Abstract
Background
The Ferula genus encompasses 180–185 species and is one of the largest genera in Apiaceae, with many of Ferula species possessing important medical value. The previous studies provided more information for Ferula, but its infrageneric relationships are still confusing. In addition, its genetic basis of its adaptive evolution remains poorly understood. Plastid genomes with more variable sites have the potential to reconstruct robust phylogeny in plants and investigate the adaptive evolution of plants. Although chloroplast genomes have been reported within the Ferula genus, few studies have been conducted using chloroplast genomes, especially for endemic species in China.
Results
Comprehensively comparative analyses of 22 newly sequenced and assembled plastomes indicated that these plastomes had highly conserved genome structure, gene number, codon usage, and repeats type and distribution, but varied in plastomes size, GC content, and the SC/IR boundaries. Thirteen mutation hotspot regions were detected and they would serve as the promising DNA barcodes candidates for species identification in Ferula and related genera. Phylogenomic analyses with high supports and resolutions showed that Talassia transiliensis and Soranthus meyeri were nested in the Ferula genus, and thus they should be transferred into the Ferula genus. Our phylogenies also indicated the monophyly of subgenera Sinoferula and subgenera Narthex in Ferula genus. Twelve genes with significant posterior probabilities for codon sites were identified in the positively selective analysis, and their function may relate to the photosystem II, ATP subunit, and NADH dehydrogenase. Most of them might play an important role to help Ferula species adapt to high-temperatures, strong-light, and drought habitats.
Conclusion
Plastome data is powerful and efficient to improve the support and resolution of the complicated Ferula phylogeny. Twelve genes with significant posterior probabilities for codon sites were helpful for Ferula to adapt to the harsh environment. Overall, our study supplies a new perspective for comprehending the phylogeny and evolution of Ferula.
Similar content being viewed by others
Background
Ferula L. is one of the genera of Apiaceae [1], which was once classified in the tribe Peucedaneae [2, 3], but now in the tribe Scandiceae [4,5,6]. This genus, encompassing about 180–185 species all over the world [7], distributes in the Mediterranean region, Siberia, Central Asia, and northern Africa [3, 8, 9], and grows mostly in mountainous regions and desert clay soils [8, 10]. The Ferula genus has been chiefly recognized by the prominent taproots, stout stems, finely divided leaves with large inflated sheaths, and strongly dorsally compressed mericarps with filamentary or prominent dorsal ribs, narrowly or broadly winged marginal ribs and the plane or slightly concave commissural face [1, 6]. However, due to the great variations in the leaf, inflorescences, and mericarps anatomy, distinguishing this genus from nearby genera was extremely difficult. Hence, the taxonomic delimitation of Ferula has long been contentious. Pimenov [11, 12] suggested that Talassia and Soranthus should be transferred into Ferula according to the anatomical characteristics of the fruit which was the presence of a sclerotic cell layer in the mesocarp of fruits. Pimenov [13], according to the type specimens and morphological features, summarized the nomenclatural combinations of Ferula in China and merged the S. meyeri and T. transiliensis into the Ferula. However, Qin and Shen [14] believed that Ferula L., Soranthus Ledeb., and Talassia Korov. should exist as separate genera in Apiaceae, based on the comparison of the external morphology, fruit anatomy, and pollen characteristics of the plants. In Flora of China [1] and The Flora of Reipublicae Popularis Sinica [15], Soranthus and Talassia were also separated from the Ferula. Therefore, the generic limits between the Ferula and its nearby genera based solely on morphological characteristics was challenging.
Before, scholars have recently used molecular data to study the taxonomy and phylogeny of Ferula and its relative nearby genera. Dowine et al. [16] summarizing the previous study results, proposed that Talassia and Soranthus were closely related to Ferula but more research is needed to resolve the relationship. Kurzyna-Młynik et al. [6] and Panahi et al. [17, 18] have placed the T. transiliensis and S. meyeri into Ferula according to the phylogenetic trees using the nuclear ribosomal DNA internal sequence data (ITS) and three plastid non-coding regions. But the support and resolution in these phylogenetic trees were weak and low, and thus the phylogenetic position of T. transiliensis and S. meyeri within Ferula genus was unresolved. So additional markers are needed to obtain a robust phylogeny.
The infrageneric taxonomic system of Ferula has been complicated. Based on habit and vegetative characteristics, Korovin [19] established the six subgenera and eight sections of this genus. And this division was adopted in The Flora of Reipublicae Popularis Sinica [15] where the Ferula species grown in China were divided into four sections and four subgenera. However, Safina and Pimenov [20,21,22] contested the infrageneric division provided by Korovin and proposed 12 new sections of Ferula genus based on mericarps morphology and anatomy. Nevertheless, subsequent molecular study did not agree with those infrageneric taxonomies, and inferred a new infrageneric classification of Ferula. Panahi et al. [17] used nrITS and three plastid non-coding regions to propose a new classification system for Ferula of four subgenera and ten sections, where the species growing in China were divided into two subgenera.
