Abstract
As a member of the miR-1 family, miR-206 is located between IL-17 and PKHD1 genes in human. This miRNA has been shown to be involved in the pathogenic processes in a variety of human disorders including cancers, amyotrophic lateral sclerosis, Alzheimer’s disease, atherosclerosis, bronchopulmonary dysplasia, coronary artery disease, chronic obstructive pulmonary disease, epilepsy, nonalcoholic fatty liver disease, Hirschsprung disease, muscular dystrophies, pulmonary arterial hypertension, sepsis and ulcerative colitis. In the current review, we summarize the role of miR-206 in both malignant and non-malignant situations and explain its possible therapeutic implications.
Similar content being viewed by others
Introduction
MicroRNAs (miRNAs) are a group of naturally happening short non-coding RNAs with 21 to 22 nucleotide long. These transcripts contribute to post-transcriptional silencing of target genes [1, 2]. A single miRNA can affect expression of thousands of mRNAs and their target genes [3, 4]. Universally, miRNAs interact with 3′UTR to inhibit translation or degrade target transcripts. The critical seed region of miRNAs is located in the nucleotides 2–7 of their 5′UTR [5, 6]. miRNAs participate in several critical regulatory functions associated with cell growth, developmental processes and differentiation. Dysregulation of miRNAs is associated with a wide array of human pathway pathologies, especially cancers [7, 8]. Disruption in miRNA levels have been reported in numerous disease processes. So, they have the potential to be developed into novel therapeutic targets [9,10,11].
miR-206 is a member of the miR-1 family. The gene encoding this miRNA is located between the IL-17 and PKHD1 genes in human [5]. The cytogenetic band of miR-206 is 6p12.2. This miRNA has been shown to participate in the pathogenesis of a variety of malignant and non-malignant conditions. Like other member of this miRNA family, miR-206 has physiological roles as well. Members of mouse mir-1 family have important functions in muscle development [12]. In the current review, we summarize the role of miR-206 in both malignant and non-malignant conditions and explain its possible therapeutic implications.
Bioinformatics step
Prediction of miR-206 target genes
miRWalk (http://mirwalk.umm.uni-heidelberg.de/), miRDB (http://www.mirdb.org/), and TargetScan databases (https://www.targetscan.org/) were used to predict the miR-206 target genes. A total of 82 mRNAs have been identified as common targets of this miRNA in these three databases (Fig. 1). Based on the results, miR-206 is predicted to target a variety of genes being involved in a wide range of cellular functions.
miR-206 target genes. A Prediction of a total of 82 common target genes in the three databases miRWalk, miRDB, and TargetScan. B 82 common genes targeted by miR-206. The interaction network was constructed by Cytoscape software (Cytoscape (3.9.1), using Java 11, Free Software Foundation, Inc., MA 02111–1307 USA)
Gene ontology (GO) and pathway enrichment analysis of miR-206 target genes
The Enrichr online database (https://maayanlab.cloud/Enrichr/) was used to perform GO analyses for the top miR-206 target genes. The target genes of miR-206 were remarkably enriched in the following GO terms: activation of cysteine-type endopeptidase activity involved in apoptotic process, collagen fibril organization, and hemopoiesis in biological process (BP), intracellular membrane-bounded organelle, actin cytoskeleton, and nucleus in cellular component (CC); and RNA polymerase binding, methyl-CpG binding, and DNA binding in molecular function (MF) (Table 1).
Additionally, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were conducted using CancerMIRNome database (http://bioinfo.jialab-ucr.org/CancerMIRNome/). This uncovered that Proteoglycans in cancer, Endocrine resistance, and AGE-RAGE signaling pathways were the top-ranked pathways for miR-206 (Fig. 2).
Literature search step
Role of miR-206 in cancers
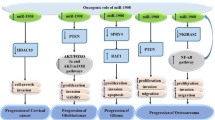
Several studies have assessed expression of miR-206 in different types of cancers and found the molecular mechanism of involvement of this miRNA in the carcinogenesis (Fig. 3).
