, Li Y, Xie L, et al. Wild-type IDH2 contributes to Epstein-Barr virus-dependent metabolic alterations and tumorigenesis. Mol Metab. 2020;36:100966.
Li Y, Liu X, Lin X, Zhao M, Xiao Y, Liu C, et al. Chemical compound cinobufotalin potently induces FOXO1-stimulated cisplatin sensitivity by antagonizing its binding partner MYH9. Signal Transduct Target Ther. 2019;4:48.
Zhen Y, Liu Z, Yang H, Yu X, Wu Q, Hua S, et al. Tumor suppressor PDCD4 modulates miR-184-mediated direct suppression of C-MYC and BCL2 blocking cell growth and survival in nasopharyngeal carcinoma. Cell Death Dis. 2013;4(10):e872.
Yu X, Zhen Y, Yang H, Wang H, Zhou Y, Wang E, et al. Loss of connective tissue growth factor as an unfavorable prognosis factor activates miR-18b by PI3K/AKT/C-Jun and C-Myc and promotes cell growth in nasopharyngeal carcinoma. Cell Death Dis. 2013;4(5):e634.
Huang Z, Cheng Y, Chiu PM, Cheung FM, Nicholls JM, Kwong DL, et al. Tumor suppressor alpha B-crystallin (CRYAB) associates with the cadherin/catenin adherens junction and impairs NPC progression-associated properties. Oncogene. 2012;31(32):3709-20.
Zhang J, Wen X, Liu N, Li YQ, Tang XR, Wang YQ, et al. Epigenetic mediated zinc finger protein 671 downregulation promotes cell proliferation and tumorigenicity in nasopharyngeal carcinoma by inhibiting cell cycle arrest. J Exp Clin Cancer Res. 2017;36(1):147.
Lian Y, Xiong F, Yang L, Bo H, Gong Z, Wang Y, et al. Long noncoding RNA AFAP1-AS1 acts AS a competing endogenous RNA of miR-423-5p to facilitate nasopharyngeal carcinoma metastasis through regulating the rho/Rac pathway. J Exp Clin Cancer Res. 2018;37(1):253.
Guan X, Yu D, HuangFu M, Huang Z, Dou T, Liu Y, et al. Curcumol inhibits EBV-positive nasopharyngeal carcinoma migration and invasion by targeting nucleolin. Biochem Pharmacol. 2021;192:14742.
Yakubov B, Chelladurai B, Schmitt J, Emerson R, Turchi JJ, Matei D. Extracellular tissue transglutaminase activates noncanonical NF-κB signaling and promotes metastasis in ovarian cancer. Neoplasia. 2013;15(6):609-19.
Guan Z, Zhang J, Wang J, Wang H, Zheng F, Peng J, et al. SOX1 down-regulates β-catenin and reverses malignant phenotype in nasopharyngeal carcinoma. Mol Cancer. 2014;13:257.
Najumudeen AK, Ceteci F, Fey SK, Hamm G, Steven RT, Hall H, et al. The amino acid transporter SLC7A5 is required for efficient growth of KRAS-mutant colorectal cancer. Nat Genet. 2021;53(1):16-26.
Jia T, Jacquet T, Dalonneau F, Coudert P, Vaganay E, Exbrayat-Heritier C, et al. FGF-2 promotes angiogenesis through a SRSF1/SRSF3/SRPK1-dependent axis that controls VEGFR1 splicing in endothelial cells. BMC Biol. 2021;19(1):173.
Acknowledgements
We thank Prof. Yong Li for providing pcDNA3.1(+) CircRNA Mini Vectors and Prof. Zheng Li for the help of mass spectrometry.
Funding
This study was supported by grants from the National Natural Science Foundation of China (82073135, 82002239, 81972776, 81803025), the Natural Science Foundation of Hunan Province (2021JJ30897, 2021JJ31127, 2021JJ41027, 2021JJ41043), Central South University Graduate Research and Innovation Project (2021zzts0310).