Besides, many species of Ferula have medical value and are extensively used in traditional medicine in folk and pharmacy. For example, F. sinkiangensis K. M. Shen and F. fukanensis K. M. Shen are used as vital traditional medicines to eliminate stagnation, resolve symptoms, disperse lumps, and kill worms [23, 24]. Other species, such as F. lehmannii Boiss., F. songarica Pall. ex Spreng., F. olivacea (Diels) H. Wolff ex Hand. -Mazz., and F. feruloides (Steud.) Korovin, also have significant pharmaceutical effects [25]. However, due to the high market value and morphological similarities, the other Ferula species are usually used as substitutes for F. sinkiangensis and F. fukanensis. Consequently, it is indispensable to develop more DNA barcodes for species authentication to ensure medicinal quality.
The plastid is an essential organelle for green plants, which is responsible for photosynthesis and offers the basic energy for plants [26]. The plastid genome (plastome) is uniparentally inherited, lacks recombination, has low nucleotide substitution rates, and contains abundant variable sites. Therefore, the plastome is a useful tool to improve the certainty of phylogenetic trees [27, 28]. The plastome generally is 115 to 165 kb in length, containing a large single-copy region (LSC), two separately inverted repeat regions (IRs), and one small single-copy region (SSC), and encodes about 110–130 unique genes [29,30,31]. Comparative analysis of plastomes reveals the variation in its structural combination and gene arrangement, which is helpful to further identify the mutational hotspots for species authentication [32, 33]. Consequently, with the processing development of next-generation sequencing and multiple bioinformatics technologies, plastomes have been broadly and successfully applied to the development of DNA barcodes and analysis of phylogenetics [34, 35]. In addition, the plastomes are used to investigate the adaptive evolution of plants. Adaptive evolution implies that the adaptability of species is enhanced during the evolutionary processes, driven by the natural selective pressure applied to the genetic variation through gene flow, recombination, and mutations [36] and causes biodiversity in each aspect of biological organization [37]. Understanding the adaptive evolution of organisms could contribute to elucidating the latent mechanism of adapting to the local environment and providing guidance for future protection [38, 39]. For example, accD, rpoA, and rpoC2 genes were positively selected in the Rehmannia species, which helped species to grow in divergent light intensity habits [40]. Furthermore, psbH, psbM, and rbcL genes may work in the growth of all Dipterocarpoideae species to adapt to a strongly illuminated environment [41]. As for Ferula genus, limited chloroplast genome data has been reported [42], and few studies, especially for focusing on endemic species in China, have been conducted using chloroplast genomes.
Here, with newly sequenced 22 plastomes of Ferula, Talassia, and Soranthus species, we analyzed 42 plastomes from the Apiaceae subfamily and aimed to (1) evaluate the infrageneric classification system of Ferula; (2) exploit promising candidate DNA markers of this genus; and (3) investigate the adaptive evolution of this genus based on plastome data. In brief, our study will enhance knowledge of the phylogeny and adaptive evolution of Ferula.
Results
Features of the plastome
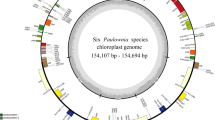
The plastomes of 22 species ranged from 160,901 bp (F. conocaula) to 167,208 bp (F. olivacea) in length (Table 1). All plastomes possessed the typical quadripartite structure with two copies of IR regions (28,922–31,989 bp) separated by the LSC region (84,904 -85,895 bp) and SSC region (17,546–17,846 bp). The total GC content was between 37.6 and 38.0%, and the IR regions were the highest (42.8–43.1%) compared to the LSC (35.5–35.7%) and SSC regions (30.6–31.1%). The rRNA genes had the highest GC content, greater than the tRNA genes and protein-coding genes. Each of these 22 plastomes contained 133 genes, consisting of 87 protein-coding genes, 37 tRNA genes, and eight rRNA genes (Fig. 1, Table 1). Of these genes, 14 genes contained one intron, and four genes contained two introns (Fig. 1, Table S1).
Repeat sequences analysis and codon usage
The total number of SSRs ranged from 65 (F. kingdon-wardii) to 80 (T. transiliensis) within the 22 plastomes (Fig. 2A). The most abundant were mononucleotide repeats (32–48), followed by dinucleotides (14–19), tetranucleotides (8–12), trinucleotides (3–5), and pentanucleotides (0–3). Only F. songarica and F. kingdon-wardii had one hexanucleotide (Fig. 2A). (T)10 was found in the intergenic region between atpH and atpI in only F. olivacea, F. paeoniifolia, and F. kingdon-wardii. We also found (ATTA)3 was distributed in the coding region of rps2 in F. olivacea and F. paeoniifolia. (G)10 or (G)11 was allocated at the intergenic region (psbZ/trnG) in F. olivacea and F. paeoniifolia, while (AAAT)3 was only found in the intergenic region (trnS /psbZ) in F. kirialovii, and (A)15 was observed in the ndhF gene only in T. transiliensis, and so on (Table S2). SSRs were distributed largely in the LSC region, less in the SSC and IR regions. Moreover, the analysis of SSRs locations uncovered that the majority of SSRs were distributed in the non-coding regions that contained the intron and the intergenic regions (Table S2). In addition, the forward, palindromic, complementary, and reverse repeats were detected in the 22 species, and the total number of repeats was 1,314. The forward repeats were the most abundant (649), while the complement repeats were the least (6) (Fig. 2B, Table S3). Among the 22 species, the F. licentiana had the most repeats (89), while F. caspica possessed the least repeats (46). In addition, we divided the repeats into four types according to length: 30–45 bp, 45–60 bp, 60–75 bp, and > 70 bp, and most of the repeats (70.32%) were 30–45 bp long (Fig. 2C).