Effect of miR-206 in human cancers. miR-206 exerts its role in the development of cancer through modulation of expression of a variety of targets. Detailed data is shown in Table 2
An experiment in bladder cancer tissues has shown over-expression of lncRNA RMRP in these tissues compared with adjacent tissues using qRT-PCR method. As revealed by MTT assay and transwell assay, RMRP induces cell proliferation, migration and invasiveness of bladder cancer cells through regulation of miR-206. The latter finding is based on the observed binding of miR-206 and RMRP in luciferase assay [13].
Similarly, in breast cancer cells, miR-206 has a tumor suppressor role possibly through down-regulation of PFKFB3. Expression of miR-206 in estrogen receptor α (ERα) positive breast cancer cells has been found to be reduced by 17β-estradiol in a dose-dependent manner. Over-expression of miR-206 could impede production of fructose-2,6-bisphosphate, diminish lactate synthesis and reduce proliferative ability and migration of breast cancer cells [14]. An independent study in breast cancer has shown association between down-regulation of miR-206 and large tumor dimension and advanced clinical stage. Over-expression of miR-206 in MCF-7 cells has suppressed cell growth through hindering G1/S transition. This effect is mediated through suppression of expression of cyclin D2. Consistent with this finding, expression levels of miR-206 have been inversely correlated with those of cyclin D2 in breast cancer tissues [15]. Expression of miR-206 has also been shown to be reduced in ERα-positive breast tumors. Besides, expression of miR-206 has been inversely related with ERα but not ERβ transcript levels in breast cancer tissues. Forced over-expression of miR-206 into MCF-7 cells has led to reduction of cell growth in both dose- and time-dependent manners, implying that miR-206 can be a target for endocrine therapy in this type of cancer [16]. Similarly, miR-206 has been shown to inhibit stemness and metastastatic ability of breast cancer cells through influencing activity of MKL1/IL11 axis [17]. Moreover, this miRNA can suppress epithelial mesenchymal transition (EMT) through influencing activity of TGF-β signals in ER-positive breast cancer cells [18].
miR-206 has also been down-regulated in the cervical cancer tissues, parallel with up-regulation of its target gene c-Met as revealed by qRT-PCR assay and immunohistochemistry. Kaplan–Meier and log-rank analyses have shown relation between down-regulation of miR-206 and shorter overall survival. Besides, down-regulation of miR-206 in cervical cancer tissues has been associated with lymph node metastasis, advanced stage and advanced histological grade indicating the role of miR-206 in the metastasis and progression of cervical cancer. In fact, miR-206 has been found to be independent prognostic marker for overall survival of patients with this type of cancer [19]. Table 2 shows summary of the role of miR-206 in malignant conditions.
Diagnostic role of miR-206 has been assessed in epithelial ovarian cancer [32], renal cell carcinoma [52] and rhabdomyosarcoma [53] (Table 3). In epithelial ovarian cancer, miR-206 levels can be used for discrimination of patients with incomplete response to platinum chemotherapy from those with complete response to this modality with area under the receiver characteristic curve (AUC) of 0.82 [32]. Most notably, expression levels of miR-206 has a high accuracy in discrimination of patients with rhabdomyosarcoma from healthy subjects with AUC value of 0.96 [53].
Role of miR-206 in non-malignant conditions
miR-206 has crucial effects in the pathophysiology of several non-malignant disorders through modulation of a variety of targets (Fig. 4).
Role of miR-206 in non-malignant disorders. Detailed data about experiments is shown in Table 4
Expression assays in amyotrophic lateral sclerosis (ALS) have shown dysregulation of circulatory levels of several miRNAs in these patients. Notably, miR-206 has been among up-regulated miRNAs in these patients. In addition, constant changes in miRNAs signature have been found to persist during progression of ALS. This finding indicates the potential of selected miRNAs such as miR-206 as longitudinal markers for this disorder [59].
Circulatory levels of a number of miRNAs such as miR-206 have also been used as possible predictors for the progression of amnestic mild cognitive impairment (aMCI) to Alzheimer's disease (AD). Notably, serum levels of miR-206 have been found to be higher in aMCI patients progressed to AD. Kaplan–Meier analysis has also demonstrated remarkable correlation between conversion of aMCI to AD and over-expression of miR-206 [60]. Over-expression of miR-206 in olfactory mucosal cells can also been used as an early diagnostic approach in AD [61]. Another experiment in animal models of AD and temporal cortex samples from AD patients has verified over-expression of miR-206. These effects are mainly mediated through modulation of BDNF expression. In fact, a neutralizing inhibitor of this miRNA could prevent the harmful effect of amyloid-β42 on BDNF and dendritic spine degeneration [62]. On the other hand, another study has shown a neuroprotective effect of miR-206-3p in AD [63]. Table 4 shows summary of the role of miR-206 in non-malignant conditions.
Diagnostic value of miR-206 has been evaluated in AD and muscular dystrophies (Table 5). In Duchene muscular dystrophy, expression levels of miR-206 can be used as a diagnostic marker with AUC value of 0.96 [71]. Similarly, this miRNA can be used as a marker for diagnosis of Becker muscular dystrophy [77].
Discussion
miR-206 is an example of miRNAs with crucial roles in the pathogenesis of a wide range of human disorders. In the context of cancer, expression assays using qRT-PCR method and functional studies have led to the supposition of miR-206 as a tumor suppressor miRNA (summarized in Table 2), although some exceptions have been demonstrated [53]. It can reduce proliferation of cancer cells and induce their apoptosis [14] via different routes. Moreover, it can regulate cell cycle progression through modulation of expression of cell cycle-related genes [15]. The activity of several oncogenic pathways is modulated by miR-206. Examples of these pathways are EGF/EGFR [21], EGFR/MAPK [21], TGF-β [18], Notch3 [26], PTEN/AKT/mTOR [29], VEGF [37], c-Met-Akt/Erk [42], Erk1/2 [42] and PI3K/AKT [43]. Most importantly, expression assays in patients with different responses to chemotherapeutic agents and functional studies in cell lines have shown that miR-206 can enhance cytotoxic effects of anti-cancer agents on cancer cells [22]. The latter finding highlights the importance of this miRNA in design of novel modalities to combat chemoresistance.
Among non-malignant conditions, dysregulation of miR-206 has been reported in amyotrophic lateral sclerosis, Alzheimer’s disease, atherosclerosis, bronchopulmonary dysplasia, coronary artery disease, chronic obstructive pulmonary disease, epilepsy, nonalcoholic fatty liver disease, Hirschsprung disease, muscular dystrophies, pulmonary arterial hypertension, sepsis and ulcerative colitis (summarized in Table 4). Thus, this miRNA can affect pathogenesis of a wide array of human disorders.
In silico studies have revealed that miR-206 can affect expression of tens of mRNAs being involved in the regulation of crucial cellular mechanisms such as activation of cysteine-type endopeptidase activity involved in apoptotic process, collagen fibril organization, hemopoiesis, regulation of tubulin deacetylation, regulation of vascular endothelial cell proliferation, regulation of gonadotropin secretion, response to prostaglandin, RNA polymerase binding, methyl-CpG binding and sequence-specific DNA binding. Therefore, it is not surprising that miR-206 influences pathoetiology of several disorders.
Notably, expression levels of miR-206 not only can be used for cancer diagnosis [53] and in determination of response to anti-cancer therapies [32], but also may be potential markers for discrimination of patients with muscular dystrophies from healthy subjects [71] or prediction of course of Alzheimer’s disease [61]. Since miRNAs can be easily tracked in the biofluids, these findings open a new era for detection of human disorders via non-invasive tools.
Conclusion
Altered expression of miR-206 in tumor tissues has been associated with malignant characteristics of cancers in terms of higher metastatic aptitude and lower survival rate, implying the role of this miRNA as a prognostic marker. Finally, forced over-expression of miR-206 in many cancer cell lines has led to reduction of malignant characteristics in cell line assays as well as animal models. Thus, this strategy can be used as a novel therapeutic approach for cancers. Meanwhile, miR-206 is involved in the pathophysiology of several non-malignant conditions, including neurodegenerative and neuropsychiatric disorders and muscular atrophies. Since efficient therapies for these kinds of disorders have not been developed yet, miR-206-targetted therapies might revolutionize this research field.
Availability of data and materials
The analyzed data sets generated during the study are available from the co‑responding author on reasonable request.
References
Jalali-Qomi S, Motovali-Bashi M, Rezaei H, Khalilian S. Experimental validation of a predicted microRNA within human FVIII gene. Molecular Biology Research Communications. 2021;10(2):45.
Kadkhoda S, Ghafouri-Fard S. The importance of miRNA-630 in human diseases with an especial focus on cancers. Cancer Cell Int. 2022;22(1):105.
Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205.
Kadkhoda S, Ghafouri-Fard S. Function of miRNA-145–5p in the pathogenesis of human disorders. Pathol Res Pract. 2022;231:153780.
Ma G, Wang Y, Li Y, Cui L, Zhao Y, Zhao B, et al. MiR-206, a key modulator of skeletal muscle development and disease. Int J Biol Sci. 2015;11(3):345.
Taheri M, Khoshbakht T, Hussen BM, Ghafouri-Fard S, Samadian M. A review on the role of miR-1246 in the pathoetiology of different cancers. Front Mol Biosci. 2022. https://doi.org/10.3389/fmolb.2021.771835.
Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358(5):502–11.
Ghafouri-Fard S, Khoshbakht T, Hussen BM, Jamal HH, Taheri M, Hajiesmaeili M. A comprehensive review on function of miR-15b-5p in malignant and non-malignant disorders. Front Oncol. 2022. https://doi.org/10.3389/fonc.2022.870996.
Rezaei H, Motovali-Bashi M, Khalilian S. Identification of Novel miRNAs in the F8 Gene Via Bioinformatics Tools. Iranian Journal of Biotechnology. 2021;19(2):e2700.
Ghafouri-Fard S, Khoshbakht T, Hussen BM, Kadkhoda S, Taheri M, Tafrishinejad A. A review on the role of miR-149-5p in the carcinogenesis. Int J Mol Sci. 2021;23(1):415.
Ghafouri-Fard S, Shaterabadi D, Abak A, Shoorei H, Bahroudi Z, Taheri M, et al. An update on the role of miR-379 in human disorders. Biomed Pharmacother. 2021;139: 111553.
Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–62.
Cao H, Liu Z, Huang P, Yue Y, Xi J. lncRNA-RMRP promotes proliferation, migration and invasion of bladder cancer via miR-206. Eur Rev Med Pharmacol Sci. 2019;23(3):1012–21.
Ge X, Lyu P, Cao Z, Li J, Guo G, Xia W, et al. Overexpression of miR-206 suppresses glycolysis, proliferation and migration in breast cancer cells via PFKFB3 targeting. Biochem Biophys Res Commun. 2015;463(4):1115–21.
Zhou J, Tian Y, Li J, Lu B, Sun M, Zou Y, et al. miR-206 is down-regulated in breast cancer and inhibits cell proliferation through the up-regulation of cyclinD2. Biochem Biophys Res Commun. 2013;433(2):207–12.
Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H. MiR-206 expression is down-regulated in estrogen receptor α–positive human breast cancer. Can Res. 2008;68(13):5004–8.
Samaeekia R, Adorno-Cruz V, Bockhorn J, Chang Y-F, Huang S, Prat A, et al. miR-206 inhibits stemness and metastasis of breast cancer by targeting MKL1/IL11 Pathwaymir-206 inhibits stemness and metastasis. Clin Cancer Res. 2017;23(4):1091–103.
Yin K, Yin W, Wang Y, Zhou L, Liu Y, Yang G, et al. MiR-206 suppresses epithelial mesenchymal transition by targeting TGF-β signaling in estrogen receptor positive breast cancer cells. Oncotarget. 2016;7(17):24537.
Chen AH, Qin YE, Tang WF, Tao J, Song HM, Zuo M. MiR-34a and miR-206 act as novel prognostic and therapy biomarkers in cervical cancer. Cancer Cell Int. 2017;17(1):1–9.
Hesari Z, Nourbakhsh M, Hosseinkhani S, Abdolvahabi Z, Alipour M, Tavakoli-Yaraki M, et al. Down-regulation of NAMPT expression by mir-206 reduces cell survival of breast cancer cells. Gene. 2018;673:149–58.
Adams BD, Cowee DM, White BA. The role of miR-206 in the epidermal growth factor (EGF) induced repression of estrogen receptor-α (ERα) signaling and a luminal phenotype in MCF-7 breast cancer cells. Mol Endocrinol. 2009;23(8):1215–30.
Wang R, Zhang T, Yang Z, Jiang C, Seng J. Long non-coding RNA FTH 1P3 activates paclitaxel resistance in breast cancer through miR-206/ABCB 1. J Cell Mol Med. 2018;22(9):4068–75.
Zhou Y, Wang M, Tong Y, Liu X, Zhang L, Dong D, et al. miR-206 promotes cancer progression by targeting full-length neurokinin-1 receptor in breast cancer. Technol Cancer Res Treat. 2019;18:1533033819875168.
Sun Q, Qi X, Zhang W, Li X. Knockdown of circRNA_0007534 suppresses the tumorigenesis of cervical cancer via miR-206/GREM1 axis. Cancer Cell Int. 2021;21(1):1–14.
Wang Y, Tian Y. miR-206 inhibits cell proliferation, migration, and invasion by targeting BAG3 in human cervical cancer. Oncol Res. 2018;26(6):923.
Song G, Zhang Y, Wang L. MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J Biol Chem. 2009;284(46):31921–7.
Parasramka MA, Dashwood WM, Wang R, Saeed HH, Williams DE, Ho E, et al. A role for low-abundance miRNAs in colon cancer: the miR-206/Krüppel-like factor 4 (KLF4) axis. Clin Epigenetics. 2012;4(1):1–10.
Meng X, Fu R. miR-206 regulates 5-FU resistance by targeting Bcl-2 in colon cancer cells. Onco Targets Ther. 2018;11:1757.
Zheng Y, Yang X, Wang C, Zhang S, Wang Z, Li M, et al. HDAC6, modulated by miR-206, promotes endometrial cancer progression through the PTEN/AKT/mTOR pathway. Sci Rep. 2020;10(1):1–12.
Chen X, Yan Q, Li S, Zhou L, Yang H, Yang Y, et al. Expression of the tumor suppressor miR-206 is associated with cellular proliferative inhibition and impairs invasion in ERα-positive endometrioid adenocarcinoma. Cancer Lett. 2012;314(1):41–53.
Dai C, Xie Y, Zhuang X, Yuan Z. MiR-206 inhibits epithelial ovarian cancer cells growth and invasion via blocking c-Met/AKT/mTOR signaling pathway. Biomed Pharmacother. 2018;104:763–70.
Yu X, Zhang X, Wang G, Wang B, Ding Y, Zhao J, et al. miR-206 as a prognostic and sensitivity biomarker for platinum chemotherapy in epithelial ovarian cancer. Cancer Cell Int. 2020;20(1):1–16.
Zhang C, Luo Y, Cao J, Wang X, Miao Z, Shao G. Exosomal lncRNA FAM225A accelerates esophageal squamous cell carcinoma progression and angiogenesis via sponging miR-206 to upregulate NETO2 and FOXP1 expression. Cancer Med. 2020;9(22):8600–11.
Wang S-H, Zhang W-J, Wu X-C, Zhang M-D, Weng M-Z, Zhou D, et al. Long non-coding RNA Malat1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget. 2016;7(25):37857.
Zhang L, Liu X, Jin H, Guo X, Xia L, Chen Z, et al. miR-206 inhibits gastric cancer proliferation in part by repressing cyclinD2. Cancer Lett. 2013;332(1):94–101.
Pang C, Huang G, Luo K, Dong Y, He F, Du G, et al. miR-206 inhibits the growth of hepatocellular carcinoma cells via targeting CDK9. Cancer Med. 2017;6(10):2398–409.
Zhang T, Liu M, Wang C, Lin C, Sun Y, Jin D. Down-regulation of MiR-206 promotes proliferation and invasion of laryngeal cancer by regulating VEGF expression. Anticancer Res. 2011;31(11):3859–63.
Yu W, Wang H, Lu B, Zhang G, Ma H, Wu Z. miR-206 inhibits human laryngeal squamous cell carcinoma cell growth by regulation of cyclinD2. Eur Rev Med Pharmacol Sci. 2015;19(14):2697–702.
Wang X, Yu B, Jin Q, Zhang J, Yan B, Yang L, et al. Regulation of laryngeal squamous cell cancer progression by the lncRNA RP11-159K7. 2/miR-206/DNMT3A axis. J Cell Mol Med. 2020;24(12):6781–95.
Liu C, Li J, Wang W, Zhong X, Xu F, Lu J. miR-206 inhibits liver cancer stem cell expansion by regulating EGFR expression. Cell Cycle. 2020;19(10):1077–88.
Chen QY, Jiao DM, Yan L, Wu YQ, Hu HZ, Song J, et al. Comprehensive gene and microRNA expression profiling reveals miR-206 inhibits MET in lung cancer metastasis. Mol Biosyst. 2015;11(8):2290–302.
Jiao D, Chen J, Li Y, Tang X, Wang J, Xu W, et al. miR-1-3p and miR-206 sensitizes HGF-induced gefitinib-resistant human lung cancer cells through inhibition of c-Met signalling and EMT. J Cell Mol Med. 2018;22(7):3526–36.
Wang T, Dong X-M, Zhang F-L, Zhang J-R. miR-206 enhances nasopharyngeal carcinoma radiosensitivity by targeting IGF1. Kaohsiung J Med Sci. 2017;33(9):427–32.
Wu K, Li J, Qi Y, Zhang C, Zhu D, Liu D, et al. SNHG14 confers gefitinib resistance in non-small cell lung cancer by up-regulating ABCB1 via sponging miR-206-3p. Biomed Pharmacother. 2019;116: 108995.
Sheng N, Xu Y-Z, Xi Q-H, Jiang H-Y, Wang C-Y, Zhang Y, et al. Overexpression of KIF2A is suppressed by miR-206 and associated with poor prognosis in ovarian cancer. Cell Physiol Biochem. 2018;50(3):810–22.
Zhang Y, Guo J, Cai E, Cai J, Wen Y, Lu S, et al. HOTAIR maintains the stemness of ovarian cancer stem cells via the miR-206/TBX3 axis. Exp Cell Res. 2020;395(2): 112218.
Liu F, Yin R, Chen X, Chen W, Qian Y, Zhao Y, et al. Over-expression of miR-206 decreases the Euthyrox-resistance by targeting MAP4K3 in papillary thyroid carcinoma. Biomed Pharmacother. 2019;114: 108605.
Wang P, Guo L, Liang Z, Lou J, Zhao J. Baicalein inhibits cell development in papillary thyroid cancer by regulating miR-206/RAP1B pathway. Trop J Pharm Res. 2020;19(7):1383–8.
Ding T, Zhu Y, Jin H, Zhang P, Guo J, Zheng J. Circular RNA circ_0057558 controls prostate cancer cell proliferation through regulating miR-206/USP33/c-Myc Axis. Front Cell Dev Biol. 2021;9: 644397.
Wang Y, Xu H, Si L, Li Q, Zhu X, Yu T, et al. MiR-206 inhibits proliferation and migration of prostate cancer cells by targeting CXCL11. Prostate. 2018;78(7):479–90.
Wei C, Wang S, Ye ZQ, Chen ZQ. miR-206 inhibits renal cell cancer growth by targeting GAK. J Huazhong Univ Sci Technol Med Sci. 2016;236(6):852–8.
Heinemann FG, Tolkach Y, Deng M, Schmidt D, Perner S, Kristiansen G, et al. Serum miR-122-5p and miR-206 expression: non-invasive prognostic biomarkers for renal cell carcinoma. Clin Epigenetics. 2018;10(1):1–9.
Miyachi M, Tsuchiya K, Yoshida H, Yagyu S, Kikuchi K, Misawa A, et al. Circulating muscle-specific microRNA, miR-206, as a potential diagnostic marker for rhabdomyosarcoma. Biochem Biophys Res Commun. 2010;400(1):89–93.
Koshizuka K, Hanazawa T, Fukumoto I, Kikkawa N, Matsushita R, Mataki H, et al. Dual-receptor (EGFR and c-MET) inhibition by tumor-suppressive miR-1 and miR-206 in head and neck squamous cell carcinoma. J Hum Genet. 2017;62(1):113–21.
Wang P, Gu J, Wang K, Shang J, Wang W. miR-206 inhibits thyroid cancer proliferation and invasion by targeting RAP1B. J Cell Biochem. 2019;120(11):18927–36.
Fu Y, Shao Z, He Q, Jiang B, Wu Y, Zhuang Z. Hsa-miR-206 represses the proliferation and invasion of breast cancer cells by targeting Cx43. Eur Rev Med Pharmacol Sci. 2015;19(11):2091–104.
Li H, Xu W, Xia Z, Liu W, Pan G, Ding J, et al. Hsa_circ_0000199 facilitates chemo-tolerance of triple-negative breast cancer by interfering with miR-206/613-led PI3K/Akt/mTOR signaling. Aging. 2021;13(3):4522.
Wang J, Tsouko E, Jonsson P, Bergh J, Hartman J, Aydogdu E, et al. miR-206 inhibits cell migration through direct targeting of the actin-binding protein coronin 1C in triple-negative breast cancer. Mol Oncol. 2014;8(8):1690–702.
Waller R, Goodall EF, Milo M, Cooper-Knock J, Da Costa M, Hobson E, et al. Serum miRNAs miR-206, 143–3p and 374b–5p as potential biomarkers for amyotrophic lateral sclerosis (ALS). Neurobiol Aging. 2017;55:123–31.
Xie B, Liu Z, Jiang L, Liu W, Song M, Zhang Q, et al. Increased serum miR-206 level predicts conversion from amnestic mild cognitive impairment to Alzheimer’s disease: a 5-year follow-up study. J Alzheimers Dis. 2017;55(2):509–20.
Moon J, Lee S-T, Kong IG, Byun J-I, Sunwoo J-S, Shin J-W, et al. Early diagnosis of Alzheimer’s disease from elevated olfactory mucosal miR-206 level. Sci Rep. 2016;6(1):1–9.
Lee ST, Chu K, Jung KH, Kim JH, Huh JY, Yoon H, et al. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann Neurol. 2012;72(2):269–77.
Shao Y, Xu T. A study on the neuroprotective effect of miR-206–3p on Alzheimer’s disease mice by regulating brain-derived neurotrophic factor. Ann Transl Med. 2022;10(2):85.
Tian N, Cao Z, Zhang Y. MiR-206 decreases brain-derived neurotrophic factor levels in a transgenic mouse model of Alzheimer’s disease. Neurosci Bull. 2014;30(2):191–7.
Gao Y, Yue J, Huang Z. LncRNA MIAT mediates ox-LDL-induced endothelial cell injury via miR-206/RAB22A axis. J Surg Res. 2021;265:303–12.
Duan J, Zhang X, Zhang S, Hua S, Feng Z. miR-206 inhibits FN1 expression and proliferation and promotes apoptosis of rat type II alveolar epithelial cells. Exp Ther Med. 2017;13(6):3203–8.
Wang M, Ji Y, Cai S, Ding W. MiR-206 suppresses the progression of coronary artery disease by modulating vascular endothelial growth factor (VEGF) expression. Med Sci Monit. 2016;22:5011.
Tang Y, Zhang Y, Chen Y, Xiang Y, Xie Y. Role of the micro RNA, miR-206, and its target PIK 3C2α in endothelial progenitor cell function–potential link with coronary artery disease. FEBS J. 2015;282(19):3758–72.
Sun Y, An N, Li J, Xia J, Tian Y, Zhao P, et al. miRNA-206 regulates human pulmonary microvascular endothelial cell apoptosis via targeting in chronic obstructive pulmonary disease. J Cell Biochem. 2019;120(4):6223–36.
Liu N, Williams AH, Maxeiner JM, Bezprozvannaya S, Shelton JM, Richardson JA, et al. microRNA-206 promotes skeletal muscle regeneration and delays progression of Duchenne muscular dystrophy in mice. J Clin Investig. 2012;122(6):2054–65.
Hu J, Kong M, Ye Y, Hong S, Cheng L, Jiang L. Serum miR-206 and other muscle-specific micro RNA s as non-invasive biomarkers for Duchenne muscular dystrophy. J Neurochem. 2014;129(5):877–83.
Wu Z, Liu Y, Huang J, Huang Y, Fan L. MiR-206 inhibits epilepsy and seizure-induced brain injury by targeting CCL2. Cytotechnology. 2019;71(4):809–18.
Zhang J, Fan J, Zeng X, Nie M, Luan J, Wang Y, et al. Hedgehog signaling in gastrointestinal carcinogenesis and the gastrointestinal tumor microenvironment. Acta Pharm Sin B. 2021;11(3):609–20.
Sharan A, Zhu H, Xie H, Li H, Tang J, Tang W, et al. Down-regulation of miR-206 is associated with Hirschsprung disease and suppresses cell migration and proliferation in cell models. Sci Rep. 2015;5(1):1–7.
Luo J, Han J, Li Y, Liu Y. Downregulated SOX9 mediated by miR-206 promoted cell apoptosis in Legg-Calvé-Perthes disease. Oncol Lett. 2018;15(1):1319–24.
Pegoraro V, Angelini C. Circulating miR-206 as a biomarker for patients affected by severe limb girdle muscle dystrophies. Genes. 2021;12(1):85.
Matsuzaka Y, Kishi S, Aoki Y, Komaki H, Oya Y, Takeda SI, et al. Three novel serum biomarkers, miR-1, miR-133a, and miR-206 for Limb-girdle muscular dystrophy, Facioscapulohumeral muscular dystrophy, and Becker muscular dystrophy. Environ Health Prev Med. 2014;19(6):452–8.
Jalali S, Ramanathan GK, Parthasarathy PT, Aljubran S, Galam L, Yunus A, et al. Mir-206 regulates pulmonary artery smooth muscle cell proliferation and differentiation. PLoS ONE. 2012. https://doi.org/10.1371/journal.pone.0046808.
Liang G, Wu Y, Guan Y, Dong Y, Jiang L, Mao G, et al. The correlations between the serum expression of miR-206 and the severity and prognosis of sepsis. Ann Palliat Med. 2020;9(5):3222–34.
Valsecchi V, Anzilotti S, Serani A, Laudati G, Brancaccio P, Guida N, et al. miR-206 reduces the severity of motor neuron degeneration in the facial nuclei of the brainstem in a mouse model of SMA. Mol Ther. 2020;28(4):1154–66.
Minacapelli CD, Bajpai M, Geng X, Van Gurp J, Poplin E, Amenta PS, et al. miR-206 as a biomarker for response to mesalamine treatment in ulcerative colitis. Inflamm Bowel Dis. 2019;25(1):78–84.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Conceptualization, study design, investigation, validation of the collected papers, designing the tables and figures, bioinformatics step, review and editing were performed by SK; Data collection, completing the tables, and Figure preparation were performed by SZH; Draft manuscript preparation, revision and supervision were performed by SGF. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Khalilian, S., Hosseini Imani, S.Z. & Ghafouri-Fard, S. Emerging roles and mechanisms of miR-206 in human disorders: a comprehensive review. Cancer Cell Int 22, 412 (2022). https://doi.org/10.1186/s12935-022-02833-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-022-02833-2