Author information
Authors and Affiliations
Contributions
Yian Wang conceived and designed the project, completed most of the experiments and analyzed the data. Qijia Yan, Yongzhen Mo, Yuhang Liu, Yumin Wang performed some of the experiments and collected tissue samples. Yian Wang wrote the manuscript. Shanshan Zhang, Can Guo, Fuyan Wang, Guiyuan Li, Zhaoyang Zeng, Wei Xiong revised the manuscript. Wei Xiong is responsible for research supervision and funding acquisition. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Central South University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
circCAMSAP1 is highly expressed in NPC. A. Scatter plot of expression of circCAMSAP1 in GSE68799 database. NPE, n = 4; NPC, n = 41. p = 0.2320. B. The RT-qPCR results showed that circCAMSAP1 was related to the clinical stage and TN stage of patients with NPC in 12 NPE and 29 NPC samples. *, p < 0.05; **, p < 0.01; ****, p < 0.0001. C. The ISH results showed that circCAMSAP1 is related to the clinical stage and TNM stage of patients with NPC in 29 NPE and 82 NPC samples. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. D. The expression of circCAMSAP1 in NPC cell lines and immortalized NPE cell line NP69 detected by RT-qPCR. E. Sanger sequencing verified that circCAMSAP1 is composed of reverse splicing of exons 2 and 3 of CAMSAP1 mRNA, 425 bp, followed by RT-qPCR.
Additional file 2: Figure S2.
circCAMSAP1 promotes proliferation, migration, and invasion of NPC cells in vitro. A. The efficiency of overexpression or knockdown of circCAMSAP1 in NPC cell lines detected by RT-qPCR. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. B-D. Wound-healing assays (B), MTT assays (C), and the original data of flow cytometry analysis (D) in NPC cells after overexpression or knockdown of circCAMSAP1. Data are shown as means ± SD. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Additional file 3: Figure S3.
LC-MS/MS identification of downstream regulatory proteins by circCAMSAP1. A. Mass spectrometry data showed circCAMSAP1 potential regulatory proteins in CNE2 cells after overexpression or knockdown of circCAMSAP1. B. The expression of SERPINH1 in NPC microarrays GSE12452 and GSE53819. C. The expression of SERPINH1 was measured in 29 NPC tissues and 12 non-cancerous NPE tissue samples by RT-qPCR. β-actin was used as an internal reference. *, p < 0.05. D. Correlation analysis of circCAMSAP1 and SERPINH1 mRNA expression in 29 NPC tissues. p < 0.0001. E. No binding between circCAMSAP1 and SERPINH1 protein confirmed by RNA pull-down. F. circCAMSAP1 was predicted to bind to and the SERPINH1 mRNA 3’UTR using the RNA22 software.
Additional file 4: Figure S4.
SERPINH1 promotes proliferation, migration, and invasion of NPC cells in vitro. A-B. The efficiency of SERPINH1 overexpression or knockdown in NPC cell lines examined by RT-qPCR and western blotting. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. C-E. Wound-healing assay (C), transwell invasion (D), and MTT assays (E) in NPC cells after overexpression or knockdown of SERPINH1. Data are shown as means ± SD. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Additional file 5: Figure S5.
circCAMSAP1 promotes proliferation, migration, and invasion of NPC cells through SERPINH1. Wound-healing assay (A) and MTT assays (B) in NPC cells after knockdown or overexpression of circCAMSAP1 and SERPINH1. Data are shown as means ± SD. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Additional file 6: Figure S6.
circCAMSAP1 promotes c-Myc expression through SERPINH1 in NPC cells. A. The expression of c-Myc after overexpression or knockdown of SERPINH1 in NPC cell lines examined by western blotting. B. The expression of c-Myc in NPC cells after overexpression or knockdown of circCAMSAP1 and SERPINH1 examined by western blotting. C. The expression of c-Myc mRNA in NPC cells after overexpression or knockdown of circCAMSAP1 and SERPINH1 examined by RT-qPCR.