We extracted and connected 53 protein-coding genes in each species to characterize the codon usage of 22 plastomes (Fig. 3, Table S4). These protein sequences encoded 21,087–21,185 codons (Table S4). Among them, Leu, Ser, and Arg were encoded by six codons indicating the highest preference, and Leu was most abundant (2,092–2,234), while the Cys was least (217–221) in all plastomes (Table S4). Additionally, relative synonymous codon usage (RSCU) values of all codons ranged from 0.31 to 2.00 in all species, and the RSCU values of about 30 codons were greater than 1(Fig. 3).
Comparison of plastomes
The borders of LSC/IRb, IRb/SSC, SSC/IRa, and IRa/LSC among the 22 plastomes were relatively conserved and similar (Fig. 4). The LSC/IRb borders were fell into rps19; IRb/SSC borders were fell into the ndhF gene, but located between the ycf1 and ndhF genes in F. sinkiangensis, F. dissecta, S. meyeri, F. kirialovii, and F. kingdon-wardii; the SSC/IRa borders were fell into ψycf1 gene and the IRa/LSC borders were located between the rpl2 gene and trnH gene.
Using the mVISTA program, we found that the plastomes of the 22 taxa were highly conserved, and the IR regions and coding regions were more conserved than the SC regions and non-coding regions (Fig. 5). Nevertheless, 13 hotspot regions were detected, including five coding regions (ycf1, ndhF, rps11, matK, and rpl22) that possessed Pi > 0.004 and eight non-coding regions (ycf15/trnV, trnH /psbA, trnG/trnR, trnR /atpA, psbI/trnS, rps15/ycf1, rps2/rpoC2, and ycf3/trnS) that had Pi > 0.010 (Fig. 6). In total, these regions could be used for DNA barcode studies in the future.
Phylogenetic relationships
We reconstructed the phylogenetic trees based on single-copy CDs of 42 species plastomes and 62 ITS sequence data (Table S5). The plastomes tree and ITS tree showed incongruent topologies, but the trees indicated that T. transiliensis and S. meyeri nested within Ferula species (Fig. 7; Fig. S1). In the plastome tree, the ML (maximum likelihood) and BI (Bayesian inference) analyses resulted in identical trees, and both analyses strongly showed that the T. transiliensis, S. meyeri, and Ferula members formed a robustly supported clade (BS > 85%, PP ≥ 0.95). Soranthus meyeri was clustered with the branch formed by F. syreitschikowii, T. transiliensis, and F. conocaula where F. syreitschikowii was the sister to a subclade consisting of the T. transiliensis and F. conocaula (BS = 90, PP = 1.00), and then they nested within the Ferula members (BS ≥ 90, PP ≥ 0.99). Furthermore, all members of Ferula formed two lineages, one lineage contained F. olivacea, F. paeoniifolia, and F. kingdon-wardii (BS = 100, PP = 1), and the other lineage contained all other Ferula species (Fig. 7). Moreover, the phylogenetic relationships among non-Ferula species conducted in our study were consistent with previous research [43].
Phylogenetic tree reconstruction of 42 taxa inferred from Maximum likelihood (ML) and Bayesian inference (BI) analyses based on the single-copy CDs. Numbers indicate Bayesian posterior probabilities (PP) and maximum likelihood bootstrap values (BS), and (*) indicates maximum support in both two analyses
Mericarp morphology
Twenty-two species had mericarps elliptic or ovate, strongly dorsally compressed, and endosperm commissural face plane or slightly concave. Mericarps had a shorter distance between dorsal and median ribs than that of median and lateral ribs. Dorsal and median ribs filiform or sometimes prominent, lateral ribs narrowly or broadly winged. A number of vittae in each furrow (1–4) and commissure (2–12) (Fig. 8, Fig. S2, Table S6).
Positive selection analysis
Fifty-two single-copy CDs genes were eventually selected for positive selection analysis. The results showed that the 12 genes (atpB, atpF, ndhA, ndhC, ndhI, ndhJ, ndhK, psbK, rpl20, rpoB, rpoC1, and rpoC2) were observed with significant posterior probabilities suggesting sites positively selected in the BEB test (Table S7). In addition, among these genes, most had one positive selective site, apart from the rpoC2 that had four positive selective sites (Fig. 9; Fig. S3).
Partial alignment of three out of twelve positively selected genes. A Partial aligned amino acids sequences of the rpoC2 gene; B partial aligned amino acids sequences of the atpF gene; C partial aligned amino acids sequences of the ndhK gene. The red blocks indicate the amino acids in Ferula, Talassia, and Soranthus with a high BEB posterior probability
Discussion
Comparison of Ferula plastomes
In this study, we implemented a comprehensive comparative analysis of 22 plastomes from Ferula, Talassia, and Soranthus. All plastomes possessed a typical circular tetrad structure with two inverted repeat regions, one SSC region, and one LSC region, which is common in other plants [44,45,46]. Additionally, the gene numbers, type and distribution of large repeats, number and type of SSRs, and codon usage were rather similar among these plastomes. This circumstance is common across other genera in the family Apiaceae [47, 48]. Therefore, these results demonstrated that the plastomes were highly conserved in terms of structure, gene number, type and distribution of large repeat, number and type of SSRs, and codon usage in Ferula, Talassia and Soranthus.
However, we noticed the obvious divergence within the size of 22 plastomes, varying from 160,901 bp (F. conocaula) to 167,208 bp (F. olivacea). Previous studies inferred that the variation of plastome size was mainly influenced by the following three factors. First, the contraction and expansion of IR regions were the most common reason for the variation of plastome size. For example, a significant expansion was detected in Pelargonium hortorum, which resulted in the plastome size increasing [49]. Second, gene losses could lead to the shrinkage of plastome size, especially within several parasitic plants [50]. Third, the indels had an important influence on the plastid genome size within some genera [51, 52]. In this study, the borders of IR/SC regions were slightly varied and gene content was highly conserved, while about 3,020 bp, 2,837 bp, and 2,190 bp deletions in F. kingdon-wardii, F. conocaula, and F. caspica were detected in ycf15/ trnV, which resulted in the plastome length of the three species being shorter than the other species. As a result, the deletions may be largely responsible for the variation of plastome size in the 22 plastomes.
The SSRs are used to be the molecular markers, in particular, in studies of biogeography and plant population genetics and the identification of species because they have high polymorphic rates [45]. Therefore, those fragments, such as (AAAT)3 only found in the intergenic region (trnS /psbZ) in F. kirialovii and (A)15 observed in the ndhF gene only in T. transiliensis, may be useful for selecting as molecular markers to differentiate between Ferula species in the future.
Promising DNA barcodes
Accurate species identification has usually been difficult for taxonomists, which was large due to restrictions on incomplete specimens and limitations of field observation of the whole plant. The developing DNA barcoding technology, discriminating species by the short DNA fragments with variable sites [53], looks forward to working out this difficulty. In animals, the mitochondrial gene cytochrome oxidase 1 has been confirmed to be reliable and valid as the DNA barcode for species identification [54, 55]. In plants, the common DNA barcodes including trnH-psbA, matK, and rbcL are insufficient to accurately identify species [56, 57]. The variation of the rbcL gene was relatively low (Pi = 0.00161) in the 22 studied plant species. As a result, this region may be restricted to accurately delimitating Ferula species.
According to the sequence variation, we chose five protein-coding regions (ycf1, ndhF, matK, rps11, and rpl22) and eight non-coding regions (ycf15/trnV, trnH /psbA, trnG/trnR, trnR /atpA, psbI/trnS, rps15/ycf1, rps2/rpoC2, and ycf3/trnS) as the potential identifiers for species in Ferula. Among them, the trnH-psbA region is a member of universal DNA barcodes [57]; ycf1 and rpl22, have been selected as the coming DNA barcodes in some plants [58, 59]. We will examine if these sequences could serve as valid DNA barcodes for species identification in the Ferula genus in future research.
Phylogenetic analyses
Same to previous results obtained by Kurzyna-Młynik et al. [6] based on nrITS data and by Panahi et al. [18] based on nrITS and three plastid DNA rps16 and rpoC1 intron, and rpoB-trnC intergenic spacer, our phylogeny based on plastome data robustly supported that T. transiliensis and S. meyeri nested in Ferula genus. This relationship also showed in our ITS-based phylogenetic tree, although the support of which was weak. Hence, transferring T. transiliensis and S. meyeri into the Ferula genus should be reasonable. And their name should be the F. transiliensis [60] and F. sibirica [11]. Additionally, our phylogenetic result with high resolution indicated that T. transiliensis and S. meyeri were more closely related to F. conocaula and F. syreitschikowii than the other Ferula species. However, due to the limited samples of Ferula acquired in our study and maternal inheritance of plastome, their phylogenetic positions within Ferula genus need to completely exploit in future studies.
The infrageneric taxonomy of Ferula was inconsistent in previous studies. Korovin et al. [19, 61] divided Ferula into six subgenera and eight sections based on vegetative features and habits. In The Flora of Reipublicae Popularis Sinica [15], the Ferula species grown in China were placed in four subgenera and four sections [15, 19]. However, Panahi et al. [17] proposed a new classification that included four subgenera and eight sections based on molecular phylogenetic results.
In our study, the 22 species were strongly divided into two lineages: one encompassed F. olivacea, F. paeoniifolia, and F. kingdon-wardii (lineage I); the other had the remaining species (lineage II). This result was further supported by species’ geographical distributions and mericarp structures. The members of lineage I are distributed in the alpine meadows and rock cranny of cliffs in Yunnan and Sichuan Provinces [1, 62]; the mericarps of these three species have very prominent dorsal and lateral ribs, and two vascular bundles were present in the dorsal and lateral ribs [63]. Whereas the members of lineage II are located in the gravelly slopes and desert gravels in Xinjiang and other provinces; their mericarps have filiform or slightly prominent dorsal and lateral ribs with one vascular bundle [15, 63]. Combining the robust phylogenetic framework and morphological characteristics, our result strongly supported the establishment of subgenera Sinoferula and subgenera Narthex [17]. But our result showed that the F. licentiana should be placed in the subgenera Narthex, and F. peaoniifolia should be added into subgenera Sinoferula. In addition, our result inferred that the infrageneric taxonomy of Ferula genus in Flora of Reipublicae Popularis Sinica [15] was inappropriate.
