Abstract
Background
The national policy for malaria treatment of the Democratic Republic of Congo recommends two first-line artemisinin-based combinations for the treatment of uncomplicated malaria: artesunate-amodiaquine and artemether-lumefantrine. This study investigated the presence of markers associated with resistance to the current first-line artemisinin-based combination therapy (ACT) in isolates of Plasmodium falciparum from treatment failure patients in the Democratic Republic of Congo.
Methods
From November 2018 to November 2019, dried blood spots were taken from patients returning to health centres for fever within 28 days after an initial malaria treatment in six sentinel sites of the National Malaria Control Programme across Democratic Republic of Congo. The new episode of malaria was first detected by a rapid diagnostic test and then confirmed by a real-time PCR assay to define treatment failure. Fragments of interest in pfk13 and pfcrt genes were amplified by conventional PCR before sequencing and the Pfmdr1 gene copy number was determined by a TaqMan real-time PCR assay.
Results
Out of 474 enrolled patients, 364 (76.8%) were confirmed positive by PCR for a new episode of P. falciparum malaria, thus considered as treatment failure. Of the 325 P. falciparum isolates obtained from 364 P. falciparum-positive patients and successfully sequenced in the pfk13-propeller gene, 7 (2.2%) isolates carried non-synonymous mutations, among which 3 have been previously reported (N498I, N554K and A557S) and 4 had not yet been reported (F506L, E507V, D516E and G538S). Of the 335 isolates successfully sequenced in the pfcrt gene, 139 (41.5%) harboured the K76T mutation known to be associated with chloroquine resistance. The SVMNT haplotype associated with resistance to amodiaquine was not found. None of the isolates carried an increased copy number of the pfmdr1 gene among the 322 P. falciparum isolates successfully analysed.
Conclusion
No molecular markers currently known to be associated with resistance to the first-line ACT in use were detected in isolates of P. falciparum from treatment failure patients. Regular monitoring through in vivo drug efficacy and molecular studies must continue to ensure the effectiveness of malaria treatment in Democratic Republic of Congo.
Similar content being viewed by others
Background
Plasmodium falciparum is the most widespread Plasmodium species and is responsible for most severe forms of malaria, and death related to malaria in sub-Saharan Africa. Plasmodium falciparum has developed resistance mechanisms against almost all existing anti-malarial drugs, and this is a major threat to malaria control worldwide. The high level of P. falciparum resistance to chloroquine (CQ) and then to sulfadoxine-pyrimethamine (SP) has led the Democratic Republic of Congo (DRC), like all endemic countries, to change its anti-malarial drug policy for the treatment of uncomplicated malaria. The DRC national policy currently supports two first-line artemisinin-based combinations for the treatment of uncomplicated P. falciparum malaria: artemether–lumefantrine (AL) and artesunate–amodiaquine (ASAQ). If one of the first-line treatments is not available or is poorly tolerated by the patient, the other can be used. In case of treatment failure (confirmed by microscopy) with both first-line artemisinin-based combinations, the patient should be given a dual therapy of quinine plus clindamycin or plus doxycycline, while dihydroartemisinin-piperaquine (DP) could be used in a one-time episode [1].
The World Health Organization (WHO) recommends regular surveillance of artemisinin-based combination therapy (ACT) efficacy to provide an early warning against the emergence and spread of resistance [2]. Numerous polymorphisms in the P. falciparum genome have been suggested to provide resistance to ACT, both to artemisinin and the associated drugs [3, 4]. Mutations in the propeller domain of the P. falciparum kelch 13 gene (pfk13) have been associated with in vivo delayed parasite clearance and in vitro artemisinin (ART) resistance in the ring stage survival assay [5, 6]. These mutations have spread within the Greater Mekong Subregion of Southeast Asia and have been classified as validated markers and candidate/associated markers of ART resistance [7]. Some of these mutations are increasingly being detected in sub-Saharan African countries, providing evidence for de novo emergence of resistance to artemisinin in Africa [8,9,10,11]. Mutations in the chloroquine resistance transporter gene (pfcrt) originally identified as a marker of CQ resistance, have also been associated with resistance to amodiaquine (AQ) [12]. The pfcrt gene is highly polymorphic in codon position 72–76, determining different haplotypes that include the key mutation K76T associated with CQ resistance, while the SVMNT haplotype has been associated with AQ resistance [13]. Additionally, an increased copy number of the gene encoding the multidrug resistance 1 transporter (pfmdr1) has been postulated to confer resistance to lumefantrine (LU) [14]. With the currently recommended artemisinin-based combinations, the absence of resolution of fever and parasitaemia or their recurrence within 28 days of treatment is considered as treatment failure, while all recurrence of fever and parasitaemia after 28 days of an initial treatment should, from an operational standpoint, be considered as re-infection and treated with first-line ACT [15]. The distinction between recrudescence and reinfection may be confirmed only by genotyping of parasites from initial and recurrent infections. This concern is addressed by therapeutic efficacy studies according to the World Health Organization (WHO) protocol [16]. Treatment failure is the inability to clear parasites from a patient’s blood or to prevent their recrudescence after the administration of an anti-malarial drug. Many factors can contribute to treatment failure, including resistance and inadequate exposure to drug due to sub-optimal dosing, poor patient compliance, poor drug quality, vomiting, poor drug absorption and drug interactions. The present study investigated the presence of molecular markers in pfcrt, pfk13 and pfmdr1 genes associated with resistance to current first-line ACT in P. falciparum from treatment failure patients in the DRC.
Methods
Study area
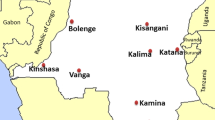
Six sentinel sites of the National Malaria Control Programme (NMCP) were selected among 26 provinces of the DRC. The NMCP sentinel sites are organized within one selected Health Zone entity, per province, and are part of a national network for malaria surveillance. The study sites included 2 of the 3 largest cities of the country and 4 other sites were selected based on their epidemiological facies and their accessibility. Thus, the following sentinel sites were selected: for the largest cities, Kingasani in Kinshasa, the capital city of the DRC and Kabondo in Kisangani; for the equatorial facies, Bolenge in Equateur province and Vanga in Kwilu province; for the tropical facies, Fungurume in Lualaba province; and for the mountainous facies, Katana in Sud-Kivu province. In each study site, 1 to 3 health centres were selected based on their accessibility and attendance for performing the study.
Study patients
From November 2018 to November 2019, patients of all ages were seen on day 0 for the initial malaria episode and, in case of the absence of resolution or the recurrence of fever within 28 days of the artemisinin-based combination treatment, the patients were encouraged to return to the health centre for follow up. Returning patients who had a positive malaria rapid diagnostic test (RDT) were enrolled after receiving informed consent. The new malaria episode in returning patients was afterward confirmed by a P. falciparum real-time PCR assay to define treatment failure. Patients who reported an incomplete ACT during the initial episode were excluded from the study.
Blood sample collection
Screening tests were performed on blood samples taken by finger prick. Malaria was detected using RDTs available on site: SD Bioline Malaria Ag P.f (Standard Diagnostics, South Korea) or CareStart Malaria Pf (Access Bio, South Korea). After enrollment, a blood sample was taken by finger prick and three spots were deposited on Whatman Grade GB003 filter paper (Whatman, GE Healthcare). Dried blood spots (DBS) were placed in an individual grip seal plastic bag containing silica gel desiccant and were then stored at – 20 °C before molecular analysis.
DNA extraction
DNA was extracted from blood spots using the QIAamp DNA Mini Kit (Qiagen, Germany) following the manufacturer’s recommended protocol for DBS. The extracted DNA was stored at – 20 °C before PCR testing.
Detection of P. falciparum by real-time PCR
A real-time PCR for the detection of P. falciparum was performed according to a modified version of a previously described procedure [17]. Briefly, the mixture contained: 1 × Taqman Universal PCR Master Mix (Applied Biosystems, USA), 200 nM P. falciparum primers and probe, 15 µM primers and 5 µM probe of the host β-globin as internal positive control and 5 µl of DNA template in a total volume of 25 µl. Assays were run on an ABI 7500 Fast real-time thermocycler (Applied Biosystems, USA) at the Laboratory of Molecular Biology of the School of Medicine, University of Kinshasa, DRC.
pfk13 and pfcrt genes polymorphisms
The target sequence was a 506-nucleotide fragment of the pfk13-propeller gene covering codon-positions 427–595, following a procedure previously described [18]. This segment of interest includes mutations associated with ART resistance [7]. For the pfcrt gene, the 152-nucleotide fragment of interest (containing codon-positions 72–76) was amplified following a previously described procedure [19]. PCR assays were run using a conventional thermal cycler Hybaid HBPXE 0.2 (Thermo Scientific, USA) at the Laboratory of Molecular Biology of the School of Medicine, University of Kinshasa, DRC. After purification using AMPure XP magnetic beads (Beckman Coulter, USA), the PCR products were added to a mix of Big Dye Terminator V3.1 for the sequencing reaction. The resulting nucleotide sequences were analysed on an ABI 3730 DNA Analyzer automated sequencer (Applied Biosystems, USA) using the Sanger method at GIGA (University of Liège’s Interdisciplinary Research Institute in the Biomedical Sciences). Sequences (forward and reverse) were aligned using Vector NTI (Thermo Fisher Scientific, US) and compared to the reference sequence PF3D7_0709000 (https://www.ncbi.nlm.nih.gov/gene/term=PF3D7_0709000, accessed March 2020) for pfcrt and PF3D7_1343700 (http://www.plasmodb.org, accessed March 2020) for pfk13 using the online Basic Local Alignment Search Tool (BLAST) [National Center for Biotechnology Information (NCBI), USA] for identifying mutations.
Estimation of pfmdr1 copy number
The Pfmdr1 copy number was assessed by a relative quantification real-time PCR using an ABI 7500 thermocycler (Applied Biosystems, US), as previously described [20]. In each run of real-time PCR, 3D7 and Dd2 clones were used as calibration controls with single and multiple copies of the pfmdr1 gene, respectively 1 copy and 4 copies, and a negative control containing no template DNA was also included. Test samples were assayed in triplicate, the copy number of pfmdr1 was determined using the comparative ΔΔCt method and calculated as 2–∆∆Ct. Samples with a spread of ∆∆Ct among the three triplicates of more than 1.5 or with a Ct > 35 were repeated and the second result used. Analyses were performed at the Laboratory of Clinical Microbiology of the University of Liege, Belgium.
Statistical analysis
Data were entered in an Excel 2010 spreadsheet by an independent data clerk. Statistical analysis was performed using SPSS V. 20.0 (IBM Corp., Armonk, USA). Samples for which the genotype could not be determined were excluded from the analysis. Categorical variables were expressed as frequencies with 95% confidence intervals (95% CI) while quantitative variables were expressed as median with interquartile range (IQR). The mutant and wild-type alleles identified in the sequenced isolates were used to generate the prevalence of mutations. Proportions in different characteristics of patients were compared by Chi-square. Tests were two-sided, p-values < 0.05 were considered significant.
Results
Baseline characteristics of patients
In total, 474 patients were enrolled in the study, their age ranged from 0 to 76 years with a median age of 10 years (IQR: 4–21 years). The male to female sex ratio was 0.74. As shown in Table 1, the distribution of patients per age range and per study site was heterogeneous (p value < 0.001) and more than half of them were female (p value < 0.001). The treatment of the initial episode of malaria was based on AL, ASAQ and DP, respectively in 273 (57.6%), 196 (41.3%) and 5 (1.1%) patients. DP was used only in Kingasani (Kinshasa) while both AL and ASAQ were used in all sites, except in Bolenge where only AL was used during the study. Real-time PCR analysis of DNA extracted from DBS samples confirmed the new episode of P. falciparum infection for 364 (76.8%; 95% CI 72.7–80.5%) patients, considered treatment failure patients.
Pfcrt 72–76 haplotype for eventual resistance to amodiaquine
Of the 364 P. falciparum isolates detected by real-time PCR, 335 were successfully sequenced among which 139 (41.5%; 95% CI 36.2–47.0%) harboured the K76T mutation known to confer resistance to CQ. The CVIET haplotype was found in 98 (70.5%) isolates harbouring the K76T mutation, followed by CVIKT (20.1%), CVINT (5.8%) and CVMNT (3.6%). The SVMNT haplotype associated with resistance to AQ was not detected. Among isolates harbouring the K76T mutation, 65/133 (48.9%) of them were found in patients treated with ASAQ while 74/198 (37.4%) occurred in patients treated with AL (p value = 0.038).
Pfk13-propeller polymorphism for eventual resistance to artemisinin
Of the 325 successfully sequenced P. falciparum isolates, 318 (97.8%; 95% CI 95.6–99.1%) were wild-type and 7 (2.2%; 95% CI 0.9–4.4%) carried non-synonymous (NS) mutations in the pfk13-propeller domain. Among these, 3 have been previously described (N498I, N554K and A557S) and 4 were newly reported (F506L, E507V, D516E and G538S). None of the mutations that have been validated or suspected to be associated with ART resistance were found in the study.
The N498I, D516E and G538S mutations were each found in isolates also harbouring the K76T mutation. Table 2 shows the distribution of mutations in the pfk13-propeller gene and the key mutation K76T in the pfcrt gene among enrolled treatment failure patients per site.
Pfmdr1 copy number for eventual resistance to lumefantrine
In total, 322 P. falciparum isolates from enrolled treatment failure patients were successfully analysed for copy number variation in the pfmdr1 gene. Using a copy number threshold of 1.5 to define multiple copies, all isolates harboured a single copy of the pfmdr1 gene.
Discussion
The present study has not detected molecular markers associated with resistance to first-line ACT in use in the DRC in treatment failure patients. Although none of the mutations associated with ART resistance in Southeast Asia have been found in this study, 7 coding substitutions that are of unknown phenotype were observed, among which 3 have been previously reported, notably N498I in Kenya [21], N554K in Comoros and A557S in Togo and DRC [22], and 4 others that had not yet been reported (F506L, E507V, D516E, G538S). Numerous pfk13-propeller mutations of unknown function are commonly reported in sub-Saharan African countries, including the DRC [18, 22, 23]. There are criteria for prioritizing further laboratory studies, notably the frequent observation of a new allele with a non-synonymous mutation, the evidence of dissemination and preliminary association with clinical data whenever possible [23]. Several independent single nucleotide polymorphisms (SNPs) could be responsible for the ART resistance in sub-Saharan Africa, as the known mutations that confer drug resistance would differ from one location to another, depending on the parasite genetics. There is the possibility that pfk13 mutations do not cause ART resistance in isolation, but could act in combination with other genetic or non-genetic factors that are different in African and Southeast Asian parasite populations [24, 25]. Since African pfk13-propeller mutations were shown to be different from those found in Southeast Asia, further molecular and biochemical studies should investigate whether other factors such as additional mutations could be associated with altered functions of the PFK13 protein, resulting in altered ART sensitivity. However, some of the validated and candidate mutations associated with ART resistance in Southeast Asia have been detected in the DRC [11, 26] and in some neighbouring countries, such as Rwanda [8, 9] and Uganda [10], providing evidence of de novo emergence of ART resistance in sub-Saharan Africa. Thus, surveillance must be strengthened to avoid the worst.
The global prevalence of the K76T mutation known to be associated with CQ resistance was 41.5%, but this was variable from one site to another, ranging from 10.0% in Fungurume to 76.9% in Katana. In 2017, a study conducted in 10 sites (including 6 sites of the present study) reported a global prevalence of the K76T mutation of 28.5% but with a high between-regions variability ranging from 1.5% in Fungurume to 89.5% in Katana [27]. In the present study, the prevalence of K76T in patients treated with ASAQ (48.9%) was higher than in those treated with AL (37.4%) with low statistical difference (p value = 0.038). The possibility that AQ could continue to contribute to selection for the K76T mutant even after discontinuance of CQ usage has been previously raised [28]. The simultaneous presence of very low and high prevalence of CQ resistance could be related to a between-regions difference of CQ pressure and also to the effect of selection for CQ resistance depending on the genetic structure of parasite populations, which have been shown to vary significantly across the country [29]. Data concerning current CQ use in the country are not available, further studies at the community level and parasite genetic studies should be conducted to explain the persistence of a high CQ resistance rate in some provinces despite the withdrawal of this molecule from the national policy of malaria treatment. The SVMNT haplotype associated with AQ resistance was not detected in the present study, which is encouraging for the DRC national policy for the continued use of AQ in ACT. This haplotype has not yet been reported in the DRC [19, 26, 27, 30] whereas it was found in neighbouring countries, such as Tanzania and Angola [31, 32].
None of the P. falciparum isolates had multiple copies of the pfmdr1 gene in this study, consistent with the general absence [33, 34] or low frequencies [26, 35, 36] of pfmdr1 copy number variation in P. falciparum from sub-Saharan Africa. The multiple copy number of the pfmdr1 gene has been postulated to confer resistance to lumefantrine [14], the drug associated to AL in one of the artemisinin-based combinations used in the DRC.
The artemisinin-based combinations used to treat the initial episode were those which were available on each study site during the investigation and best tolerated by the patient. In practice, AL tends to be primarily used in urban areas because patients have more options to obtain it from private pharmacies, while ASAQ is used in rural areas. The use of DP as a first-line ACT in five patients in Kinshasa constituted a case of non-adherence to treatment guidelines often observed in urban areas [1]. The study assessed molecular markers of anti-malarial resistance in treatment failure patients. Many other factors not assessed by the present study can contribute to treatment failure, notably those related to poor exposure to drugs. The non-homogeneous distribution of enrolled treatment failure patients across different characteristics suggests that some of these factors may be dependent on age, gender and region. When possible, treatment failure must be confirmed by microscopy, as histidine rich protein-2 (HRP-2)-based RDT may remain positive for weeks after successful treatment due to persistent antigenaemia, even without recrudescence [37, 38]. In this study, a P. falciparum real-time PCR assay was used afterwards to confirm P. falciparum malaria in patients returning to health centres for fever within 28 days of an initial malaria treatment.
The study contributes to the ongoing surveillance of resistance to anti-malarial drugs in use in the DRC as recommended to track the emergence and spread of P. falciparum resistance to different molecules used in malaria management. However, although the study was carried out in six provinces of the DRC with varied geography and malaria endemicity, the results were not representative of either any one province or the entire country.
Conclusion
No molecular marker currently known to be associated with resistance to first-line ACT components in use in the DRC was detected in P. falciparum isolated from treatment failure patients. The findings are supportive of the DRC’s current malaria treatment policy, however, the appearance of new mutants with as-yet unknown functions calls for further investigation. In addition, the other factors, apart from the drug resistance, that could contribute to treatment failure must be assessed in order to ensure the efficacy of the treatment of malaria in the DRC.
Availability of data and materials
All data generated and analysed during this study are included in this published article.
Abbreviations
- ACT:
-
Artemisinin-based combination therapy
- AL:
-
Artemether-lumefantrine
- AQ:
-
Amodiaquine
- ASAQ:
-
Artesunate-amodiaquine
- CQ:
-
Chloroquine
- DBS:
-
Dried blood spot
- DNA:
-
Deoxyribonucleic acid
- DP:
-
Dihydroartemisinin-piperaquine
- DRC:
-
Democratic Republic of Congo
- IQR:
-
Interquartile range
- LU:
-
Lumefantrine
- NMCP:
-
National Malaria Control Programme
- PCR:
-
Polymerase chain reaction
- pfcrt :
-
Plasmodium falciparum Chloroquine resistance transporter
- pfk13 :
-
Plasmodium falciparum Kelch 13
- pfmdr1 :
-
Plasmodium falciparum Multidrug resistance1
- RDT:
-
Rapid diagnostic test
- SNP:
-
Single nucleotide polymorphisms
- SP:
-
Sulfadoxine-pyrimethamine
- WHO:
-
World Health Organization
References
USAID. President’s Malaria Initiative: Democratic Republic of the Congo - Malaria Operational Plan FY 2018. https://www.pmi.gov/docs/default-source/default-document-library/malaria-operational-plans/fy-2018/fy-2018-democratic-republic-of-the-congo-malaria-operational-plan.pdf?sfvrsn=5. Accessed 12 May 2020.
WHO. Methods for surveillance of antimalarial drug efficacy. Geneva: World Health Organization; 2009.
Vestergaard LS, Ringwald P. Responding to the challenge of antimalarial drug resistance by routine monitoring to update national malaria treatment policies. Am J Trop Med Hyg. 2007;77:153–9.
Cui L, Mharakurwa S, Ndiaye D, Rathod PK, Rosenthal PJ. Antimalarial drug resistance: literature review and activities and findings of the ICEMR Network. Am J Trop Med Hyg. 2015;93:57–68.
Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–23.
Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois A-C, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–5.
WHO. Status report on artemisinin resistance and ACT efficacy. Geneva, World Health Organization/Global Malaria programme; August 2018. https://www.who.int/malaria/publications/atoz/artemisinin-resistanceaugust2018/en/. Accessed 20 April 2019.
Uwimana A, Legrand E, Stokes BH, Ndikumana J-LM, Warsame M, Umulisa N, et al. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat Med. 2020;10:1603–8.
Tacoli C, Gai P, Bayingana C, Sifft K, Geus D, Ndoli J, et al. Artemisinin resistance-associated K13 polymorphisms of Plasmodium falciparum in Southern Rwanda, 2010–2015. Am J Trop Med Hyg. 2016;95:1090–3.
Ikeda M, Kaneko M, Tachibana SI, Balikagala B, Sakurai-Yatsushiro M, Yatsushiro S, et al. Artemisinin-resistant Plasmodium falciparum with high survival rates, Uganda, 2014–2016. Emerg Infect Dis. 2018;24:718–26.
Kayiba NK, Yobi DM, Tshibangu-Kabamba E, Tuan VP, Yamaoka Y, Devleesschauwer B, et al. Spatial and molecular mapping of Pfkelch13 gene polymorphism in Africa in the era of emerging Plasmodium falciparum resistance to artemisinin: a systematic review. Lancet Infect Dis. 2020;S1473-3099(20)30493-X. (online ahead of print).
Ecker A, Lehane AM, Clain J, Fidock DA. PfCRT and its role in antimalarial drug resistance. Trends Parasitol. 2012;28:504–14.
Beshir K, Sutherland CJ, Merinopoulos I, Durrani N, Leslie T, Rowland M, et al. Amodiaquine resistance in Plasmodium falciparum malaria in Afghanistan is associated with the pfcrt SVMNT allele at codons 72 to 76. Antimicrob Agents Chemother. 2010;54:3714–6.
Price RN, Uhlemann A-C, Van Vugt M, Brockman A, Hutagalung R, Nair S, et al. Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria. Clin Infect Dis. 2006;42:1570–7.
WHO. Guidelines for the treatment of malaria 3rd Edn. Geneva, World Health Organization 2015. https://extranet.who.int/iris/restricted/handle/10665/162441. Accessed 15 July 2020.
WHO. Tools for monitoring antimalarial drug efficacy. Geneva, World Health Organization. https://www.who.int/malaria/areas/drug_resistance/efficacy-monitoring-tools/en/. Accessed 15 June 2020.
Cnops L, Jacobs J, Esbroeck MV. Validation of a four-primer real-time PCR as a diagnostic tool for single and mixed Plasmodium infections. Clin Microbiol Infect. 2011;17:1101–7.
Yobi DM, Kayiba NK, Mvumbi DM, Boreux R, Bontems S, Kabututu PZ, et al. The lack of K13-propeller mutations associated with artemisinin resistance in Plasmodium falciparum in Democratic Republic of Congo (DRC). PLoS ONE. 2020;15:e0237791.
Mvumbi DM, Boreux R, Sacheli R, Lelo M, Lengu B, Nani-Tuma S, et al. Assessment of pfcrt 72–76 haplotypes eight years after chloroquine withdrawal in Kinshasa, Democratic Republic of Congo. Malar J. 2013;12:459.
Price RN, Uhlemann A-C, Brockman A, McGready R, Ashley E, Phaipun L, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–47.
De Laurent ZR, Chebon LJ, Ingasia LA, Akala HM, Andagalu B, Ochola-Oyier LI, et al. Polymorphisms in the K13 gene in Plasmodium falciparum from different malaria transmission areas of Kenya. Am J Trop Med Hyg. 2018;98:1360–6.
Ménard D, Khim N, Beghain J, Adegnika AA, ShafiulAlam M, Amodu O, et al. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med. 2016;374:2453–64.
WWARN K13 Genotype-Phenotype Study Group. Association of mutations in the Plasmodium falciparum (Pf3D7_1343700) with parasite clearance rates after artemisinin-based treatments – a WWARN individual patient data meta-analysis. BMC Med. 2019;17:1.
Kamau E, Campino S, Amenga-Etego L, Drury E, Ishengoma D, Johnson K, et al. K13 propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis. 2015;211:1352–5.
Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, et al. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2014;347:428–31.
Mvumbi DM, Bobanga TL, Kayembe J-MN, Mvumbi GL, Situakibanza HN-T, Benoit-Vical F, et al. Molecular surveillance of Plasmodium falciparum resistance to artemisinin-based combination therapies in the Democratic Republic of Congo. PLoS ONE. 2017;12:e0179142.
Yobi DM, Kayiba NK, Mvumbi DM, Boreux R, Kabututu PZ, Situakibanza HNT, et al. Molecular surveillance of anti-malarial drug resistance in Democratic Republic of Congo: high variability of chloroquinoresistance and lack of amodiaquinoresistance. Malar J. 2020;19:121.
Frank M, Lehners N, Mayengue PI, Gabor J, Dal-Bianco M, Kombila DU, et al. A thirteen-year analysis of Plasmodium falciparum populations reveals high conservation of the mutant pfcrt haplotype despite the withdrawal of chloroquine from national treatment guidelines in Gabon. Malar J. 2011;10:304.
Verity R, Aydemir O, Brazeau NF, Watson OJ, Hathaway NJ, Mwandagalirwa MK, et al. The impact of antimalarial resistance on the genetic structure of Plasmodium falciparum in the DRC. Nat Commun. 2020;11:2107.
Meshnick SR, Janko M, Tshefu AK, Taylor SM, Emch M, Antonia AL. A cross-sectional survey of Plasmodium falciparum pfcrt mutant haplotypes in the Democratic Republic of Congo. Am J Trop Med Hyg. 2014;90:1094–7.
Alifrangis M, Dalgaard MB, Lusingu JP, Vestergaard LS, Staalsoe T, Jensen ATR, et al. Occurrence of the Southeast Asian/South American SVMNT haplotype of the chloroquine resistance transporter gene in Plasmodium falciparum in Tanzania. J Infect Dis. 2006;193:1738–41.
Gama BE, Pereira-Carvalho GA, Lutucuta Kosi FJ, Almeida de Oliveira NK, Fortes F, Rosenthal PJ, et al. Plasmodium falciparum isolates from Angola show the SVMNT haplotype in the pfcrt gene. Malar J. 2010;9:174.
Menard S, Morlais I, Tahar R, Sayang C, Mayengue P, Iriart X, et al. Molecular monitoring of Plasmodium falciparum drug susceptibility at the time of the introduction of artemisinin-based combination therapy in Yaoundé, Cameroon: Implications for the future. Malar J. 2012;11:113.
Davlantes E, Dimbu PR, Ferreira CM, Florinda Joao M, Pode D, Félix J, et al. Efficacy and safety of artemether–lumefantrine, artesunate–amodiaquine, and dihydroartemisinin–piperaquine for the treatment of uncomplicated Plasmodium falciparum malaria in three provinces in Angola, 2017. Malar J. 2018;17:144.
Witkowski B, Nicolau M-L, Soh PN, Iriart X, Menard S, Alvarez M, et al. Plasmodium falciparum isolates with increased pfmdr1 copy number circulate in West Africa. Antimicrob Agents Chemother. 2010;54:3049–51.
Nguetse CN, Adegnika AA, Agbenyega T, Ogutu BR, Krishna S, Kremsner PG, et al. Molecular markers of anti-malarial drug resistance in Central, West and East African children with severe malaria. Malar J. 2017;16:217.
Kyabayinze DJ, Tibenderana JK, Odong GW, Rwakimari JB, Counihan H. Operational accuracy and comparative persistent antigenicity of HRP2 rapid diagnostic tests for Plasmodium falciparum malaria in a hyperendemic region of Uganda. Malar J. 2008;7:221.
Grandesso F, Nabasumba C, Nyehangane D, Page A-L, Bastard M, De Smet M, et al. Performance and time to become negative after treatment of three malaria rapid diagnostic tests in low and high malaria transmission settings. Malar J. 2016;15:496.
Acknowledgements
The study was conducted in collaboration with the National Malaria Control Programme (NMCP). We would like to thank all patients and their guardians, as well as the staff of all collecting sites for their participation in this study. We are grateful to Anna Rosanas and Pieter Guetens of the Institute of Tropical Medicine, Antwerp, Belgium for the P. falciparum Dd2 clone used as calibration controls with multiple copies of the pfmdr1 gene.
Funding
This work was supported by the Académie de Recherche et d’Enseignement Supérieur (ARES), Belgium.
Author information
Authors and Affiliations
Contributions
DMY, NKK, DMM, PDM, GLM and MPH conceived and wrote the study protocol. DMY and NKK collected the samples and data. DMY and RB carried out the molecular analyses. DMY, DMM, RB and MPH analysed molecular data. DMY wrote the first draft of the manuscript and all the authors were involved in editing and approval of the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The protocol and the informed consent form were approved by the Ethics Committee of the Faculty of Medicine, University of Kinshasa (Approval No.: ESP/CE/111/2018). All participants involved in the study signed an informed consent form. For participants aged < 18 years old, the consent form was approved and signed by their parent or guardian.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
: Pfk13 and pfcrt sequences for wild and mutant isolates.
Additional file 2:
: Minimal data set containing raw data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yobi, D.M., Kayiba, N.K., Mvumbi, D.M. et al. Assessment of Plasmodium falciparum anti-malarial drug resistance markers in pfk13-propeller, pfcrt and pfmdr1 genes in isolates from treatment failure patients in Democratic Republic of Congo, 2018–2019. Malar J 20, 144 (2021). https://doi.org/10.1186/s12936-021-03636-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-021-03636-y