Abstract
Background
Klebsiella spp. are important pathogens associated with bacteremia among admitted children and is among the leading cause of death in children < 5 years in postmortem studies, supporting a larger role than previously considered in childhood mortality. Herein, we compared the antimicrobial susceptibility, mechanisms of resistance, and the virulence profile of Klebsiella spp. from admitted and postmortem children.
Methods
Antimicrobial susceptibility and virulence factors of Klebsiella spp. recovered from blood samples collected upon admission to the hospital (n = 88) and postmortem blood (n = 23) from children < 5 years were assessed by disk diffusion and multiplex PCR.
Results
Klebsiella isolates from postmortem blood were likely to be ceftriaxone resistant (69.6%, 16/23 vs. 48.9%, 43/88, p = 0.045) or extended-spectrum β-lactamase (ESBL) producers (60.9%, 14/23 vs. 25%, 22/88, p = 0.001) compared to those from admitted children. blaCTX-M-15 was the most frequent ESBL gene: 65.3%, 9/14 in postmortem isolates and 22.7% (5/22) from admitted children. We found higher frequency of genes associated with hypermucoviscosity phenotype and invasin in postmortem isolates than those from admitted children: rmpA (30.4%; 7/23 vs. 9.1%, 8/88, p = 0.011), wzi-K1 (34.7%; 8/23 vs. 8%; 7/88, p = 0.002) and traT (60.8%; 14/23 vs. 10.2%; 9/88, p < 0.0001), respectively. Additionally, serine protease auto-transporters of Enterobacteriaceae were detected from 1.8% (pic) to 12.6% (pet) among all isolates. Klebsiella case fatality rate was 30.7% (23/75).
Conclusion
Multidrug resistant Klebsiella spp. harboring genes associated with hypermucoviscosity phenotype has emerged in Mozambique causing invasive fatal disease in children; highlighting the urgent need for prompt diagnosis, appropriate treatment and effective preventive measures for infection control.
Similar content being viewed by others
Background
Klebsiella spp. are among the most frequently reported pathogens associated with opportunistic infections in immunocompromised individuals and patients hospitalized for prolonged periods [1], and commonly implicated in nosocomial urinary tract infections and sepsis in infants from sub-Saharan Africa [2, 3]. Given the importance of Klebsiella spp. in nosocomial infection and the continuous increase and spread of community-acquired multi-drug resistant (MDR) Klebsiella pneumoniae, including Extended-Spectrum β-Lactamase (ESBL) producers, the associated high mortality is causing a grave concern [2, 4].
A new K. pneumoniae, called hypervirulent (hvKp) is a global public health concern because of its invasiveness (e.g. causing pyogenic liver abscess, bacteremia, pneumonia), the capacity to form metastasis and for being more virulent than the classical K. pneumoniae (cKp), affecting healthy individuals in the community settings [5, 6]. The hvKp can be distinguished from the classical ones by specific biomarkers, namely, the presence of siderophores (aerobactin [iuc] system), hypermucoviscosity (encoded by rmpA, rmpA2) and salmochelin (iro) [6,7,8,9].
The virulence genes of both cKp and hvKp includes: siderophores for the iron acquisition (e.g. enterobactin (ent), adhesins for adherence (eg. fimH, mrkD), invasins for invasion and escape of the immune system defence (eg. traT), and protectins (eg. magA responsible for the hypermucoviscous phenotype) [10,11,12,13].
Recently, postmortem studies conducted in urban (Maputo city) and rural (Manhiça District) Mozambique to ascertain causes of death in stillbirths and young children using minimally invasive tissue sampling (MITS) approaches found Klebsiella spp. to be among the top pathogens assigned in the chain of events leading to death [14,15,16].
Data on the molecular epidemiology and characterization of Klebsiella spp. are scarce in Africa including Mozambique [17,18,19]. However, data from an urban hospital in the city of Beira, central Mozambique showed high frequency (100% of isolates) of ESBL-producing K. pneumoniae among hospitalized children with urinary tract infections [20]. Additionally, in our previous work we documented the occurrence of community-acquired Klebsiella spp. strains harboring blaCTX-M-15 among children living in the Manhiça District, southern Mozambique [21, 22]. Herein, we described and compared the molecular characterization, antimicrobial susceptibility of Klebsiella spp. isolates recovered from bloodstream infections and postmortem blood specimens collected in children under 5 years of age and we characterize the MDR associated Klebsiella spp. from admitted children with bloodstream infections who have died. In this prospective study, we documented the occurrence of hypermucoviscous, multidrug-resistant Klebsiella spp. with an ESBL-producing phenotype, harboring blaCTX-M-15, and causing invasive disease associated with poor outcomes in infants and young children in a rural hospital in Southern Mozambique.
Methods
Clinical isolates and study area
We analyzed 111 Klebsiella spp. isolates recovered from bloodstream infection (n = 88) of children under 5 years old admitted at the Manhiça District Hospital (January 2001 and December 2019) and from postmortem blood specimens (n = 23) of deceased children under 5 years old enrolled in the ongoing Child Health and Mortality Prevention Surveillance (CHAMPS) project (December 2016 and December 2019) [23]. The CHAMPS Network was established in seven countries in south Asia and sub-Saharan Africa including Mozambique (Manhiça site) to collect standardized, population-based, longitudinal data on mortality among children under 5 years and stillbirths, to improve the accuracy of causes of death determination using a novel approach of minimally invasive tissue sampling (MITS) [24].
In Mozambique, CHAMPS is being conducted by the Centro de Investigação em Saúde de Manhiça (CISM) in the district of Manhiça, a rural area of Maputo Province in southern Mozambique, with an estimated population of approximately 201,845 inhabitants. In this site, a continuous demographic surveillance system (DSS) has been running since 1996 with regular update of vital events; and currently covering the entire district of Manhiça [25].