The adaptation evolution of Ferula plastome
Ferula species mostly grow in high-temperature, strong-bright, and drought environments, and thus we speculated several genes were probably under a special evolutionary process [1]. As we expected, 12 genes with significant posterior probabilities for codon sites were identified by the BEB test in our study. Researchers proposed that codon sites with higher posterior probabilities could be considered as positively selected sites, and genes in possession of positively selected sites may evolve under various selection pressure [64]. Therefore, 12 genes detected in our study may have undergone positive selection pressures. The 12 genes comprised two ATP subunit genes (atpB and atpF), five NADH dehydrogenase genes (ndhA, ndhC, ndhI, ndhJ, and ndhK), one gene (psbK) associated with photosystem II, one gene (rpl20) about large subunit of ribosome, and three RNA polymerase subunits genes (rpoB, rpoC1, and rpoC2). Among them, the largest proportion of genes (ndhA, ndhC, ndhI, ndhJ, and ndhK) are related to the NADH-dehydrogenase subunits. NADH-dehydrogenase subunits were fundamental to the electron transport chain for the generation of ATP, and photosynthesis of plants [65, 66]. Wang et al. [67] found that NADH could induce the PSI cycle electron to divert the electrons to avoid plants being injured and provide the ∆pH for CO2 assimilation for a certain period of time under high-temperature stress. Therefore, these genes under positive selection helped Ferula species refrain from injury and thrive in drought and intense light environments. Additionally, several codon sites with significant posterior probabilities were found in rpo genes (rpoB, rpoC1, and rpoC2). The rpoB gene encodes the β-subunit of RNA Polymerase in plastomes [68], and the rpoC2 gene encodes another subunit of RNA Polymerase which is responsible for the expression of photosynthetic genes [69]. The previous research indicated that RNA polymerase could not only keep the essential metabolic process to survive, but also regulate the process of gene transcription and expression, for facilitating species to respond to the changing environment conditions [70, 71]. Moreover, via implementing comparative experiments, Gao et al. [72] revealed that the rpoC2 gene underwent strong positive selection in the sun-loving rice species, and this phenomenon inferred that this gene was important for sun-loving rice species to adapt to the sunlight habitat. Hence, those rpo genes under positive selection in our analysis may contribute to adapting the bright environments for Ferula species. Furthermore, the atpF gene, encoding one of the subunits of H+-ATP synthase, played the crucial role in electron transportation, and photorespiration for plants [73]. In a previous study, this gene was positively selected in two evergreen Quercus species comparing with two deciduous Quercus species, which could help the evergreen species to resist the stress from cold and drought [74]. Generally, the Ferula species grow and develop in early spring and live in the arid desert areas [15, 75], thereby the atpF gene may be significant in environment adaptation of Ferula species. In brief, these positively selected genes have been beneficial to the development and reproduction of Ferula species, and played an important role in adapting to the harsh environment where Ferula species grow.
Conclusion
In our study, we sequenced and assembled 22 plastomes of Ferula, Talassia, and Soranthus species. Based on the comparative analysis of plastomes, we observed conservation in genome structure, gene number, codon usage, and repeats types and distribution, but variation in plastomes size, GC content, and the SC/IR boundaries. Thirteen mutation hotspot regions were detected and has potential as DNA barcodes for species identification in Ferula and related genera. Based on the phylogenetic analysis for Ferula using 22 plastomes and 62 ITS sequences, we agreed with some previous studies that Talassia and Soranthus should be placed into Ferula. Our result also supported the monophyly of subgenera Sinoferula and subgenera Narthex. The phylogeny reconstructed by the plastomes highlighted the strength of the plastome that possessed the more variable sites and greatly resolved the phylogeny of studied species. In addition, twelve genes with significant posterior probabilities for codon sites helped Ferula species to adapt to their harsh environments. Our study offers a new perspective for further study in phylogeny and evolution of Ferula species.
Methods
Plant materials and DNA extraction
Fresh leaves from adult plants of the 22 species were collected from each yield site. Then, the leaves were immediately dried using silica gel for DNA extracting. The total genomic DNA was extracted from the dried leaf tissue using a plant DNA extraction kit (Cwbio Biosciences, Beijing, China). The formal identification of those samples collected was undertaken by Associate Professor Songdong Zhou (Sichuan University). The Voucher specimens were deposited at the herbarium of Sichuan University (Chengdu, China), and their deposition numbers were listed in the Additional file 11: Table S8. The newly sequenced 22 ITS have been submitted to NCBI (Additional file 8: Table S5).
Plastome genome sequencing and assembling
The raw reads of 22 newly sequenced species were generated from the Illumina HiSeq X Ten platform (paired-end, 150 bp) at Novogene (Tianjin, China). The raw reads were filtered using fastP version v0.15.0 (-n 10 and -q 15) to yield clean reads [76]. Then clean reads were used to assemble plastomes using NOVOPlasty v2.6.2 [77] with default parameters and the rbcL gene (MK749921.1) of F. bungeana downloaded from NCBI as seed. The assembled genomes were initially annotated by the PGA [78], and then adjusted manually in Geneious v9.0.2 [79]. Using the same method, the plastomes of non-Ferula obtained from the NCBI were re-annotated. Finally, the plastid genome maps were drawn using Chloroplot [80].
Repeat sequences and codon usage
The Perl script MISA (http://pgrc.ipk-gatersleben.de/misa/) was used to analyze simple sequence repeats (SSRs) in the plastome sequences. The parameters of SSRs were set as follows: 10, 5, 4, 3, 3, and 3, in response to mono-, di-, tri-, tetra-, penta-, and hexanucleotides, respectively. The REPuter online program [81] was used to search repeat sequences including (F) forward, (P) palindromic, (R) reverse, and (C) complementary repeats. The parameters were as follows: (1) a repeat size of over 30 bp; (2) two repeats with more than 90% sequence identity; and (3) Hamming distance = 3. Then, the protein-coding genes were extracted from the 22 plastid genomes for codon analysis by the CodonW v1.4.2 program [82].
Genome structure and sequence diversity
The IR region contraction and expansion at the border of the plastome were analyzed by the online program IR scope [83]. The size and position of the gene were then manually adjusted. The sequence identity of whole plastomes was detected and visualized by the online program m-VISTA [84] in Shuffle-LAGAN mode, with the F. sinkiangensis as a reference. Nucleotide diversities of the coding genes and intergenic regions were calculated by DnaSP v5 [85].
Phylogenetic analysis
To investigate the phylogeny of Ferula, 42 plastomes and 62 nuclear ITS sequences were used to reconstruct the phylogenetic tree (Table S5). Chamaesium jiulongense X. L. Guo & X. J. He, Bupleurum commelynoideum de Boiss. were selected as the outgroups to root the phylogenetic tree according to the results of Zhou et al. [86]. For plastome data, 80 single-copy protein-coding sequences (CDs) commonly shared by the 42 plastomes were extracted using Phylosuite v.1.2.2 [87] and then respectively aligned by MAFFT v7.221 [88]. These alignments were concatenated as a super matrix by Phylosuite v.1.2.2 [87]. The nrITS sequences were aligned by MAFFT v7.221 [88].
The prepared data sets of CDs and nrITS were then subjected to Maximum-Likelihood (ML) analyses and Bayesian Inference (BI). For ML analysis, the phylogenetic trees were generated by RAxML 8.2.8 [89] with the GTRGAMMA model, as suggested in the RAxML manual, and 1,000 bootstrap replicates. The BI analysis was conducted using MrBayes v.3.2.5 [90], with the TVM + I + G and GTR + I + 0 substitution models determined by Modeltest v3.7 [91] for plastomes and ITS, respectively. Markov chain Monte Carlo (MCMC) algorithm was run for one million generations, with one tree sampled every 100 generations. The first 25% of trees were discarded as burn-in, and the remaining trees were used to build the consensus tree. The phylogenetic tree was displayed and edited in FigTree v1.4.2 [92].
Positive selected analysis
The Optimized Branch-Site model [93] and the Bayesian Empirical Bayes (BEB) [64] method were used to identify genes that were positively selected in Ferula species compared to the non-Ferula species. Single-copy protein-coding regions of 42 plastomes were extracted and then aligned using the ClustalW [94] with the amino acid codons. Then the alignments of sequences were trimmed. Finally, the trimmed alignments were used to implement the positive selection analysis by the CODEML algorithm in the PAML package [95] in EasyCodeml [96] with the branch-site model and the Ferula clade designed as the foreground branch. The BEB method was used to compute the posterior probabilities of amino acid sites to confirm whether these sites were selected positively and with high posterior probabilities [64]. The likelihood-ratio tests (LRT) were implemented based on Lan et al. [97], as a result, if the gene was with a p-value < 0.5, it would be certified as the positively selected gene. We then used Jalview v.2.11.1.7 [98] to view the amino acid sequences of positively selected genes.
Morphological observations of mericarps
The whole structures of dorsal and commissural side views, and anatomical structures including transverse section, rib shape, and vittae of mericarps in 12 species were observed and photographed via a stereomicroscope (SMZ25, Nikon Corp., Tokyo, Japan). These mature mericarps were selected randomly and measured by the KaryoType [99]. Mericarp terminology is based on Kljuykov et al. [100].
Availability of data and materials
Twenty-two annotated plastomes have been submitted to NCBI (https://www.ncbi.nlm.nih.gov) with accession numbers: OP324722-OP324743; newly sequenced 22 nrITS have been submitted to NCBI (https://www.ncbi.nlm.nih.gov) with accession numbers: OP341492-OP341513 (Additional file 8: Table S5).
Abbreviations
- BEB:
-
Bayes empirical bayes
- BI:
-
Bayesian inference
- bp:
-
Base pair
- BS:
-
Branch support
- CDS:
-
Protein-coding sequences
- IR:
-
Inverted repeat
- ITS:
-
Internal transcribed spacer
- LRT:
-
Likelihood ratio test
- LSC:
-
Large single copy
- MCMC:
-
Markov chain Monte Carlo
- ML:
-
Maximum Likelihood
- Pi:
-
Nucleotide diversity
- PP:
-
Posterior probability
- rRNA:
-
Ribosomal RNA
- RSCU:
-
Relative synonymous codon usage
- SSC:
-
Small single copy
- SSR:
-
Simple sequence repeat
- tRNA:
-
Transfer RNA
References
Sheh ML, Watson MF. Ferula Linnaeus. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Beijing: Science Press & St. Louis: Missouri Botanic Garden Press; 2005. p. 174–80.
Drude CGO. Umbelliferae. In: Engler A, Prantl K, editors. Die natu¨rlichen Pflanzenfamilien. Leipzig: Wilhelm Engelmann; 1897-1898. p. 63–250.
Pimenov MG, Leonov MV. The genera of Umbelliferae. Kew: Royal Botanical Gardens; 1993.
Downie SR, Katz-Downie DS, Watson MF. A phylogeny of the flowering plant family Apiaceae based on chloroplast DNA rpl16 and rpoC1 intron sequences: towards a suprageneric classification of subfamily Apioideae. Amer J Bot. 2000;87(2):273–92.
Downie SR, Watson MF, Spalik K, Katz-Downie DS. Molecular systematics of Old World Apioideae (Apiaceae): relationships among some members of tribe Peucedaneae sensu lato, the placement of several island-endemic species, and resolution within the apioid superclade. Canad J Bot. 2000;78(4):506–28.
Kurzyna-Młynik R, Oskolski AA, Downie SR, Kopacz R, Wojewódzka A, Spalik K. Phylogenetic position of the genus Ferula (Apiaceae) and its placement in tribe Scandiceae as inferred from nrDNA ITS sequence variation. Pl Syst Evol. 2008;274(1):47–66.
Kadereit JW, Bittrich V. Flowering plants. Eudicots: Aapiales, Gentianales (except Rubiaceae). In: The families and genera of vascular plants. Berlin: Springer; 2018.
Wu ZY, Lu AM, Tang YC, Chen ZD, Li DZ. The families and genera of Angiosperms in China: a comprehensive analysis. Beijing: Science Press; 2003.
Ajani Y, Ajani M. A new species of Ferula (Umbellıferae) from Southern Iran. Edinb J Bot. 2008;65(3):425–31.
Hamzeloomoghadam H, Esmaeili S, Fotoohi F, Naghibi F, Pirani A, Hajimehdipoor H. In vitro evaluation for cytotoxic activity of three Ferula species. Int J Pharm Sci Res. 2013;4(7):2673–6.
Pimenov MG, Kirillina NA. The carpology of Soranthus, Ladyginia, Eriosynaphe and Schumannia in connection with the problem of the taxonomic limits of the genus Ferula (Apiaceae). Botanicheskii Zhurnal. 1980;65:1756–66.
Pimenov MG. Glaucooselinum section of the genus Ferula. Biol Nauki (Mosc). 1983;12:74–9.
Pimenov MG. Updated checklist of Chinese Umbelliferae: nomenclature, synonymy, typification, distribution. Turczaninowia. 2017;20(2):106–239.
Qin XM, Shen GM. The taxonomic studies on Xinjiang Ferula and its Close Genera. Desert Research. 1990;4(4):24–33.
Sheh ML, Ferula L. In: Shan RH, Sheh ML, editors. Flora Reipublicae Popularis Sinica. Beijing: Science Press; 1992. p. 85–117.
Downie SR, Spalik K, Katz-Downie DS, Reduron JP. Major clades within Apiaceae subfamily Apioideae as inferred by phylogenetic analysis of nrDNA ITS sequences. Plant Div Evol. 2010;128(1):111–36.
Panahi M, Banasiak Ł, Piwczyński M, Puchałka R, Kanani MR, Oskolski AA, et al. Taxonomy of the traditional medicinal plant genus Ferula (Apiaceae) is confounded by incongruence between nuclear rDNA and plastid DNA. Bot J Linn Soc. 2018;188(2):173–89.
Panahi M, Banasiak Ł, Piwczyński M, Puchałka R, Oskolski AA, Spalik K, et al. Phylogenetic relationships among Dorema, Ferula and Leutea (Apiaceae, Scandiceae, Ferulinae) inferred from nrDNA ITS and cpDNA noncoding sequences. Taxon. 2015;64(4):770–83.
Korovin EP. Generis Ferula (Tourn.) L. monographia illustrata. Tashkent: Academiae Scientiarum UzRSS; 1947.
Safina LK, Pimenov MG. The Carpoanatomical features of the species of the genus Ferula of the Subgenus peucedanides (Umbelliferae) in Connection with the Systematics of the genus. Bot Zhurn. 1983;68:730–9.
Safna LK, Pimenov MG. Feruly Kazakhstana. Alma-ata: Nauka Kazakhskoi SSR; 1984. p. 110.
Safina LK, Pimenov MG. Carpology of the species of type subgenus of the Ferula and some problems of their systematics. Feddes Repertorium. 1990;101(3–4):135–51.
Commission SP. Pharmacopoeia of the People’s Republic of China Part I. Beijing: China Medical Science and Technology Press; 2020.
Zhang YL, Kaisa S, Wang GP. Advances in Studies of Ferula fukanensis. J Xinjiang Univ (Nat Sci Ed). 2007;24:156–9.
Yang XW. Bioactive material basis of medicinal plants in genus Ferula. Mod Chin Med. 2018;20(02):123–44.
Downie SR, Ramanath S, Katz-Downie DS, Llanas E. Molecular systematics of Apiaceae subfamily Apioideae: phylogenetic analyses of nuclear ribosomal DNA internal transcribed spacer and plastid rpoC1 intron sequences. Am J Bot. 1998;85(4):563–91.
Chen Q, Hu H, Zhang D. DNA barcoding and Phylogenomic analysis of the Genus Fritillaria in China based on complete chloroplast genomes. Front Plant Sci. 2022;13: 764255.
Zhang YM, Han LJ, Yang CW, Yin ZL, Tian X, Qian ZG, et al. Comparative chloroplast genome analysis of medicinally important Veratrum (Melanthiaceae) in China: insights into genomic characterization and phylogenetic relationships. Plant Divers. 2022;44(1):70–82.
Han H, Qiu R, Liu Y, Zhou X, Gao C, Pang Y, et al. Analysis of chloroplast genomes provides insights into the evolution of Agropyron. Front Genet. 2022;13: 832809.
Li ZX, Guo XL, Price M, Zhou SD, He XJ. Phylogenetic position of Ligusticopsis (Apiaceae, Apioideae): evidence from molecular data and carpological characters. Aob Plants. 2022;14(2):plac008.
Fan XR, Wang WC, Wagutu GK, Li W, Li XL, Chen YY. Fifteen complete chloroplast genomes of Trapa species (Trapaceae): insight into genome structure, comparative analysis and phylogenetic relationships. BMC Plant Biol. 2022;22(1):230.
Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 2007;104(49):19369–74.
Liu CK, Lei JQ, Jiang QP, Zhou SD, He XJ. The complete plastomes of seven Peucedanum plants: comparative and phylogenetic analyses for the Peucedanum genus. BMC Plant Biol. 2022;22(1):1–14.
Li HT, Yi TS, Gao LM, Ma PF, Zhang T, Yang JB, et al. Origin of angiosperms and the puzzle of the Jurassic gap. Nature Plants. 2019;5(5):461–70.
Xu SQ, Sun MY, Mei Y, Gu Y, Huang D, Wang JH. The complete chloroplast genome sequence of the medicinal plant Abrus pulchellus subsp. cantoniensis: genome structure, comparative and phylogenetic relationship analysis. J Plant Res. 2022;135(3):443–52.
Scott-Phillips TC, Laland KN, Shuker DM, Dickins TE, West SA. The niche construction perspective: a critical appraisal. Evolution. 2014;68(5):1231–43.
Hall BK, Hallgrímsson B, Strickberger MW. Strickberger’s Evolution: The Integration of Genes, Organisms and Populations. Burlington, MA: Jones and Bartlett Learning; 2008.
Montero-Mendieta S, Tan K, Christmas MJ, Olsson A, Vilà C, Wallberg A, et al. The genomic basis of adaptation to high-altitude habitats in the eastern honey bee (Apis cerana). Mol Ecol. 2019;28(4):746–60.
Liu NN, Liu HM, Yan J, Li XG, Li Y, Wang TJ, et al. Geometric morphology and population genomics provide insights into the adaptive evolution of Apis cerana in Changbai Mountain. BMC Genomics. 2022;23(1):1–16.
Zeng S, Tao Z, Han K, Yang Y, Zhao J, Liu ZL. The complete chloroplast genome sequences of six Rehmannia species. Genes. 2017;8(3):103.
Yu Y, Han Y, Peng YM, Tian ZZ, Zeng P, Zong H, et al. Comparative and phylogenetic analyses of eleven complete chloroplast genomes of Dipterocarpoideae. Chin Med. 2021;6(1):1–15.
Fan CZ, Wang GP, Qiu YJ, Zhao YQ, Zhang JZ, Song JY, et al. The complete chloroplast genome sequence of Ferula sinkiangensis K. M. Shen, a precious and endangered traditional Chinese medicine. Mitochondrial DNA B. 2021;6(6):1670–2.
Wen J, Xie DF, Price M, Ren T, Deng YQ, Gui LJ, et al. Backbone phylogeny and evolution of Apioideae (Apiaceae): New insights from phylogenomic analyses of plastome data. Mol Phylogenet Evol. 2021;161: 107183.
Li Y, Dong Y, Liu Y, Yu X, Yang M, Huang Y. Comparative analyses of Euonymus chloroplast genomes: genetic structure, screening for loci with suitable polymorphism, positive selection genes, and phylogenetic relationships within Celastrineae. Front Plant Sci. 2021;11: 593984.
Li Y, Wang TR, Kozlowski G, Liu MH, Yi LT, Song YG. Complete chloroplast genome of an endangered species Quercus litseoides, and Its comparative, evolutionary, and phylogenetic study with other Quercus section Cyclobalanopsis species. Genes. 2022;13(7):1184.
Huang Y, Li J, Yang ZR, An WL, Xie CZ, Liu SS, et al. Comprehensive analysis of complete chloroplast genome and phylogenetic aspects of ten Ficus species. BMC plant Biol. 2022;22(1):253.
Gou W, Jia SB, Price M, Guo XL, Zhou SD, He XJ. Complete plastid genome sequencing of eight species from Hansenia, Haplosphaera and Sinodielsia (Apiaceae): comparative analyses and phylogenetic implications. Plants. 2020;9(11):1523.
Ren T, Li ZX, Xie DF, Gui LJ, Peng C, Wen J, et al. Plastomes of eight Ligusticum species: characterization, genome evolution, and phylogenetic relationships. BMC Plant Biol. 2020;20(1):519.
Palmer JD, Nugent JM, