Abstract
Background
Humans living in regions with high falciparum malaria transmission intensity harbour multi-strain infections comprised of several genetically distinct malaria haplotypes. The number of distinct malaria parasite haplotypes identified from an infected human host at a given time is referred to as the complexity of infection (COI). In this study, an amplicon-based deep sequencing method targeting the Plasmodium falciparum apical membrane antigen 1 (pfama1) was utilized to (1) investigate the relationship between P. falciparum prevalence and COI, (2) to explore the population genetic structure of P. falciparum parasites from malaria asymptomatic individuals participating in the 2007 Demographic and Health Survey (DHS) in the Democratic Republic of Congo (DRC), and (3) to explore selection pressures on geospatially divergent parasite populations by comparing AMA1 amino acid frequencies in the DRC and Mali.
Results
A total of 900 P. falciparum infections across 11 DRC provinces were examined. Deep sequencing of both individuals, for COI analysis, and pools of individuals, to examine population structure, identified 77 unique pfama1 haplotypes. The majority of individual infections (64.5%) contained polyclonal (COI > 1) malaria infections based on the presence of genetically distinct pfama1 haplotypes. A minimal correlation between COI and malaria prevalence as determined by sensitive real-time PCR was identified. Population genetic analyses revealed extensive haplotype diversity, the vast majority of which was shared across the sites. AMA1 amino acid frequencies were similar between parasite populations in the DRC and Mali.
Conclusions
Amplicon-based deep sequencing is a useful tool for the detection of multi-strain infections that can aid in the understanding of antigen heterogeneity of potential malaria vaccine candidates, population genetics of malaria parasites, and factors that influence complex, polyclonal malaria infections. While AMA1 and other diverse markers under balancing selection may perform well for understanding COI, they may offer little geographic or temporal discrimination between parasite populations.
Similar content being viewed by others
Background
Malaria caused an estimated 429,000 deaths worldwide in 2015, with the overwhelming majority of deaths occurring in sub-Saharan Africa [1]. In regions of holoendemic malaria transmission, individuals are routinely exposed to malaria parasites and subsequently develop naturally acquired partial immunity to malaria clinical disease despite harbouring malaria parasites [2,3,4,5,6,7,8,9,10]. Individuals with asymptomatic or chronic malaria have been identified as important reservoirs for malaria transmission and represent a major challenge for malaria control and elimination strategies [11,12,13,14,15].
Early molecular studies revealed that genetically diverse Plasmodium falciparum strains circulate in malaria endemic regions and that this genetic heterogeneity contributes to the ability of P. falciparum to evade the host immune response and develop resistance to anti-malarial drugs [16,17,18,19,20,21,22]. It has been suggested that multiclonal malaria infections can influence clinical outcomes in a manner that is dependent on transmission intensity [23], and may negatively impact an individual’s response to anti-malarial drug treatment [24]. Further, multiclonal P. falciparum infections increase the likelihood of inter-strain genetic recombination during the sexual stage in the anopheline vector, resulting in the generation of genetically diverse P. falciparum strains and facilitating parasite evolution [25,26,27,28,29]. Multiclonal P. falciparum infections can occur either via multiple mosquito bites each with a different strain of P. falciparum or via a single mosquito bite containing multiple P. falciparum strains [4, 30, 31]. The number of distinct P. falciparum strains present within a single individual is defined as the complexity of infection (COI) [32]. The relationship between COI and malaria transmission intensity is complex. On one hand, recent studies have shown a positive correlation between the intensity of malaria transmission and P. falciparum COI, with malaria holoendemic regions typically experiencing higher P. falciparum COIs compared to areas with seasonal or low malaria endemicity [6, 33,34,35,36,37,38,39]. Thus, COI has been proposed as a method for measuring changes in malaria transmission intensity after the implementation of malaria control programmes [33, 35, 40,41,42]. Conversely, other studies have demonstrated a lack of correlation between malaria transmission intensity and P. falciparum COI [43,44,45]. Additional studies into the relationship between malaria transmission intensity and P. falciparum COI are, therefore, needed to better understand the relationship between malaria parasite genetic diversity and transmission dynamics and the potential utility of COI as a measure of change in malaria prevalence.
Several genetic tools and strategies have been employed to detect multiclonal P. falciparum infections, including targeting size polymorphisms of the merozoite surface proteins (MSP1, MSP2) and GLURP [5, 8, 46,47,48,49]. Some PCR based methods rely on DNA sequence length polymorphisms, which can be visualized via gel or capillary electrophoresis and the COI defined as the number of distinct bands present. However, these methods lack the sensitivity to identify distinct P. falciparum strains that differ by only a few nucleotides in length or that contain single nucleotide polymorphisms (SNPs). Also, these methods have poor sensitivity in terms of detecting less abundant strains [50,51,52,53], and differing methods can result in high variability in the number of strains detected between laboratories [54]. Novel approaches based on DNA deep sequencing technologies provide increased capabilities to detect minor variant P. falciparum strains as well as discriminate SNPs and small indels. These deep sequencing technologies provide a more accurate determination of the COI within an individual or population thereby improving subsequent population genetic analyses [4, 6, 34, 50, 55, 56].
In the Democratic Republic of Congo, malaria is a leading cause of morbidity and mortality with over 95% of malaria infections due to P. falciparum [57]. The DRC Ministry of Health estimates that 97% of the population in the DRC live in areas where malaria transmission occurs 8–12 months out of the year [57]. The 2007 DRC demographic and Health Survey (DHS) and subsequent studies reported over one-third (33.5%) of adults (15–59 years) were positive for malaria by real-time PCR (qPCR) [58, 59]. Several studies have explored the complex malaria spatial epidemiology and population genetics in the DRC [59,60,61,62,63,64,65,66,67]. For instance, a recent spatial and genetic analysis revealed P. falciparum parasite populations are dispersed across seven geographical areas, likely due to movement of human populations between provinces in the DRC and the region [61]. Additionally, Taylor et al. report spatial and genetic clustering of P. falciparum sulfadoxine drug resistance between western and eastern DRC [65]. Further studies to examine P. falciparum haplotype diversity are, therefore, warranted to inform malaria control strategies and to monitor changes in malaria parasite population structure in response to malaria control efforts in the DRC.
In this study, a PCR amplicon-based deep sequencing approach was utilized to target the extensive allelic diversity of the P. falciparum apical membrane antigen 1 (pfama1) gene in order to (1) examine the relationship between P. falciparum COI and P. falciparum prevalence as determined previously by real-time PCR [59], (2) to investigate the P. falciparum population genetic structure at both the individual and population level in the DRC, and (3) to explore AMA1 amino acid frequencies and potential selection pressures between geographically distinct malaria parasite populations in the DRC and Mali. The authors hypothesized that P. falciparum COI would be positively correlated with P. falciparum prevalence in a region, and that similar pfama1 haplotypes would be identified at the individual and population level in the DRC and Mali. In order to investigate pfama1 diversity at both the individual and population level, individual samples (representing a malaria infection in a single person), and pooled samples (representing population cluster samples) were targeted in this study. Pooling samples is a cost-effective approach to amplicon-based deep sequencing as it reduces the number of PCR reactions and library preparations, and this pooled approach has been utilized in several malaria population genetic studies [68,69,70,71]. This dual sample type (individual and population cluster) approach allows for the examination of COI using the individual samples and also powers spatial population genetic analyses combining the individual samples and the pooled population cluster samples.