In addition to the DSS, CISM has been conducting morbidity surveillance which includes invasive bacterial disease surveillance in the pediatric population (children < 15 years of age) at the Manhiça District Hospital (MDH) (a 150 beds referral district hospital of the Manhiça district) and other 5 peripheral health facilities since January 1997, documenting all outpatients and inpatients visits using standard questionnaires. Blood cultures were routinely performed as part of the clinical practice upon hospital admission for all children under 2 years old and for older children under 15 years with axillary temperature ≥ 39 °C or who meet, according to the admitting clinician, specific severity criteria [4, 26]. Bacterial cultures were performed at the CISM Microbiology laboratory for Klebsiella spp. and other bacterial isolation and identification using conventional microbiology and biochemical methods [4]. Clinically relevant isolates were stored at − 70 °C in a biobank (in a STGG medium – 2% of Skim milk + 3% tryptic soy broth + 0.5% glucose + 10% glycerol) for further characterization [27]. Klebsiella bacteremia was defined as Klebsiella positive blood culture collected from children upon admission at the MDH.
Under the health demographic surveillance system (HDSS) platform, CISM has been conducting a mortality surveillance in the context of the CHAMPS network, since 2016. CHAMPS uses the MITS approach on deaths reported from the community or health facility, to track the definitive/immediate causes of deaths in stillbirths and children under 5 years old, as described elsewhere [14, 23]. Tissue samples, including blood and cerebrospinal fluid (CSF) are investigated using conventional microbiology and molecular techniques for pathogens detection, including Klebsiella [23, 28, 29]. The causes of death were determined by a panel of experts that comprises at least 1 of each of the following: clinician (eg, pediatricians, neonatologists, and obstetricians), pathologist, epidemiologist, and microbiologist. The Panelists receive all available information from linked maternal data, child clinical data, individual demographic data, verbal autopsy, microbiology, molecular testing, clinical diagnostics (human immunodeficiency virus [HIV], tuberculosis, malaria), photographs of the deceased from the MITS procedure, and histopathology findings in the form of a determination of cause of death package and assign the underlying, antemortem, and immediate (and other contributing) causes of death [14]. Postmortem Klebsiella is defined as Klebsiella isolates recovered from blood samples collected through MITS in deceased children enrolled in the CHAMPS.
Antimicrobial susceptibility testing and empirical therapy
The antimicrobial susceptibility phenotypes for ampicillin (AMP), amikacin (AMK), amoxicillin-clavulanic acid (AUG), piperacillin-tazobactam (TZP), cefuroxime (CXM), ceftriaxone (CRO), ceftazidime (CAZ), aztreonam (ATM), ertapenem (ETP), imipenem (IPM), meropenem (MEM), nalidixic acid (NA), ciprofloxacin (CIP), chloramphenicol (CHL), tobramycin (TOB), gentamicin (GEN), tetracycline (TET) and trimethoprim-sulfamethoxazole (SXT) were determined for all isolates by conventional disk diffusion on Mueller Hinton agar (BD, Heidelberg, Germany) using commercially available disks (BD, Heidelberg, Germany). The selection of the antibiotics tested was done according to their use for local empirical treatment and those recommended by the Clinical Laboratory Standard Institute (CLSI) guidelines [30]. The interpretative category of resistance was determined according to the CLSI guidelines (29th Edition) [30]. All isolates resistant to three or more unrelated families of antibiotics were defined as MDR [31]. In the MDH, children are treated empirically with ampicillin or ampicillin + gentamicin and amikacin or ceftriaxone.
Detection of ESBL phenotype and genotype
All ceftriaxone, aztreonam or ceftazidime resistant isolates were then screened for ESBL production by disk synergy assay with the addition of a beta-lactamase inhibitor disc, as previously described [32]. Briefly, AMP, CAZ, and CRO disk were placed 2 cm distant and AUG disk was placed on the center closer to the other disks on Muller-Hinton agar plate inoculated with 0.5McF of Klebsiella isolates. The tests were considered positive if lightly growth-inhibitory zone (phantom zone) was observed between the disks. Isolates with ESBL phenotype were assessed by a multiplex polymerase chain reaction (PCR) for blaCTX-M, blaTEM, blaSHV genes, using universal primers (Table 1). The multiplex PCR was performed in 25 μL reaction volumes, including 0.5 μM of each primer, 12,5 μL of 2X master mix (Fermentas, Thermo Scientific, MA, USA), 3 μL of genomic DNA and nuclease free water. Cycling conditions included an initial denaturation step of 5 min at 95 °C, followed by 30 amplification cycles (denaturation at 94 °C for 1 min, annealing at 57 °C for 1 min and elongation at 72 °C for 2 min) and a final extension step at 72 °C for 10 min.
PCR amplicons were visualised on 2% ultrapure agarose gel (Invitrogen, CA, USA) stained with 25 μM ethidium bromide solution (Invitrogen, CA, USA).
Amplicons of the expected size, for blaCTX-M, blaTEM, blaSHV genes were purified using QIAquick PCR purification kit (QIAGEN, Hilden, Germany), and shipped to a commercial sequencing services company (Macrogen Europe BV, Amsterdam, the Netherlands) for Sanger sequencing in both directions using the same sets of primers for each gene (Table 1). Sequence analysis and alignments were performed using the BioEdit software (v7.2.5) and the BLAST Program (http://blast.ncbi.nlm.nih.gov/Blast.cgi), which was also used to determine the allele correspondent for each gene (blaCTX-M, blaTEM, blaSHV).
Detection of virulence genes
The virulence genes associated with protectins and invasins (rmpA, wzi-K1, and traT), adhesins (fimH, mrkD, and ycfM), toxins (hylA and hylC) and siderophores (iutA, fyuA, and entB) were assessed by 4 multiplexed PCRs for each group of genes, optimized in this study and using primers previously described (Table 1). All genes, except those encoding Serine Protease Auto-transporters of Enterobacteriaceae (SPATEs), were amplified in 25 μL reaction volumes, including 0.5 μM of each primer, 12,5 μL of 2X master mix (PCR Master Mix 2x, Thermo Scientific, MA, USA), 3 μL of genomic DNA and nuclease free water. The cycling conditions included an initial denaturation step at 95 °C for 2 min, followed by 35 amplification cycles with denaturation at 94 °C for 30 s, annealing for 1.5 min (temperatures are described in Table 1) an elongation at 72 °C for 1.5 min and a final extension step at 72 °C for 10 min.
The detection of SPATEs genes (sat, sepA, sigA, piC, and pet) were assessed by a multiplex PCR previously described [39]. Briefly the genes were amplified in 25 μL reaction volume, consisting of 25 μM of each primer, 12.5 μL of the QIAGEN master mix (QIAGEN multiplex PCR kit; QIAGEN, Courtaboeuf, France), 2.5 μL of Q-solution (QIAGEN, Courtaboeuf, France) and 3 μL of genomic DNA.
One PCR product of the expected size per SPATE gene (sat, sepA, sigA, piC, and pet) was also shipped for Sanger sequencing with the corresponding sets of primers (described in Table 1) for confirmation.
Hypermucoviscous phenotype
To verify the concordance with the genotype and the expression of the hypermucoviscous phenotype, all rmpA and wzi-K1 positive isolates were tested by string tests as previously described elsewhere [8].
Statistical analysis
PCR and antibiotic susceptibility data were entered in an Excel spreadsheet and checked for consistency by an independent laboratory technician. Data were analyzed using Stata software version 14.0 (STATA Corporation, College Station, Tx, USA). Frequencies and proportions were used to describe categorical variables, and differences of proportions were compared using the chi-square or Fisher’s exact test where appropriate, with a p-value of < 0.05 being considered significant.
Surviving children or Srv. Bacteremia were defined as children hospitalized at the MDH who tested positive for Klebsiella spp. in blood culture upon admission and that were alive at discharge (n = 41) and dead children or Dth. Bacteremia were defined as children hospitalized at the MDH who tested positive for Klebsiella spp. in blood culture upon admission who died and had clinical information available n = 23).
Results
Description of studied isolates
From the study period (January 2000 – December 2019), 52,968 children younger than 5 years were admitted to the hospital in the context of invasive bacterial disease surveillance and had blood culture collected, yielding a positivity rate of 8.1% (n = 4270) for any bacteria. Klebsiella spp. accounted for 2.1% (88/4270) of the total positive; in addition, between December 2016 and December 2019, 226 deceased children were enrolled in the CHAMPS, and Klebsiella spp. were isolated in 23 blood postmortem specimens. In general, 72.7% (64/88) and 87% (20/23) were K. pneumoniae, 5.7% (5/88) and 13% (3/23) were K. oxytoca, from admitted children and post-mortem blood, respectively, and the remaining 21.6% (19/88) were classified as Klebsiella spp. Most isolates from admitted children and postmortem isolates were recovered from males, 55.7% (49/88) and 60.9% (14/23), respectively. Klebisella isolates were also recovered from neonates and children between 12 and 23 months (29.5%, 26/88) upon admission and from postmortem children between 23 and 59 months (47.8%, 11/23) (Fig. 1).
Antimicrobial susceptibility and ESBL genes of Klebsiella spp.
Overall, resistance ranged from 51.4% (57/111) for amoxicillin-clavulanic acid, 53.1% (54/111) for ceftriaxone, 54.1% (60/111) for gentamicin to 92.7% (103/111) for ampicillin, with 66.7% (74/111) of isolates being MDR. Ceftriaxone resistance was more frequent among isolates recovered from postmortem compared to those from admitted children (69.6%, 16/23 vs. 48.9%, 43/88, p = 0.045). Eight percent (7/88) of isolates from admitted children had reduced susceptibility to meropenem, while 34.8% (8/23) and 13% (3/23) of the postmortem isolates showed full resistance to ertapenem and meropenem, respectively (Table 2), and 30.4% (7/23) of postmortem isolates had intermediate resistance to ertapenem.
ESBL phenotype was higher among postmortem isolates (60.9%, 14/23) compared to those from admitted children (25%, 22/88), p = 0.001. Among beta-lactamase genes detected in postmortem isolates and in isolates from admitted children, respectively, blaCTX-M accounted for 71.4% (10/14) and 41% (9/22), p = 0.075, blaSHV accounted for 42.8% (6/14) and 27.2% (6/22), p = 0.33, and blaTEM accounted for 57.1% (8/14) and 9.1% (2/22), p = 0.002. blaCTX-M-15 was found in 90% (9/10) of postmortem blaCTX-M isolates and 55.6% (5/9) of blaCTX-M isolates from admitted children, whereas blaSHV-1 was detected in 66.7% (4/6) and 50% (3/6) of postmortem and blaSHV isolates from admitted children, respectively. All blaTEM isolates were TEM-1 variant.
Virulence profile of Klebsiella isolates
Genes associated with the hypermuscoviscous phenotype or hypervirulence were more commonly found in the postmortem isolates compared to isolates from admitted children (Table 3): wzi-K1 (34.7%; 8/23 vs. 8%; 7/88, p = 0.002), rmpA (30.4%; 7/23 vs. 9.1%; 8/88, p = 0.011), and invasins, traT (60.8%, 14/23 vs. 10.2%, 9/88, p < 0.0001), respectively. Those three genes were simultaneously detected among 17.3% (4/23) of postmortem isolates, of which 2/3 of these isolates were MDR. The virulence genes were not in accordance with the phenotype, as 2 out of 25 rmpA and wzi-K1 positive isolates expressed the hypermucoviscous phenotype, and both were wzi-K1 positive.
The SPATE encoding genes, sat, sepA, piC, pet and sigA were detected in 9.9% (11/111), 2.7% (3/111), 1.8% (2/111), 12.6% (14/111) and 3.6% (4/111) of Klebsiella isolates, respectively, with similar distribution between postmortem isolates and isolates from admitted children (Table 3).
Among MDR isolates from admitted and post-mortem children, 73.3% (11/15) and 75.9% (44/58) carried siderophores genes, 73.3% (11/15) and 82.7% (48/58) carried adhesion genes, 80% (12/15) and 31% (18/58) carried invasins/protectins, 33.3% (5/15) and 24.1% (14/58) carried SPATES and 13.3% (2/15) and 22.4% (13/58) carried toxin genes, respectively. Almost all isolates from both admitted (100%, 7/7) and postmortem children (75%, 6/8) or (87.5%, 7/8 vs. 71.4, 5/7) carrying wzi-K1 and rmpA genes were MDR.
Outcome of Klebsiella infections in bacteremic patients
Of the admitted children, outcome data was available for 75 out of 88 children (85.2%) with Klebsiella bacteremia, yielding an in-hospital case fatality rate (CFR) of 30.7% (23/75), which was particularly high (43.5%; 10/23) among younger than 2 years (between 12 and 23 months), and 11 children were transferred or left the hospital.
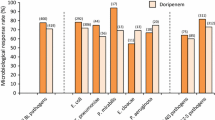
Resistance to chloramphenicol (56.5%, 13/23 vs. 51.2%, 21/41 p = 0.06) occurred more frequently among isolates from children who died when compared to those who recovered, as shown in Fig. 2A. Similarly, the postmortem isolates more often had a wzi-K1, rmpA, traT genes (Fig. 2B). Also, isolates from admitted children with poor outcome were likely to harbor hylC and fimH genes (Fig. 2A and B).
Isolates from admitted children who died were frequently ampicillin resistant (100%, 23/23), trimethoprim-sulfamethoxazole resistant (69.6%, 16/23), amoxicillin-clavulanic acid and chloramphenicol resistant (56.5%, 13/23), gentamicin and ceftriaxone resistant (52.2%, 12/23), or harboring entB (69.6%, 16/23), mrkD (65.2%, 15/23), or ycfM (52.2%, 12/23) genes. Almost all patients who died received oral aminoglycosides as empirical treatment, except three patients who took only cephalosporin, trimethoprim-sulfamethoxazole or chloramphenicol (Table 4). Furthermore, these patients died in the first 48 h (56.5%, 13/23) of hospitalization, except for those who had adequate antibiotherapy, but had an array of genes, including rmpA, hylC, pic, saT, sepA and fimH, who died after 6 days or more of hospitalization.
Among children with clinical information, 65.3% (49/75) carried MDR isolates, of which 26.5% (13/49) died. Additionally, 12 out of 75 carried isolates that were MDR and harbored SPATE genes, of which 50% of them died; and 40% of children carrying isolates that harbored toxin or invasins and protectin genes and were simultaneously MDR (15/75 and 10/75), died as well.
From the post-mortem children, clinical information was available for 21 out of 23 children, among which Klebsiella spp. was in the chain of event of death as the immediate cause of death in approximately one third of children; and in-hospital associated mortality was 95% (20/21), with an average of hospital admission stay of 4 days (Table S1). The clinical presentation of those patients was available in 18 out of 21 children, with difficulty of breathing (55.6%, 10/18) being the most common followed by fever (44.4%, 8/18), cough (38.9%, 7/18), loss of appetite (33.3%, 6/18) and diarrhea (27.8%, 5/18). Sepsis was the immediate cause of death for almost 43% (9/21) of post-mortem children, while respiratory infections, including pneumonia was the second major immediate cause of death among post-mortem children accounting for one third of the children. All post-mortem isolates were resistant to at least one antibiotic and 59% (13/22) were MDR, of which, 38% (5/13) carried rmpA gene and 23% (2/13) carried both rmpA and capsular serotype K1 gene (wzi-K1).
Discussion
This is the first report comparing the antimicrobial susceptibility profile, its associated mechanisms of resistance, and the virulence repertoire between Klebsiella spp. isolates from postmortem studies and isolates recovered from children < 5 years of age with severe disease. The finding of a higher prevalence of MDR strains harboring both ESBL and genes associated with the hypermucoviscous phenotype in postmortem isolates may suggest the potential virulence and contribution of specific strains of Klebsiella to life-threatening infections, and thus, its potential role in childhood mortality in Mozambique.
The high CFR observed among children admitted with bacteremia may be explained by the high proportion (66%) of MDR strains. Most MDR isolates were also resistant to ceftriaxone due to the presence of blaCTX-M-15 in more than 25% of isolates limiting the therapeutic options, which is consistent with our previous observations of the poor outcome of Klebsiella associated bacteremia in children younger than 14 years of age [4, 22, 40]. The low resistance to chloramphenicol among postmortem isolates may be explained by the fact that this antibiotic was used as a drug of choice for empirical therapy [4] and is no longer a choice for routine clinical management. On the other hand, the presence of hylC and sat among the fatal strains from bacteremic children may also help to explain the Klebsiella associated mortality, as they have been described in E.coli showing cytotoxic effect on erythrocytes and epithelial cells, respectively, by disrupting the membrane or autophagy and cell detachment [41,42,43]. The excess of Klebsiella associated mortality is similar to previous studies showing mortality rates up to 38.5% in other countries [44, 45], including in the neighboring country, South Africa (26–35%) [46].