Additional file 7: Figure S7.
circCAMSAP1 inhibits c-Myc ubiquitination degradation through SERPINH1 in NPC cells. A. Western blotting analysis of the stability of c-Myc protein in SERPINH1 overexpressed NPC cell lines after treatment with CHX. B. Western blotting analysis of the ubiquitination of c-Myc protein after knockdown of SERPINH1 in NPC cell lines. C. The phosphorylation level of c-Myc T58 and S62 phosphorylation sites was measured by western blotting in NPC cell lines after overexpression or knockdown of SERPINH1. D. The phosphorylation level of c-Myc T58 and S62 phosphorylation sites was measured by western blotting in NPC cell lines after overexpression or knockdown of circCAMSAP1 and SERPINH1.
Additional file 8: Figure S8.
circCAMSAP1 promotes c-Myc to play the role of transcription factor through SERPINH1 in NPC cells. A. The nuclear expression of c-Myc was examined in NPC cell lines after overexpression of SERPINH1 in NPC cell lines by western blotting. B. The transcript activity of c-Myc was measured in NPC cells after overexpression or knockdown of SERPINH1. ***, p < 0.001; ****, p < 0.0001.
Additional file 9: Figure S9.
circCAMSAP1 promotes the metastasis and proliferation of NPC through SERPINH1/c-Myc in vivo. A. H&E staining, ISH and immunohistochemistry detect the overexpression and knockdown efficiency of circAMSAP1 and the expression of SERPINH1 and c-Myc in lung metastasis model in nude mice (left) and nude mouse subcutaneous tumor model (right). Scale bar = 20 μm. B-C. The expression of circCAMSAP1, SERPINH1, and c-Myc and the correlation analysis in nude mouse lung metastasis model and subcutaneous tumor model. *, p < 0.05; **, p < 0.01; ****, p < 0.0001.
Additional file 10: Figure S10.
c-Myc combined with SRSF10 to form a positive feedback to promote the expression of circCAMSAP1. A. The efficiency of c-Myc overexpression or knockdown in NPC cell lines detected by RT-qPCR. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. B. The efficiency of c-Myc overexpression or knockdown in NPC cell lines detected by western blotting. C. The expression of CAMSAP1 mRNA was examined by RT-qPCR in NPC cells after overexpression or knockdown of c-Myc. *, p < 0.05; **, p < 0.01; ***, p < 0.001. D. The expression of CAMSAP1 mRNA was analyzed in NPC microarrays GSE12452 and GSE53819. E. Bioinformatics analysis of the potential binding sites of c-Myc in the promoter region of CAMSAP1. F. The expression of SRSF10 was analyzed in NPC microarrays GSE12452 and GSE53819. G. The efficiency of overexpression or knockdown of SRSF10 in NPC cell lines by RT-qPCR. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Additional file 11: Table S1.
Clinicopathological data for 29 NPC tissues and 12 non-neoplastic NPE tissue samples used for RT-qPCR.
Additional file 12: Table S2.
Clinicopathological data of 82 paraffin-embedded NPC tissues and 29 non-neoplastic NPE tissues for in situ hybridization.
Additional file 13: Table S3:
Probes for siRNAs, ISH, FISH, RNA pull-down and circRIP, and primers for circRIP, RIP, ChIP experiment, the overexpression vectors, the luciferase reporter vectors, and RT-qPCR.
Additional file 14: Table S4.
List of antibodies for immunohistochemistry, western blotting, immunofluorescence, RNA pull-down, Co-immunoprecipitation, or ChIP analyses.
Additional file 15: Table S5.
The top 20 circRNAs according to their RPM values in 41 NPC tissues from the GSE68799 database.
Additional file 16: Table S6.
Mass spectrometry identification of proteins positively regulated by circCAMSAP1.
Additional file 17: Table S7.
Functional and signaling pathway enrichment of the differentially expressed proteins possibly regulated by circCAMSAP1.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, Y., Yan, Q., Mo, Y. et al. Splicing factor derived circular RNA circCAMSAP1 accelerates nasopharyngeal carcinoma tumorigenesis via a SERPINH1/c-Myc positive feedback loop. Mol Cancer 21, 62 (2022). https://doi.org/10.1186/s12943-022-01502-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12943-022-01502-2