Overall, a total of 77 unique pfama1 haplotypes were identified across DRC provinces. The vast majority of individual malaria infections were polyclonal (COI > 1), and no correlation was found between COI and malaria prevalence at sites/regions. Population genetic analyses revealed extensive genetic diversity of P. falciparum parasites based on the pfama1 gene and similar amino acid frequencies between malaria parasite populations in the DRC and Mali. Herein, this manuscript highlights the utility of combining individual and pooled amplicon-based deep sequencing methods for population genetic analyses layered onto the infrastructure and sample collection process of a routine Demographic and Health Survey. This manuscript also describes the spatial and genetic diversity of pfama1 haplotypes circulating in the DRC and Mali to improve the understanding of malaria transmission dynamics that could potentially inform future malaria control and elimination efforts in the region.
Methods
Ethics statement
Participants included in the Demographic and Health Survey (DHS) provided verbal informed consent as described previously [59]. Study enrollment and blood sample collection protocols were approved by the Ethics Committees of the institutions involved in the DHS and sample collection including Macro International, the School of Public Health of the University of Kinshasa, and the Institutional Review Board of the University of North Carolina.
Democratic Republic of Congo Demographic Health Survey Sample Collection
The 2007 Democratic Republic of the Congo Demographic and Health Survey (DHS) was conducted to collect health indicator data from across the DRC. Within 300 clusters, survey teams went from house to house and enrolled women aged 15–49 years, and men aged 15–59. The age distribution was constant across sites. The survey in urban Kinshasa occurred during the rainy season (January 31–March 8, 2007). The remainder of the country was surveyed during the dry season (May–August, 2007) [58, 60]. Genomic DNA was extracted from dried blood spot (DBS) samples on filter paper for malaria species-specific 18S ribosomal RNA based qPCR detection of P. falciparum, Plasmodium ovale, and Plasmodium malariae parasites [59]. The samples used herein were randomly selected from samples obtained during the 2007 DHS.
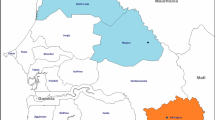
In the present study, 115 individual samples positive for asymptomatic P. falciparum infection were identified based on the following criteria: (1) positive for P. falciparum and negative for P. ovale and P. malariae by species-specific qPCR and (2) geographically representative of the eleven DRC provinces. Eighty-four population cluster samples that represented pooled asymptomatic P. falciparum samples of 2–25 individuals were also chosen based on geographical proximity to the individual samples (Additional file 1). Individual and population cluster samples were selected in order to compare parasite haplotypes from individual people (Fig. 1a) and the parasite population at large (Fig. 1b) from all DRC provinces. The presence of P. falciparum parasites was further confirmed for all individual samples based on the detection of P. falciparum lactate dehydrogenase (pfldh) gene by qPCR as described [72].
Individual (a) and population cluster (b) samples with analysable sequence reads locations. Blood samples were collected from all seven provinces including Kinshasa (box inset). Individual samples (blue circles, n = 79) represent genomic DNA isolated from a single person and population cluster samples (green circles, n = 73) are pooled genomic DNA samples from multiple (2–25) people
Amplicon-based deep sequencing of the P. falciparum apical membrane antigen 1 (pfama1) gene
To increase assay sensitivity, heminested primers were designed using Primer3 [73] to amplify a region of the pfama1 gene (GenBank Reference XM_001347979.1) (Fig. 2). The first round PCR primers, Ama1OF and Ama1R, and the second round PCR primers, Ama1F and Ama1R primers, amplify a 266 base pair (bp) and 236 bp region, respectively (Additional file 2). In order to pool PCR amplicons during sequencing library preparation, a 10-nucleotide Multiplex Identifier (MID) barcode sequence was added to the 5′ end of the Ama1F primer [74] (Additional file 2). PCR amplicons were visualized on 1% agarose gels stained with ethidium bromide and purified via the PureLink Pro 96 PCR Purification Kit (Invitrogen, Carlsbad, CA). PCR amplicon concentration was determined in duplicate and averaged using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions.
Schematic of the Plasmodium falciparum apical membrane antigen 1 (pfama1) gene. The first 72 nucleotides contain the signal sequence (ss) followed by Domain I (nucleotide 73–960), Domain II (nucleotide 961–1326), Domain III (nucleotide 1327–1638), and the transmembrane domain (tm, nucleotide 1639–1869). The heminested primer binding sites (OF = Ama1OF, F = Ama1F, R = Ama1R) are indicated by arrows located in Domain I. Nucleotide sites are based on the 3D7 reference strain (GenBank XM_001347979.1)
Purified PCR amplicons containing barcodes were pooled into sets (up 24 amplicons each based on the 24 unique MIDs) at equal concentration. Each pool was then used to generate a sequencing library with a unique index to allow for the unambiguous identification of the sequences for every amplicon. Specifically, ten nanograms of pooled purified PCR amplicons was ligated with the appropriate index and the DNA concentration of each indexed amplicon pool was determined using the Agilent High Sensitivity D1000 ScreenTape Assay on the 2200 TapeStation (Agilent Technologies, Santa Clara, CA) according to manufacturer’s instruction in order to confirm equal DNA concentrations across the 18 indexed amplicon pools. Six sequencing control samples were run as template in duplicate with the pfama1 PCR assay and included in downstream sequencing library preparations. Sequencing control samples contained P. falciparum DNA from V1S, RO33, Dd2, 7g8, and K1 strains at 5, 10, 15, 30, and 40 percent, respectively (BEI Resources/MR4, Manassas, VA).
Ion Torrent library preparation was conducted following the “Preparing Short Amplicon (< 350) Libraries Using the Ion Plus Fragment Library Kit” manual (Life Technologies, MAN0006846, revision 3.0) for each of the 18 indexed PCR pools. DNA concentrations of the 18 resulting libraries were determined using the Agilent High Sensitivity D1000 ScreenTape Assay according to manufacturer’s protocol. Equal concentrations of each library were pooled and split across two Ion 318 Chips (Life Technologies, Carlsbad, CA) utilizing 400 bp chemistry on the Ion Torrent PGM platform (Life Technologies) at the University of North Carolina Chapel Hill Microbiome Core Facility. Deep sequence data extraction, processing, and analyses were performed using the SeekDeep targeted amplicon bioinformatics pipeline [75,76,77,78,79]. A workflow diagram is provided to outline the methods and provide additional details (Additional file 3).
In order to compare AMA1 amino acid frequencies between the DRC and Mali, 506 pfama1 sequences (GenBank FJ898536–FJ899041) previously published by Takala et al. were retrieved [80]. Pfama1 sequences from Mali were trimmed and aligned to pfama1 sequences from the DRC using Geneious (v 9.1.5) [81].
Population genetics analyses
Molecular population genetic analyses were conducted using MEGA version 7 [82] and DnaSP (v5.10.1) [83, 84]. Standard nucleotide and haplotype diversity calculations weighting pools by the number of contained individuals [85] were performed in Python. Population pairwise (FST) comparisons were calculated between DRC provinces using the Analysis of Molecular Variance (AMOVA) tool in the Arlequin (v3.5.2.2) population genetics data analysis program [86]. Network (v5.0.0.1), DNA Alignment (v1.3.3.2), and Network Publisher (v2.1.1.2) add-ons were used to generate a median-joining (MJ) network diagram in order to visualize phylogenetic relationships between pfama1 haplotypes [87]. Isolation by Distance (IBD) analysis was performed using the Mantel Test in GenAlEx (v.6.503) [88, 89].
Statistical analyses and data visualization
Statistical analyses were performed in GraphPad Prism (v6), SPSS (v22), R [90], and Microsoft Excel. ArcGIS (ESRI, v.10.4.1.5686) was used to generate maps and the DRC province boundary map was obtained from the DHS Programme Spatial Data Repository [91]. All permutation testing used 10,000 replicates.
Results
Amplicon-based deep sequencing of individual and population cluster samples
Real-time PCR (qPCR) was performed for all 115 individual samples to confirm the presence of P. falciparum based on the lactate dehydrogenase (pfldh) gene. Conventional PCR based on the pfama1 gene was performed on all individual samples regardless of pfldh qPCR results and on all geographical cluster samples (n = 84). Table 1 summarizes the results of the pfldh qPCR and pfama1 PCR for both sample types.
A total of 11,511,315 pfama1 deep sequencing reads were obtained using the Ion Torrent PGM platform. Using the SeekDeep targeted amplicon analysis pipeline, reads with missing barcodes, short reads (< 50 bp), poor quality, and chimeric reads were filtered out, resulting in 4,879,911 remaining reads. The IonTorrent PGM platform is based on the “sequencing by synthesis” principle (detecting H ion release on a semiconductor matrix when a base is added during synthesis), which can result in variable quality reads particularly within homopolymer repeats. Thus, sequencing read quality varies in the proportion of reads that are poor quality or truncated and is mainly dependent on the quality of the input library as well as the specific run rather than one sequencing platform over another. These sequencing reads were subsequently de-multiplexed (separated by input amplicon based on index and MID), clustered according to samples and replicates, and haplotypes estimated using a 2.5% minimum haplotype frequency cutoff. Based on these criteria, a total of 3,754,497 reads was obtained for downstream haplotype analysis. Analysable deep sequencing reads of the target pfama1 region were generated for 79 (68.7%) of the individual samples, 73 (86.9%) of the population cluster samples, and six sequencing control samples. Analysis of the six sequencing control samples revealed similar haplotype frequencies between the expected haplotype percentage and the actual haplotype percentage determined by the SeekDeep targeted amplicon analysis pipeline (Additional file 4), demonstrating the sensitivity of a targeted amplicon based deep sequencing approach to detect mixed haplotype infections. No false haplotypes (i.e. haplotypes that were not included in the control template) were detected from the control sample sequencing reads.
Overall, a total of 77 unique pfama1 haplotypes from both the individual samples and population cluster samples were identified (Table 2; Additional files 5, 6). A total of 60 pfama1 haplotypes were identified in individual samples and 55 haplotypes were identified in the population cluster samples (Table 2; Additional file 6). Thirty-eight pfama1 haplotypes were shared between the individual and population cluster samples and 25 most frequent haplotypes were detected in both confirming their general equivalence (R = 0.71) (Additional file 6). Twenty-two and seventeen pfama1 haplotypes were unique to the individual samples and population cluster samples, respectively. Only relatively low frequency haplotypes in the population were not detected in both individual and pooled samples (frequency average 0.1%; maximum 1.0%) (Additional file 6).
The majority of P. falciparum infections in individual samples were polyclonal (64.5%) defined as a COI > 1 (Fig. 3a). The mean COI for individual samples was 2.38 and ranged from 1 to 9 haplotypes (Table 2). Not surprisingly, given they represent multiple patient samples, 84.9% of the population cluster samples were polyclonal (Fig. 3b). Comparison of the demographic characteristics between the sample types with analysable sequence reads revealed the pooled subjects tended to be slightly younger, to live in rural areas, and to be male than the subjects tested individually (Additional file 7). In order to explore the relationship between COI and malaria transmission intensity in the DRC, COI from individual samples were compared to P. falciparum prevalence obtained via real-time PCR from the 2007 DHS samples reported in a separate study [59]. As shown in Fig. 4, there was a non-significant weak linear trend of increasing COI with prevalence but the overall variance was high with little accounted for by this model (Pearson coefficient of correlation, r = 0.168, p = 0.139 by permutation) (Fig. 4, grey). A nonparametric Spearman rank correlation was also performed and demonstrated no significant relationship between observed COI and P. falciparum prevalence by qPCR (rs = 0.126, p = 0.268 by permutation). Difference in observed COI and bifurcated prevalence based on the mean (0.4413) was also tested. These high and low prevalence groups had a mean of 2.75 and 2.00, respectively, with suggestive significance (Wilcoxon two sample test, p = 0.0861 and p = 0.0353 by permutation).
Comparison of Plasmodium falciparum complexity of Infection (COI) and prevalence. The relationship between the observed (uncorrected, in grey) P. falciparum COI from individual samples and P. falciparum prevalence by qPCR showed a small positive correlation that was not significant (Pearson coefficient of correlation, r = 0.168; p = 0.139). A permutation model was used to account for strains that share the same pfama1 haplotype and average corrected COI values (red) are shown. No significant correlation between corrected COI and P. falciparum prevalence by qPCR was observed (Pearson coefficient of correlation, r = 0.169, p = 0.135)
Pfama1 amplicon deep sequencing, like all single locus methods, will underestimate the true COI when strains share the same haplotype. A permutation model was utilized to correct for COI when strains share the same pfama1 haplotype. Based on the observed pfama1 haplotype frequencies, the probability of true COI of 2, 3, 4, 5, 6, 7, 8, 9, and 10 being undercalled (missing one or more strains) is 4.5, 12.7, 24, 37, 50, 62, 73, 82 and 88 percent, respectively. While these values demonstrate that the highest COIs are usually underestimated, these high COIs represent a low proportion of the observed samples in this study. Using both the probability of undercalling and the observed distribution, the corrected COIs were simulated and the average correction plotted (Fig. 4, red). Compared to the original uncorrected COI, there was minimal change in the parametric correlation (Pearson coefficient of correlation, r = 0.169, p = 0.135 by permutation) and no change in the nonparametric correlation (Spearman, r = 0.126, p = 0.268 by permutation). The mean corrected COI showed an increased spread of 2.98 versus 2.12 between the high and low prevalence groups, respectively. Despite this increased difference, the p value increased slightly (p = 0.0398 by permutation) due to increased variance resulting from modeling the imprecision of the COI measurements.
Population genetic analyses
Population genetics analysis methods were utilized to explore pfama1 haplotypes from all sample types as well as between individual samples and population cluster samples (Table 2). Overall, 33 polymorphic sites (S) from 77 pfama1 haplotypes and high haplotype diversity (Hd) were found in both sample types (Table 2). Comparison between individual and population cluster samples show similar haplotype diversity (Hd) and nucleotide diversity (π). This was supported by the lack of a statistical difference between the two types of sampling at the province level (Wilcoxon Rank Sign Test, p = 0.16 for Hd; p = 0.60 for π). However, there were fewer polymorphic sites (S) in the population cluster samples compared to the individual samples (Table 2) and these additional sites represent low prevalence rare variants within the population. This emphasizes that pooled samples should not be utilized for analyses that target or depend on the assessment of low frequency variants.
The population genetic data were examined based on DRC province for both individual and population cluster samples (Table 3). Both sample types showed similar numbers of polymorphic sites, nucleotide diversity, and haplotype diversity across the 11 provinces. However, Bas Congo appeared to differ from other provinces with the lowest haplotype diversity (0.568) and nucleotide diversity (0.0277). The overall frequencies were tested for significant outliers. Bas Congo was identified as an outlier in terms of haplotype diversity (p = 0.004; Dixon’s Q-test) but not in terms of nucleotide diversity (p = 0.6). However, this overall deviation was only supported by the pooled samples (n = 30; p = 0.004) and not the individuals (n = 4; p = 0.6). Given this difference and that this province was less deeply sampled than on average it is not clear if this is a significant deviation.
To further explore haplotype diversity between DRC provinces, population fixation index (FST) was determined between DRC provinces from haplotypes identified in both sample types (Table 4). Overall, the average FST value is 0.0008, ranging from 0 to 0.011656. Low FST values between provinces indicate pfama1 haplotypes are panmictic and, therefore, not isolated based on province in the DRC. Isolation by distance (IBD) analysis of individual samples showed no correlation of genetic distance and spatial distance (R2 = 7.7 × 10−5), indicating that genetically similar pfama1 haplotypes are not found closer together spatially. A Median-Joining Network Diagram was also constructed using the 60 haplotypes found in individual samples to examine whether related haplotypes are spatially clustered in the DRC (Fig. 5). There was no clustering of related pfama1 sequences based on DRC province, suggesting that genetically related pfama1 haplotypes do not appear to be spatially restricted in the DRC.
Comparison of pfama1 amino acid frequencies between geographically divergent malaria populations in the DRC and Mali
In order to explore the heterogeneity in amino acid frequencies between malaria populations from disparate geographical locations, the 77 pfama1 amino acid sequences from the DRC were compared to 506 previously published pfama1 sequences from Mali (FJ898536–FJ899028) [92]. Trimming the Mali pfama1 sequences to match the 162 bp region sequenced in the DRC samples resulted in 58 distinct Mali pfama1 sequences. The trimmed sequences were then aligned with the DRC pfama1 sequences and 32 (55%) of the DRC pfama1 sequences were found to be 100% identical over a 162 bp region to the Mali pfama1 sequences previously identified. Nucleic acid sequence identity between the DRC and Mali pfama1 sequences ranged from 91.4 to 100%. Analysis of the DRC and Mali pfama1 sequence heterogeneity at the amino acid level (Fig. 6) revealed highly similar amino acid frequencies between the two parasite populations despite both geographical and temporal separation suggesting balancing selection at a continental scale.
Comparison of AMA1 amino acid frequencies at variable sites between malaria populations in the DRC and Mali. The reference 3D7 amino acid sequence and corresponding amino acid position number are shown on the x-axis (GenBank XP_001348015.1). For each amino acid residue, the first bar represents the amino acid frequencies from the DRC pfama1 haplotypes and the second bar represents the amino acid frequencies from pfama1 haplotypes in Mali [92]. The amino acid frequency within this region of AMA1 is similar between the two geographical locations
Discussion
In this study, an amplicon-based deep sequencing was utilized to investigate the diversity of pfama1 genes from asymptomatic malaria infections at both the individual and population cluster level from across the DRC and in Mali. Overall, a total of 77 unique pfama1 haplotypes were identified and the majority of individual infections in the DRC were polyclonal (64.5%). Population genetic analyses revealed pfama1 haplotypes are not isolated based on distance or province within the DRC. These results align with a previous study in the DRC, which found a lack of spatial restriction of malaria parasite populations. This diversity, however, may not be due to the extensive movement of P. falciparum parasites with their human hosts between provinces and neighbouring countries [61]. Rather, more likely, potential explanations for the extensive pfama1 haplotype diversity identified in the DRC in this study include human host immune selection that maintains the antigenic diversity of pfama1 (balancing selection) and spatially restrictive protein–protein interactions [92, 93].
In order to more fully explore pfama1 diversity between geographically divergent malaria endemic regions, haplotype frequencies were compared at the amino acid level in parasite populations from the DRC and Mali. Highly similar amino acid frequencies were observed between parasite populations in the DRC and Mali (Fig. 6), suggesting analogous selective pressures could be maintaining pfama1 haplotype diversity between the two regions across the continent more so than parasite movement. A previous study to investigate the diversity of the circumsporozoite protein (CS), another hypervariable surface antigen, also showed shared amino acid frequencies between two geographically separated malaria parasite populations [93]. Highly diverse regions under balancing selection, such as AMA1, while excellent markers for COI, may therefore be poorly suited to discriminate geographically distinct malaria parasite populations or serve as a marker for malaria parasite diversity.
In contrast to several recent studies [6, 33,34,35,36], this study found a minimal positive correlation between COI and malaria prevalence that was not significant (Fig. 4). While additional samples could have increased the power in this study, other studies have also reported no correlation between COI and P. falciparum prevalence [43,44,45]. Potential explanations for these discrepancies include differing methodologies for detection of P. falciparum strains and varying malaria transmission intensity by region. Previous studies that reported significant correlations between COI and malaria prevalence typically compare low and high malaria transmission areas [34,35,36,37, 39]. This study was conducted in the DRC, which experiences high malaria transmission year round. Therefore, the lack of a significant association between COI and P. falciparum prevalence in this study compared to other studies could be because due to the high stable malaria transmission across the DRC. Additional research studies including larger sample sizes and additional markers are needed to further explore the potential relationship between COI and malaria prevalence and how population diversity indices could be utilized to monitor changes in malaria transmission intensity in the DRC and other malaria endemic regions. However, given the wide variance observed in the correlation between COI and prevalence, it may not be a reliable surrogate in differentiating malaria transmission levels within the DRC.
Deep sequencing technologies have enhanced ability to detect low frequency, minor variant P. falciparum haplotypes and characterize malaria COI from a variety of sample types including dried blood spots [4, 6, 34, 50, 55, 56]. Amplicon-based deep sequencing was utilized in this study to detect polyclonal P. falciparum infections for several reasons, including its cost-effectiveness compared to whole genome sequencing and the ability to utilize barcoding and pool several dozen samples thereby increasing sample size. The SeekDeep bioinformatics pipeline is designed for analysis of haplotype frequency from amplicon-based deep sequencing data and has been used successfully in several studies investigating malaria population genetics globally [76, 78, 79].
Pfama1 was chosen for amplicon-based deep sequencing based on several factors. First, pfama1 is a highly polymorphic gene, containing several single nucleotide polymorphisms (SNPs), likely maintained via balancing selection due to immune pressure in the human host [80, 94,95,96]. Previous studies in malaria endemic regions have identified over 60 polymorphic sites within pfama1 [96,97,98,99]. Similarly, sequencing of human samples from a malaria endemic region in Mali identified over 200 unique pfama1 haplotypes [80]. The P. falciparum AMA1 antigen is also a highly-studied malaria vaccine antigen candidate. Vaccine studies have demonstrated that AMA1 based vaccine protection against clinical malaria is extremely strain-specific and, therefore, a clear understanding of AMA1 diversity is critical to develop an effective malaria vaccine based on this polymorphic antigen [100,101,102,103,104,105]. The results from this study provide further evidence of the extensive heterogeneity of pfama1 haplotypes in the DRC and surrounding malaria endemic regions.
This study has several important limitations that may have restricted the ability to detect minor variants and calculate COI in the malaria parasite population circulating in the DRC. These limitations include: possible pfama1 sequence polymorphisms in primer binding sites, malaria parasite nucleic acid degradation stored on dried blood spots, and pfama1 haplotype frequency below the limit of detection of the PCR assay or 2.5% cut off for sequencing analysis. In addition, this study focused on a subset of asymptomatic malaria samples collected as part of the 2007 DHS in the DRC. The inclusion of more malaria positive samples, including symptomatic as well as asymptomatic malaria infections, would provide a more comprehensive description of the P. falciparum population genetic structure in the DRC. Another potential limitation is that this study targeted a region in the highly polymorphic pfama1 gene as surrogate for the entire P. falciparum genome. As such, the true genetic heterogeneity of P. falciparum parasites circulating in the DRC is underestimated. Further, as the number of polymorphic sites (S) was unexpectedly higher in the individual samples compared to the pooled population cluster samples, it is important to note that pooled sampling likely missed some variants occurring at low frequency within one or a few individuals within the population (Table 2). As such, it is critical to consider whether samples were pooled prior to amplicon-deep sequencing when designing studies to detect low frequency variants and for cross comparisons between individuals and pools while choosing statistics minimally influenced by rare variants or haplotypes, particularly in low malaria prevalence areas. However, targeted deep sequencing shows great improvement in COI estimates over traditional methods [50], particularly for pfama1 given its high 0.95 haplotype diversity. To account for the chance of strains sharing the same AMA1 haplotype, a permutation-based model incorporating undercall probability was used to simulate corrected COIs (Fig. 4). Given the high heterozygosity of pfama1 and the observed COIs, the corrections showed minimal differences compared to the observed (uncorrected) COI results (Fig. 4). This would not be the case if the average COIs in this study were higher, as COIs > 5 were estimated to be undercalled for the majority of observed measures. As deep sequencing technologies become increasingly more cost effective and less labour-intensive, future studies targeting P. falciparum strain diversity in malaria endemic regions could include whole genome deep sequencing.
Conclusion
This study describes the use of amplicon-based deep sequencing for the detection and relative quantification of P. falciparum haplotypes and characterization of COI in the DRC and the spatial epidemiology and population genetic structure of malaria parasites from both individual and population cluster samples across eleven DRC provinces. Highly similar AMA1 amino acid frequencies between parasite populations were identified in the DRC and Mali, suggesting analogous selective pressures maintain pfama1 diversity in geographically divergent locations and therefore limit the use of pfama1 as a marker to discriminate parasite populations (or other markers known to be under balancing selection). Given the P. falciparum recent speciation bottleneck and limited diversity compared to other species, selection of more appropriate genetic markers of diversity may be a challenge. Sensitive detection methods, such as amplicon-based deep sequencing, can improve the understanding of malaria strain diversity as it relates to potential malaria vaccine antigen candidates and monitor for changes in parasite genetic diversity.
References
World Health Organization. World Malaria Report. Geneva: World Health Organization; 2016.
Langhorne J, Ndungu FM, Sponaas AM, Marsh K. Immunity to malaria: more questions than answers. Nat Immunol. 2008;9:725–32.
Doolan DL, Dobano C, Baird JK. Acquired immunity to malaria. Clin Microbiol Rev. 2009;22:13–36.
Auburn S, Campino S, Miotto O, Djimde AA, Zongo I, Manske M, et al. Characterization of within-host Plasmodium falciparum diversity using next-generation sequence data. PLoS ONE. 2012;7:e32891.
Branch OH, Takala S, Kariuki S, Nahlen BL, Kolczak M, Hawley W, et al. Plasmodium falciparum genotypes, low complexity of infection, and resistance to subsequent malaria in participants in the Asembo Bay Cohort Project. Infect Immun. 2001;69:7783–92.
Galinsky K, Valim C, Salmier A, de Thoisy B, Musset L, Legrand E, et al. COIL: a methodology for evaluating malarial complexity of infection using likelihood from single nucleotide polymorphism data. Malar J. 2015;14:4.
Kang JM, Moon SU, Kim JY, Cho SH, Lin K, Sohn WM, et al. Genetic polymorphism of merozoite surface protein-1 and merozoite surface protein-2 in Plasmodium falciparum field isolates from Myanmar. Malar J. 2010;9:131.
Kiwanuka GN. Genetic diversity in Plasmodium falciparum merozoite surface protein 1 and 2 coding genes and its implications in malaria epidemiology: a review of published studies from 1997 to 2007. J Vector Borne Dis. 2009;46:1–12.
Manske M, Miotto O, Campino S, Auburn S, Almagro-Garcia J, Maslen G, et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012;487:375–9.
Smith T, Beck HP, Kitua A, Mwankusye S, Felger I, Fraser-Hurt N, et al. Age dependence of the multiplicity of Plasmodium falciparum infections and of other malariological indices in an area of high endemicity. Trans R Soc Trop Med Hyg. 1999;93(Suppl 1):15–20.
Laishram DD, Sutton PL, Nanda N, Sharma VL, Sobti RC, Carlton JM, et al. The complexities of malaria disease manifestations with a focus on asymptomatic malaria. Malar J. 2012;11:29.
Bottius E, Guanzirolli A, Trape JF, Rogier C, Konate L, Druilhe P. Malaria: even more chronic in nature than previously thought; evidence for subpatent parasitaemia detectable by the polymerase chain reaction. Trans R Soc Trop Med Hyg. 1996;90:15–9.
Alves FP, Gil LH, Marrelli MT, Ribolla PE, Camargo EP, Da Silva LH. Asymptomatic carriers of Plasmodium spp. as infection source for malaria vector mosquitoes in the Brazilian Amazon. J Med Entomol. 2005;42:777–9.
Greenwood BM. Asymptomatic malaria infections—do they matter? Parasitol Today. 1987;3:206–14.
Chen I, Clarke SE, Gosling R, Hamainza B, Killeen G, Magill A, et al. “Asymptomatic” malaria: a chronic and debilitating infection that should be treated. PLoS Med. 2016;13:e1001942.
Ferreira MU, da Silva Nunes M, Wunderlich G. Antigenic diversity and immune evasion by malaria parasites. Clin Diagn Lab Immunol. 2004;11:987–95.
Bruce MC, Donnelly CA, Alpers MP, Galinski MR, Barnwell JW, Walliker D, et al. Cross-species interactions between malaria parasites in humans. Science. 2000;287:845–8.
Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–82.
Bushman M, Morton L, Duah N, Quashie N, Abuaku B, Koram KA, et al. Within-host competition and drug resistance in the human malaria parasite Plasmodium falciparum. Proc Biol Sci. 2016;283:20153038.
Dzikowski R, Deitsch KW. Genetics of antigenic variation in Plasmodium falciparum. Curr Genet. 2009;55:103–10.
Escalante AA, Lal AA, Ayala FJ. Genetic polymorphism and natural selection in the malaria parasite Plasmodium falciparum. Genetics. 1998;149:189–202.
Escalante AA, Smith DL, Kim Y. The dynamics of mutations associated with anti-malarial drug resistance in Plasmodium falciparum. Trends Parasitol. 2009;25:557–63.
Mahdi Abdel Hamid M, Elamin AF, Albsheer MM, Abdalla AA, Mahgoub NS, Mustafa SO, et al. Multiplicity of infection and genetic diversity of Plasmodium falciparum isolates from patients with uncomplicated and severe malaria in Gezira State, Sudan. Parasit Vectors. 2016;9:362.
Muhindo Mavoko H, Kalabuanga M, Delgado-Ratto C, Maketa V, Mukele R, Fungula B, et al. Uncomplicated clinical malaria features, the efficacy of artesunate-amodiaquine and their relation with multiplicity of infection in the Democratic Republic of Congo. PLoS ONE. 2016;11:e0157074.
Hastings IM, D’Alessandro U. Modelling a predictable disaster: the rise and spread of drug-resistant malaria. Parasitol Today. 2000;16:340–7.
Mackinnon MJ, Marsh K. The selection landscape of malaria parasites. Science. 2010;328:866–71.
Jiang H, Li N, Gopalan V, Zilversmit MM, Varma S, Nagarajan V, et al. High recombination rates and hotspots in a Plasmodium falciparum genetic cross. Genome Biol. 2011;12:R33.
Mu J, Myers RA, Jiang H, Liu S, Ricklefs S, Waisberg M, et al. Plasmodium falciparum genome-wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nat Genet. 2010;42:268–71.
de Roode JC, Pansini R, Cheesman SJ, Helinski ME, Huijben S, Wargo AR, et al. Virulence and competitive ability in genetically diverse malaria infections. Proc Natl Acad Sci USA. 2005;102:7624–8.
Arez AP, Pinto J, Palsson K, Snounou G, Jaenson TG, do Rosario VE. Transmission of mixed Plasmodium species and Plasmodium falciparum genotypes. Am J Trop Med Hyg. 2003;68:161–8.
Druilhe P, Daubersies P, Patarapotikul J, Gentil C, Chene L, Chongsuphajaisiddhi T, et al. A primary malarial infection is composed of a very wide range of genetically diverse but related parasites. J Clin Invest. 1998;101:2008–16.
Bendixen M, Msangeni HA, Pedersen BV, Shayo D, Bodker R. Diversity of Plasmodium falciparum populations and complexity of infections in relation to transmission intensity and host age: a study from the Usambara Mountains, Tanzania. Trans R Soc Trop Med Hyg. 2001;95:143–8.
Volkman SK, Neafsey DE, Schaffner SF, Park DJ, Wirth DF. Harnessing genomics and genome biology to understand malaria biology. Nat Rev Genet. 2012;13:315–28.
Daniels RF, Schaffner SF, Wenger EA, Proctor JL, Chang HH, Wong W, et al. Modeling malaria genomics reveals transmission decline and rebound in Senegal. Proc Natl Acad Sci USA. 2015;112:7067–72.
Nkhoma SC, Nair S, Al-Saai S, Ashley E, McGready R, Phyo AP, et al. Population genetic correlates of declining transmission in a human pathogen. Mol Ecol. 2013;22:273–85.
Vafa M, Troye-Blomberg M, Anchang J, Garcia A, Migot-Nabias F. Multiplicity of Plasmodium falciparum infection in asymptomatic children in Senegal: relation to transmission, age and erythrocyte variants. Malar J. 2008;7:17.
Kateera F, Nsobya SL, Tukwasibwe S, Mens PF, Hakizimana E, Grobusch MP, et al. Malaria case clinical profiles and Plasmodium falciparum parasite genetic diversity: a cross sectional survey at two sites of different malaria transmission intensities in Rwanda. Malar J. 2016;15:237.
Mohd Abd Razak MR, Sastu UR, Norahmad NA, Abdul-Karim A, Muhammad A, Muniandy PK, et al. Genetic diversity of Plasmodium falciparum populations in malaria declining areas of Sabah, East Malaysia. PLoS One. 2016;11:e0152415.
Nabet C, Doumbo S, Jeddi F, Konate S, Manciulli T, Fofana B, et al. Genetic diversity of Plasmodium falciparum in human malaria cases in Mali. Malar J. 2016;15:353.
Volkman SK, Ndiaye D, Diakite M, Koita OA, Nwakanma D, Daniels RF, et al. Application of genomics to field investigations of malaria by the international centers of excellence for malaria research. Acta Trop. 2012;121:324–32.
Chang HH, Park DJ, Galinsky KJ, Schaffner SF, Ndiaye D, Ndir O, et al. Genomic sequencing of Plasmodium falciparum malaria parasites from Senegal reveals the demographic history of the population. Mol Biol Evol. 2012;29:3427–39.
Niang M, Thiam LG, Loucoubar C, Sow A, Sadio BD, Diallo M, et al. Spatio-temporal analysis of the genetic diversity and complexity of Plasmodium falciparum infections in Kedougou, southeastern Senegal. Parasit Vectors. 2017;10:33.
Agyeman-Budu A, Brown C, Adjei G, Adams M, Dosoo D, Dery D, et al. Trends in multiplicity of Plasmodium falciparum infections among asymptomatic residents in the middle belt of Ghana. Malar J. 2013;12:22.
Alam MS, Elahi R, Mohon AN, Al-Amin HM, Kibria MG, Khan WA, et al. Plasmodium falciparum genetic diversity in Bangladesh does not suggest a hypoendemic population structure. Am J Trop Med Hyg. 2016;94:1245–50.
Duah NO, Matrevi SA, Quashie NB, Abuaku B, Koram KA. Genetic diversity of Plasmodium falciparum isolates from uncomplicated malaria cases in Ghana over a decade. Parasit Vectors. 2016;9:416.
Ferreira MU, Liu Q, Kaneko O, Kimura M, Tanabe K, Kimura EA, et al. Allelic diversity at the merozoite surface protein-1 locus of Plasmodium falciparum in clinical isolates from the southwestern Brazilian Amazon. Am J Trop Med Hyg. 1998;59:474–80.
Konate L, Zwetyenga J, Rogier C, Bischoff E, Fontenille D, Tall A, et al. Variation of Plasmodium falciparum msp1 block 2 and msp2 allele prevalence and of infection complexity in two neighbouring Senegalese villages with different transmission conditions. Trans R Soc Trop Med Hyg. 1999;93(Suppl 1):21–8.
Schleiermacher D, Rogier C, Spiegel A, Tall A, Trape JF, Mercereau-Puijalon O. Increased multiplicity of Plasmodium falciparum infections and skewed distribution of individual msp1 and msp2 alleles during pregnancy in Ndiop, a Senegalese village with seasonal, mesoendemic malaria. Am J Trop Med Hyg. 2001;64:303–9.
Snounou G. Genotyping of Plasmodium spp. Nested PCR. Methods Mol Med. 2002;72:103–16.
Juliano JJ, Porter K, Mwapasa V, Sem R, Rogers WO, Ariey F, et al. Exposing malaria in-host diversity and estimating population diversity by capture-recapture using massively parallel pyrosequencing. Proc Natl Acad Sci USA. 2010;107:20138–43.
Hastings IM, Nsanzabana C, Smith TA. A comparison of methods to detect and quantify the markers of antimalarial drug resistance. Am J Trop Med Hyg. 2010;83:489–95.
Juliano JJ, Kwiek JJ, Cappell K, Mwapasa V, Meshnick SR. Minority-variant pfcrt K76T mutations and chloroquine resistance, Malawi. Emerg Infect Dis. 2007;13:872–7.
Liu S, Mu J, Jiang H, Su XZ. Effects of Plasmodium falciparum mixed infections on in vitro antimalarial drug tests and genotyping. Am J Trop Med Hyg. 2008;79:178–84.
Farnert A, Arez AP, Babiker HA, Beck HP, Benito A, Bjorkman A, et al. Genotyping of Plasmodium falciparum infections by PCR: a comparative multicentre study. Trans R Soc Trop Med Hyg. 2001;95:225–32.
Gandhi K, Thera MA, Coulibaly D, Traore K, Guindo AB, Doumbo OK, et al. Next generation sequencing to detect variation in the Plasmodium falciparum circumsporozoite protein. Am J Trop Med Hyg. 2012;86:775–81.
Robinson T, Campino SG, Auburn S, Assefa SA, Polley SD, Manske M, et al. Drug-resistant genotypes and multi-clonality in Plasmodium falciparum analysed by direct genome sequencing from peripheral blood of malaria patients. PLoS ONE. 2011;6:e23204.
Democratic Republic of Congo Malaria Operational Plan FY 2014. President’s Malaria Initiative, USAID, CDC, DRC NMCP 2014.
The Demographic and Health Surveys Program. Democratic Republic of the Congo: demographic and health survey 2007 key findings. Calverton: The Demographic and Health Surveys Program, USAID, Macro International Inc.; 2007.
Taylor SM, Messina JP, Hand CC, Juliano JJ, Muwonga J, Tshefu AK, et al. Molecular malaria epidemiology: mapping and burden estimates for the Democratic Republic of the Congo, 2007. PLoS ONE. 2011;6:e16420.
Messina JP. Disease ecology in the Democratic Republic of the Congo: integration of spatial analysis with population surveillance. Chapel Hill: University of North Carolina at Chapel Hill, Department of Geography; 2011.
Carrel M, Patel J, Taylor SM, Janko M, Mwandagalirwa MK, Tshefu AK, et al. The geography of malaria genetics in the Democratic Republic of Congo: a complex and fragmented landscape. Soc Sci Med. 2015;133:233–41.
Messina JP, Emch M, Muwonga J, Mwandagalirwa K, Edidi SB, Mama N, et al. Spatial and socio-behavioral patterns of HIV prevalence in the Democratic Republic of Congo. Soc Sci Med. 2010;71:1428–35.
Messina JP, Mwandagalirwa K, Taylor SM, Emch M, Meshnick SR. Spatial and social factors drive anemia in Congolese women. Health Place. 2013;24:54–64.
Messina JP, Taylor SM, Meshnick SR, Linke AM, Tshefu AK, Atua B, et al. Population, behavioural and environmental drivers of malaria prevalence in the Democratic Republic of Congo. Malar J. 2011;10:161.
Taylor SM, Antonia AL, Parobek CM, Juliano JJ, Janko M, Emch M, et al. Plasmodium falciparum sulfadoxine resistance is geographically and genetically clustered within the DR Congo. Sci Rep. 2013;3:1165.
Taylor SM, van Eijk AM, Hand CC, Mwandagalirwa K, Messina JP, Tshefu AK, et al. Quantification of the burden and consequences of pregnancy-associated malaria in the Democratic Republic of the Congo. J Infect Dis. 2011;204:1762–71.
Wilson PE, Kazadi W, Kamwendo DD, Mwapasa V, Purfield A, Meshnick SR. Prevalence of pfcrt mutations in Congolese and Malawian Plasmodium falciparum isolates as determined by a new Taqman assay. Acta Trop. 2005;93:97–106.
Brazeau NF, Hathaway N, Parobek CM, Lin JT, Bailey JA, Lon C, et al. Longitudinal pooled deep sequencing of the Plasmodium vivax K12 kelch gene in Cambodia reveals a lack of selection by artemisinin. Am J Trop Med Hyg. 2016;95:1409–12.
Juliano JJ, Parobek CM, Brazeau NF, Ngasala B, Randrianarivelojosia M, Lon C, et al. Pooled amplicon deep sequencing of candidate Plasmodium falciparum transmission-blocking vaccine antigens. Am J Trop Med Hyg. 2016;94:143–6.
Ngondi JM, Ishengoma DS, Doctor SM, Thwai KL, Keeler C, Mkude S, et al. Surveillance for sulfadoxine-pyrimethamine resistant malaria parasites in the Lake and Southern Zones, Tanzania, using pooling and next-generation sequencing. Malar J. 2017;16:236.
Taylor SM, Parobek CM, DeConti DK, Kayentao K, Coulibaly SO, Greenwood BM, et al. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in Sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis. 2015;211:680–8.
Pickard AL, Wongsrichanalai C, Purfield A, Kamwendo D, Emery K, Zalewski C, et al. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob Agents Chemother. 2003;47:2418–23.
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115.
Using Multiplex Identifier (MID) Adaptors for the GS FLX Titanium Chemistry—Basic MID Set. http://454.com/downloads/my454/documentation/technical-bulletins/TCB-09004_UsingMultiplexIdentifierAdaptorsForTheGSFLXTitaniumSeriesChemistry-BasicMIDSet.pdf.
Hathaway NJ, Parobek CM, Juliano JJ, Bailey JA. SeekDeep: single-base resolution clustering for amplicon deep sequencing. Nucleic Acids Res. 2017. https://doi.org/10.1093/nar/gkx1201 (Epub ahead of print).
Lin JT, Hathaway NJ, Saunders DL, Lon C, Balasubramanian S, Kharabora O, et al. Using amplicon deep sequencing to detect genetic signatures of Plasmodium vivax relapse. J Infect Dis. 2015;212:999–1008.
Hathaway N. SeekDeep Targeted Amplicon Analysis. 2015. http://baileylab.umassmed.edu/SeekDeep.
Mideo N, Bailey JA, Hathaway NJ, Ngasala B, Saunders DL, Lon C, et al. A deep sequencing tool for partitioning clearance rates following antimalarial treatment in polyclonal infections. Evol Med Public Health. 2016;2016:21–36.
Parobek CM, Lin JT, Saunders DL, Barnett EJ, Lon C, Lanteri CA, et al. Selective sweep suggests transcriptional regulation may underlie Plasmodium vivax resilience to malaria control measures in Cambodia. Proc Natl Acad Sci USA. 2016;113:E8096–105.
Takala SL, Coulibaly D, Thera MA, Batchelor AH, Cummings MP, Escalante AA, et al. Extreme polymorphism in a vaccine antigen and risk of clinical malaria: implications for vaccine development. Sci Transl Med. 2009;1:2ra5.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Rozas J. DNA sequence polymorphism analysis using DnaSP. Methods Mol Biol. 2009;537:337–50.
Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2.
Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987.
Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–7.
Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48.
Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012;28:2537–9.
Peakall R, Smouse P. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–95.
R Core Team. A language and environment for statistical computing. Vienna: R Foundation for Statistical, Computing; 2017.
Democratic Republic of Congo 2007 DHS Boundaries, spatial data repository. The demographic and health surveys program. http://spatialdata.dhsprogram.com/boundaries/#view=table&countryId=CD.
Takala SL, Plowe CV. Genetic diversity and malaria vaccine design, testing and efficacy: preventing and overcoming ‘vaccine resistant malaria’. Parasite Immunol. 2009;31:560–73.
Aragam NR, Thayer KM, Nge N, Hoffman I, Martinson F, Kamwendo D, et al. Diversity of T cell epitopes in Plasmodium falciparum circumsporozoite protein likely due to protein-protein interactions. PLoS ONE. 2013;8:e62427.
Arnott A, Wapling J, Mueller I, Ramsland PA, Siba PM, Reeder JC, et al. Distinct patterns of diversity, population structure and evolution in the AMA1 genes of sympatric Plasmodium falciparum and Plasmodium vivax populations of Papua New Guinea from an area of similarly high transmission. Malar J. 2014;13:233.
Osier FH, Weedall GD, Verra F, Murungi L, Tetteh KK, Bull P, et al. Allelic diversity and naturally acquired allele-specific antibody responses to Plasmodium falciparum apical membrane antigen 1 in Kenya. Infect Immun. 2010;78:4625–33.
Polley SD, Chokejindachai W, Conway DJ. Allele frequency-based analyses robustly map sequence sites under balancing selection in a malaria vaccine candidate antigen. Genetics. 2003;165:555–61.
Cortes A, Mellombo M, Masciantonio R, Murphy VJ, Reeder JC, Anders RF. Allele specificity of naturally acquired antibody responses against Plasmodium falciparum apical membrane antigen 1. Infect Immun. 2005;73:422–30.
Cortes A, Mellombo M, Mueller I, Benet A, Reeder JC, Anders RF. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infect Immun. 2003;71:1416–26.
Polley SD, Conway DJ. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics. 2001;158:1505–12.
Thera MA, Doumbo OK, Coulibaly D, Laurens MB, Ouattara A, Kone AK, et al. A field trial to assess a blood-stage malaria vaccine. N Engl J Med. 2011;365:1004–13.
Spring MD, Cummings JF, Ockenhouse CF, Dutta S, Reidler R, Angov E, et al. Phase 1/2a study of the malaria vaccine candidate apical membrane antigen-1 (AMA-1) administered in adjuvant system AS01B or AS02A. PLoS ONE. 2009;4:e5254.
Drew DR, Hodder AN, Wilson DW, Foley M, Mueller I, Siba PM, et al. Defining the antigenic diversity of Plasmodium falciparum apical membrane antigen 1 and the requirements for a multi-allele vaccine against malaria. PLoS ONE. 2012;7:e51023.
Dutta S, Dlugosz LS, Drew DR, Ge X, Ababacar D, Rovira YI, et al. Overcoming antigenic diversity by enhancing the immunogenicity of conserved epitopes on the malaria vaccine candidate apical membrane antigen-1. PLoS Pathog. 2013;9:e1003840.
Terheggen U, Drew DR, Hodder AN, Cross NJ, Mugyenyi CK, Barry AE, et al. Limited antigenic diversity of Plasmodium falciparum apical membrane antigen 1 supports the development of effective multi-allele vaccines. BMC Med. 2014;12:183.
Bailey JA, Pablo J, Niangaly A, Travassos MA, Ouattara A, Coulibaly D, et al. Seroreactivity to a large panel of field-derived Plasmodium falciparum apical membrane antigen 1 and merozoite surface protein 1 variants reflects seasonal and lifetime acquired responses to malaria. Am J Trop Med Hyg. 2015;92:9–12.
Authors’ contributions
Designed the study and experiments: RM, SM, JJ, AS, JB. Performed the experiments: RM, OK. Analysed and interpreted data: RM, NH, SM, JJ, AS, JB, Contributed reagents/materials/analysis tools: NH, KM, AT, SM, ST, JJ, AS, JB. All authors read and approved the final manuscript.
Acknowledgements
We thank Dr. Jeremie Muwonga (Laboratoire National de Reference SIDA et IST (LNRS), Kinshasa, DRC), Dr. Benjamin Atua (Programme National de Lutte contre le Paludisme (PNLP), Kinshasa, DRC), and the late Dr. Augustin Okenge (Programme National de Lutte Contre le SIDA, Kinshasa, DRC) for logistic and moral support during sample collection. We also thank Dr. Mark Janko (University of North Carolina, Chapel Hill, NC, USA) for his help with sample selection and geographic information support. We are grateful to Dr. Jane Messina (University of Oxford, Oxford, UK) for her guidance and prior work on the malaria molecular and spatial epidemiology using dried blood spots collected for the 2007 DHS in the DRC. We also wish to express our gratitude to the DHS administrators and the DHS participants without whom this study would not have been possible.
The following reagents were obtained through BEI Resources, NIAID, NIH: Genomic DNA from P. falciparum, Strain V1/S (MRA-176G) contributed by Dennis E. Kyle, genomic DNA from P. falciparum, strain RO-33 (MRA-200G), strain Dd2 (MRA-150G), strain 7G8 (MRA-152G), and strain K1 (MRA-159G) contributed by David Walliker.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Data generated or analysed during this study are included in this published article and its Additional files or are available from the corresponding author upon request.
Consent for publication
Not applicable.
Disclaimer
The contents of this publication are the sole responsibility of the authors and do not necessarily reflect the views, assertions, opinions or policies of the Uniformed Services University of the Health Sciences (USUHS), the Department of Defense (DoD), or the Departments of the Army, Navy, or Air Force. Mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. Government.
Ethics approval and consent to participate
Included in “Methods”.
Funding
This work was supported in part by the US National Institutes of Health (SM, 5R01AI107949; JJ, 5R01AI089819; AS, R01AI104423; JB, R01AI099473), the Uniformed Services University of the Health Sciences (AS, R0878N7), and the Naval Medical Research Center (AS, HT904-13-1-0022). The funding bodies had no role in the study design, data analysis and interpretation, or preparation of the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
12936_2017_2137_MOESM1_ESM.xlsx
Additional file 1. Population cluster sample characteristics. Asterisks (*) indicate population cluster samples that failed initial pfama1 PCR amplification (n=5). Diamonds (♦) indicate a population cluster sample that was not successfully sequenced (n=6).
12936_2017_2137_MOESM3_ESM.pdf
Additional file 3. A workflow diagram outlines the steps from nested PCR to bioinformatic analyses (left boxes) and provides corresponding background, reasoning, and details at each step in the process (right boxes).
12936_2017_2137_MOESM4_ESM.pdf
Additional file 4. Six internal quality control samples were PCR amplified and deep sequenced in duplicate. Expected (first column) and actual (sequencing control samples 1-6, averaged across duplicates) haplotype percentages are similar. The average percent error between duplicates was 4.4% (range 0.4-13.6%).
12936_2017_2137_MOESM6_ESM.xlsx
Additional file 6. Comparison of haplotype frequencies between all sample types, individual samples, and population cluster samples.
12936_2017_2137_MOESM7_ESM.docx
Additional file 7. Comparison of demographic factors between sample sequenced individually (n=79) and pooled population cluster samples (N=821) with analysable sequence reads.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Miller, R.H., Hathaway, N.J., Kharabora, O. et al. A deep sequencing approach to estimate Plasmodium falciparum complexity of infection (COI) and explore apical membrane antigen 1 diversity. Malar J 16, 490 (2017). https://doi.org/10.1186/s12936-017-2137-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-017-2137-9