As previously reported in this community and elsewhere [10, 17, 22, 47, 48], an important public health concern is related to the emergence and widespread dissemination of ESBL-producing Klebsiella spp., challenging the appropriate management of MDR Klebsiella infections, that were mostly treated with ceftriaxone. The major dilemma is related to the finding of strains with reduced susceptibility to carbapenems (meropenem and ertapenem), often used as a second-line treatment options, as per international guidelines, but not foreseen in our setting [49]. This is alarming as these antibiotics are not routinely used in our study community and are rare in Mozambique in general, even though, the isolates remained susceptible to imipenem. Our findings provide a clear indication that an appropriate action plan is required to contain the spread of antibiotic resistance, mainly because the isolates remained susceptible to antibiotics that are expensive and unavailable in most of the rural communities, constraining their use even when the available antibiotherapy is ineffective [50].
The high frequency of genes involved in the synthesis of capsule (rmpA and wzi-K1), known to inhibit and evade phagocytosis by host cells and to neutralize the antibacterial activity of host defenses [13], in postmortem isolates may suggest potential virulence and fatality of the strains recovered in deceased children. The presence of these genes, mainly, rmpA give an indication that hipervirulent Klebsiella may be circulating in our setting, as this gene is one of the markers for hvKp [8, 9, 51]. However, this should be interpreted with caution, as the other markers were not investigated. The wzi-K1 gene indicate the circulation of the K1 capsular serotype.
The high prevalence of siderophores was expected among pathogens causing invasive disease as these are essential for iron acquisition and survival of Klebsiella when interacting with host cells [12]; in addition to the high prevalence of adhesins that are critical for the first step of bacterial colonization /adhesion, with several variants that have been described [10, 11].
As of this writing, to our best knowledge this is the first report on the occurrence of the studied SPATEs in Klebsiella spp., suggesting that they potentially play an important role in the pathogenicity of Klebsiella, as previously described [52,53,54]. In addition, the sequences of SPATEs reported here are similar to those reported in other Enterobacteriaceae species as previously reported and may play the same role [52, 54].
There are some limitations in this study, one of these is that our study compared Klebsiella spp. in admitted patients and postmortem samples from different study periods; none of the children with bacteremic Klebsiella isolates was included in the postmortem study, even in the period when the IBD surveillance and CHAMPS overlapped. In deep molecular characterization including phylogenetic analysis, serotyping and multi-locus sequence typing are underway to better understand the molecular epidemiology of isolates from bacteremia and postmortem samples. This data cannot be generalized for the Mozambican or African population, as the samples were from only one geographic setting.
Conclusion
Our data calls for the urgent need for more effective alternative antibiotics for management of fatal virulent MDR blaCTX-M-15 and blaSHV-1 Klebsiella strains and the implementation of appropriate measures to contain AMR (e.g. prudent use of antibiotics for empirical treatment, reduce the antibiotic usage in the agricultural activity, improve hospital infection control, rationalize the antibiotic use in the community). Continuous monitoring and surveillance for antimicrobial resistance (AMR) are crucial to support defining local and international guidelines for effective alternative options for anti-biotherapy or guide the design and development of new intervention tools (e.g. vaccines with broad coverage spectra), which requires the description of serotypes circulating.
Availability of data and materials
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ESBL:
-
Extended Spectrum Beta-Lactamase
- MDR:
-
Multidrug Resistance
- AMP:
-
Ampicillin
- AMK:
-
Amikacin
- AUG:
-
Amoxicillin-clavulanic acid
- TZP:
-
Piperacillin-tazobactam
- CXM:
-
Cefuroxime
- CRO:
-
Ceftriaxone
- CAZ:
-
Ceftazidime
- ATM:
-
Aztreonam
- ETP:
-
Ertapenem
- IPM:
-
Imipenem
- MEM:
-
Meropenem
- NA:
-
Nalidixic acid
- CIP:
-
Ciprofloxacin
- CHL:
-
Chloramphenicol
- TOB:
-
Tobramycin
- GEN:
-
Gentamicin
- TET:
-
Tetracycline
- SXT:
-
Trimethoprim-sulfamethoxazole
- CISM:
-
Centro de Investigação em Saúde de Manhiça
- PCR:
-
Polymerase Chain Reaction
- hvKP :
-
hypervirulent Klebsiella pneumonia
- cKP :
-
Classical Klebsiella pneumonia
- MITS:
-
Minimally Invasive Tissue Sampling
- CHAMPS:
-
Child Health and Mortality Prevention Surveillance
- DSS:
-
Demographic Surveillance System
- MDH:
-
Manhiça District Hospital
- HDSS:
-
Health Demographic Surveillance System
- CSF:
-
Cerebrospinal Fluid
- CLSI:
-
Clinical Laboratory Standard Institute
- SPATE:
-
Serine Protease Autotransporters of Enterobacteriaceae
- IBD:
-
Invasive Bacterial Surveillance
- CFR:
-
Case Fatality Rate
References
Hendrik TC, Voorintholt AF, Vos MC. Clinical and molecular epidemiology of extended-spectrum beta-lactamase- producing klebsiella spp: a systematic review and meta-analyses. PLoS One. 2015;10:1–23.
Morkel G, Bekker A, Marais BJ, Kirsten G, van Wyk J, Dramowski A. Bloodstream infections and antimicrobial resistance patterns in a south African neonatal intensive care unit. Paediatr Int Child Health. 2014;34(2):108–14. https://doi.org/10.1179/2046905513Y.0000000082.
Lochan H, Pillay V, Bamford C, Nuttall J, Eley B. Bloodstream infections at a tertiary level paediatric hospital in South Africa. BMC Infect Dis. 2017;17:1–9.
Sigaúque B, Roca A, Mandomando I, Morais L, Quintó L, Sacarlal J, et al. Community-acquired bacteremia among children admitted to a rural hospital in Mozambique. Pediatr Infect Dis J. 2009;28(2):108–13. https://doi.org/10.1097/INF.0b013e318187a87d.
Kim D, Park BY, Choi MH, Yoon EJ, Lee H, Lee KJ, et al. Antimicrobial resistance and virulence factors of Klebsiella pneumoniae affecting 30 day mortality in patients with bloodstream infection. J Antimicrob Chemother. 2019;74:190–9. https://doi.org/10.1093/jac/dky397.
Choby JE, Howard-Anderson J, Weiss DS. Hypervirulent Klebsiella pneumoniae – clinical and molecular perspectives. J Intern Med. 2020;287(3):283–300. https://doi.org/10.1111/joim.13007.
Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3). https://doi.org/10.1128/CMR.00001-19.
Shon AS, Bajwa RPS, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Virulence. 2013;4(2):107–18. https://doi.org/10.4161/viru.22718.
Russo TA, Olson R, Fang C-T, Stoesser N, Miller M, MacDonald U, et al. Identification of biomarkers for differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. jcm.asm.org 1. J Clin Microbiol. 2018;56(9):776–94. https://doi.org/10.1128/JCM.00959-18.
Wasfi R, Elkhatib WF, Ashour HM. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci Rep. 2016;6(1):38929. https://doi.org/10.1038/srep38929.
El Fertas-Aissani R, Messai Y, Alouache S, Bakour R. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol Biol. 2013;61(5):209–16. https://doi.org/10.1016/j.patbio.2012.10.004.
Clegg S, Murphy CN. Epidemiology and virulence of Klebsiella pneumoniae. Microbiol Spectr. 2016;4(1):1–17. https://doi.org/10.1128/microbiolspec.UTI-0005-2012.
Khaertynov KS, Anokhin VA, Rizvanov AA, Davidyuk YN, Semyenova DR, Lubin SA, et al. Virulence factors and antibiotic resistance of Klebsiella pneumoniae strains isolated from neonates with sepsis. Front Med. 2018;5:1–9.
Salzberg NT, Sivalogan K, Bassat Q, Taylor AW, Adedini S, El Arifeen S, et al. Mortality surveillance methods to identify and characterize deaths in child health and mortality prevention surveillance network sites. Clin Infect Dis. 2019;69(Supplement_4):S262–73. https://doi.org/10.1093/cid/ciz599.
Castillo P, Hurtado JC, Martínez MJ, Jordao D, Lovane L, Ismail MR, et al. Validity of a minimally invasive autopsy for cause of death determination in maternal deaths in Mozambique: an observational study. PLoS Med. 2017;14:1–15.
Bassat Q, Castillo P, Martínez MJ, Jordao D, Lovane L, Hurtado JC, et al. Validity of a minimally invasive autopsy tool for cause of death determination in pediatric deaths in Mozambique: an observational study. PLoS Med. 2017;14:1–16.
Elmowalid GA, Ahmad AAM, Hassan MN, Abd El-Aziz NK, Abdelwahab AM, Elwan SI. Molecular detection of new SHVβ-lactamase variants in clinical Escherichia coli and Klebsiella pneumoniae isolates from Egypt. Comp Immunol Microbiol Infect Dis. 2018;60:35–41. https://doi.org/10.1016/j.cimid.2018.09.013.
Dramowski A, Aucamp M, Bekker A, Mehtar S. Infectious disease exposures and outbreaks at a south African neonatal unit with review of neonatal outbreak epidemiology in Africa. Int J Infect Dis. 2017;57:79–85. https://doi.org/10.1016/j.ijid.2017.01.026.
Sekyere JO, Govinden U, Essack S. The molecular epidemiology and genetic environment of Carbapenemases detected in Africa. Microb Drug Resist. 2016;22(1):59–68. https://doi.org/10.1089/mdr.2015.0053.
van der Meeren BT, Chhaganlal KD, Pfeiffer A, Gomez E, Ferro JJ, Hilbink M, et al. Extremely high prevalence of multi-resistance among uropathogens from hospitalised children in Beira. Mozambique South African Med J. 2013;103(6):382–6. https://doi.org/10.7196/SAMJ.5941.
Mandomando I, Jaintilal D, Pons MJ, Vallès X, Espasa M, Mensa L, et al. Antimicrobial susceptibility and mechanisms of resistance in Shigella and Salmonella isolates from children under five years of age with diarrhea in rural Mozambique. Antimicrob Agents Chemother. 2009;53(6):2450–4. https://doi.org/10.1128/AAC.01282-08.
Pons MJ, Vubil D, Guiral E, Jaintilal D, Fraile O, Soto SM, et al. Characterisation of extended-spectrum β-lactamases among Klebsiella pneumoniae isolates causing bacteraemia and urinary tract infection in Mozambique. J Glob Antimicrob Resist. 2015;3(1):19–25. https://doi.org/10.1016/j.jgar.2015.01.004.
Taylor AW, Blau DM, Bassat Q, Onyango D, Kotloff KL, El Arifeen S, et al. Initial findings from a novel population-based child mortality surveillance approach: a descriptive study. Lancet Glob Health. 2020;8(7):e909–19. https://doi.org/10.1016/S2214-109X(20)30205-9.
Raghunathan PL, Madhi SA, Breiman RF. Illuminating Child Mortality: Discovering Why Children Die. Clin Infect Dis. 2019;69(Supplement_4):S257–9. https://doi.org/10.1093/cid/ciz562.
Sacoor C, Nhacolo A, Nhalungo D, Aponte JJ, Bassat Q, Augusto O, et al. Profile: Manhiça health research Centre (Manhiça HDSS). Int J Epidemiol. 2013;42(5):1309–18. https://doi.org/10.1093/ije/dyt148.
Roca A, Sigauque B, Quinto L, Mandomando I, Valles X, Espasa M, et al. Invasive pneumococcal disease in children <5 years of age in rural Mozambique. Tropical Med Int Health. 2006;11:1422–31. https://doi.org/10.1111/j.1365-3156.2006.01697.x.
Sigaúque B, Kobayashi M, Vubil D, Nhacolo A, Chaúque A, Moaine B, et al. Invasive bacterial disease trends and characterization of group B streptococcal isolates among young infants in southern Mozambique, 2001-2015. PLoS One. 2018;13(1):e0191193. https://doi.org/10.1371/journal.pone.0191193.
Diaz MH, Waller JL, Theodore MJ, Patel N, Wolff BJ, Benitez AJ, et al. Development and Implementation of Multiplex TaqMan Array Cards for Specimen Testing at Child Health and Mortality Prevention Surveillance Site Laboratories. Clin Infect Dis. 2019;(69 Supplement_4):S311–21. https://doi.org/10.1093/cid/ciz571.
Rakislova N, Fernandes F, Lovane L, Jamisse L, Castillo P, Sanz A, et al. Standardization of Minimally Invasive Tissue Sampling Specimen Collection and Pathology Training for the Child Health and Mortality Prevention Surveillance Network. Clin Infect Dis. 2019;(69 Supplement_4):S302–10. https://doi.org/10.1093/cid/ciz565.
CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 2019. https://clsi.org/media/2663/m100ed29_sample.pdf. Accessed 30 Jan 2020.
Mandomando I, Sigaúque B, Morais L, Espasa M, Vallès X, Sacarlal J, et al. Antimicrobial drug resistance trends of bacteremia isolates in a rural hospital in southern Mozambique. Am J Trop Med Hyg. 2010;83(1):152–7. https://doi.org/10.4269/ajtmh.2010.09-0578.
Nepal K, Pant ND, Neupane B, Belbase A, Baidhya R, Shrestha RK, et al. Extended spectrum beta-lactamase and metallo beta-lactamase production among Escherichia coli and Klebsiella pneumoniae isolated from different clinical samples in a tertiary care hospital in Kathmandu, Nepal. Ann Clin Microbiol Antimicrob. 2017;16:1–7.
Jouini A, Vinue L, Slama KB, Saenz Y, Klibi N, Hammami S, et al. Characterization of CTX-M and SHV extended-spectrum -lactamases and associated resistance genes in Escherichia coli strains of food samples in Tunisia. J Antimicrob Chemother. 2007;60(5):1137–41. https://doi.org/10.1093/jac/dkm316.
Cabrera R, Ruiz J, Marco F, Oliveira I, Arroyo M, Aladueña A, et al. Mechanism of resistance to several antimicrobial agents in Salmonella clinical isolates causing traveler’s diarrhea. Antimicrob Agents Chemother. 2004;48(10):3934–9. https://doi.org/10.1128/AAC.48.10.3934-3939.2004.
Yeh K-M, Kurup A, Siu LK, Koh YL, Fung C-P, Lin J-C, et al. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J Clin Microbiol. 2007;45(2):466–71. https://doi.org/10.1128/JCM.01150-06.
Mamlouk K, Ben BIB, Gautier V, Vimont S, Picard B, Ben Redjeb S, et al. Emergence and outbreaks of CTX-M β-lactamase-producing Escherichia coli and Klebsiella pneumoniae strains in a Tunisian hospital. J Clin Microbiol. 2006;44(11):4049–56. https://doi.org/10.1128/JCM.01076-06.
Bingen E, Picard B, Brahimi N, Mathy S, Desjardins P, Elion J, et al. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant Pool of highly virulent B2 group strains. J Infect Dis. 1998;177(3):642–50. https://doi.org/10.1086/514217.
Restieri C, Garriss G, Locas M-C, Dozois CM. Autotransporter-encoding sequences are phylogenetically distributed among Escherichia coli clinical isolates and reference strains. Appl Environ Microbiol. 2007;73(5):1553–62. https://doi.org/10.1128/AEM.01542-06.
Boisen N, Ruiz-Perez F, Scheutz F, Krogfelt KA, Nataro JP. Short report: high prevalence of serine protease autotransporter cytotoxins among strains of enteroaggregative Escherichia coli. Am J Trop Med Hyg. 2009;80(2):294–301. https://doi.org/10.4269/ajtmh.2009.80.294.
Chew KL, Lin RTP, Teo JWP. Klebsiella pneumoniae in Singapore: Hypervirulent Infections and the Carbapenemase Threat. Front Cell Infect Microbiol. 2017;7:1–9.
Liévin-Le Moal V, Comenge Y, Ruby V, Amsellem R, Nicolas V, Servin AL. Secreted autotransporter toxin (sat) triggers autophagy in epithelial cells that relies on cell detachment. Cell Microbiol. 2011;13(7):992–1013. https://doi.org/10.1111/j.1462-5822.2011.01595.x.
Guyer DM, Radulovic S, Jones FE, Mobley HLT. Sat, the secreted autotransporter toxin of uropathogenic Escherichia coli, is a vacuolating cytotoxin for bladder and kidney epithelial cells. Infect Immun. 2002;70(8):4539–46. https://doi.org/10.1128/IAI.70.8.4539-4546.2002.
Fernandez-Prada C, Tall BD, Elliott SE, Hoover DL, Nataro JP, Venkatesan MM. Hemolysin-positive Enteroaggregative and cell-detaching Escherichia coli strains cause Oncosis of human monocyte-derived macrophages and apoptosis of murine J774 cells. Infect Immun. 1998;66(8):3918–24. https://doi.org/10.1128/IAI.66.8.3918-3924.1998.
Gutiérrez-Gutiérrez B, Salamanca E, de Cueto M, Hsueh PR, Viale P, Paño-Pardo JR, et al. Effect of appropriate combination therapy on mortality of patients with bloodstream infections due to carbapenemase-producing Enterobacteriaceae (INCREMENT): a retrospective cohort study. Lancet Infect Dis. 2017;17(7):726–34. https://doi.org/10.1016/S1473-3099(17)30228-1.
Palacios-Baena ZR, Gutiérrez-Gutiérrez B, De Cueto M, Viale P, Venditti M, Hernández-Torres A, et al. Development and validation of the INCREMENT-ESBL predictive score for mortality in patients with bloodstream infections due to extended-spectrum-β-lactamase-producing Enterobacteriaceae. J Antimicrob Chemother. 2017;72(3):906–13. https://doi.org/10.1093/jac/dkw513.
Buys H, Muloiwa R, Bamford C, Eley B. Klebsiella pneumoniae bloodstream infections at a south African children’s hospital 2006-2011, a cross-sectional study. BMC Infect Dis. 2016;16(1):1–10. https://doi.org/10.1186/s12879-016-1919-y.
Moradigaravand D, Martin V, Peacock SJ, Parkhill J. Evolution and epidemiology of multidrug-resistant Klebsiella pneumoniae in the United Kingdom and Ireland. MBio. 2017;8(1):1–13. https://doi.org/10.1128/mBio.01976-16.
Wyres KL, Nguyen TNT, Lam MMC, Judd LM, van Vinh CN, Dance DAB, et al. Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from south and Southeast Asia. Genome Med. 2020;12(1):11. https://doi.org/10.1186/s13073-019-0706-y.
Maddy. Antibiotic prescribing and resistance: Views from low-and middle-income prescribing and dispensing professionals Report to the World Health Organization, researched and compiled by students and staff of the Antimicrobial Resistance Centre at the London Sc. 2017. doi:t http://www.who.int/antimicrobial-resistance/LSHTMAntibiotic-Prescribing-LMIC-Prescribing-and-Dispensing-2017.pdf.
Sigaúque B, Sevene E, Manjate A, Modi B, Macuamule C, Ghid P, et al. Situation Analysis and Recommendations: Antibiotic Use and resistance in Mozambique. 2015. www.cddep.orh/GARP. Accessed 2 Mar 2020.
Harada S, Doi Y. Hypervirulent Klebsiella pneumoniae : a call for consensus definition and international collaboration. J Clin Microbiol. 2018;56(9):776–94. https://doi.org/10.1128/JCM.00959-18.
Ruiz-Perez F, Nataro JP. Bacterial serine proteases secreted by the autotransporter pathway: classification, specificity, and role in virulence. Cell Mol Life Sci. 2014;71(5):745–70. https://doi.org/10.1007/s00018-013-1355-8.
Pokharel P, Habouria H, Bessaiah H, Dozois CM. Serine protease autotransporters of the Enterobacteriaceae (SPATEs): out and about and chopping it up. Microorganisms. 2019;7(12):594. https://doi.org/10.3390/microorganisms7120594.
Ruiz-Perez F, Wahid R, Faherty CS, Kolappaswamy K, Rodriguez L, Santiago A, et al. Serine protease autotransporters from Shigella flexneri and pathogenic Escherichia coli target a broad range of leukocyte glycoproteins. Proc Natl Acad Sci U S A. 2011;108(31):12881–6. https://doi.org/10.1073/pnas.1101006108.
Acknowledgments
The authors thank the CISM and MDH staff for collecting and processing data; and the District Health Authorities for their collaboration in the research activities on-going in the Manhiça district. Special thanks for the CISM Microbiology and Molecular Biology laboratory technicians for sample processing, particularly Sultuane Givá, Samira Sirage, Mariano Sitaúbe, Esperança Lázaro and Efraim Honwana. We are indebted to the children and mothers/caretakers for participating in the study.
Disclaimer
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of [the Centers for Disease Control and Prevention/the Agency for Toxic Substances and Disease Registry].
Funding
CISM is supported by the Government of Mozambique and the Spanish Agency for International Development Cooperation (AECID). CHAMPS is funded by the Bill & Melinda Gates Foundation under the Grant OPP1126780 to Robert Breiman, subcontract SC00003286. ISGlobal receives support from the Spanish Ministry of Science and Innovation through the “Centro de Excelencia Severo Ochoa 2019-2023” Program (CEX2018–000806-S), and support from the Generalitat de Catalunya through the CERCA Program.
Author information
Authors and Affiliations
Contributions
AM contributed to the literature review, data analysis, conceptualization and writing up the manuscript. AM did also all the tables and Fig. 1. MG and AJ contributed equally, and the sequence of names were decided according to published papers. AJ did the Fig. 2A and B. NN contributed to all data acquisition. NB, AS, RV, SM, AC, JC, JO, HM, TN, RB, CW, DB, and QB reviewed critically the manuscript. IM contributed to the conceptualization, critically reviewed the manuscript and designed the study. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The protocol of invasive bacterial disease surveillance (IBD) was reviewed and approved by the Mozambican National Bioethics Committee for Health and Institutional Review Boards of Hospital Clinic of Barcelona, Spain, the U.S. Centers for Disease Control and Prevention, and the University of Maryland, School of Medicine. The CHAMPS was reviewed and approved by the CISM’s Institutional Ethics Committee for Health – CIBS-CISM and the CNBS (Ref: 285/CNBS/16 of September 05, 2016).
The isolates included in this research were obtained from participants who were enrolled in the IBD and CHAMPS, and only if the parents, caretaker or legal guardians accepted to participate and sign an informed consent.
The IBD and CHAMPS protocol and its methods were performed in accordance with the Declaration of Helsinki and Good Clinical and Laboratory Practices.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Clinical picture of post-mortem patients recruited in the CHAMPS Study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Massinga, A.J., Garrine, M., Messa, A. et al. Klebsiella spp. cause severe and fatal disease in Mozambican children: antimicrobial resistance profile and molecular characterization. BMC Infect Dis 21, 526 (2021). https://doi.org/10.1186/s12879-021-06245-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-021-06245-x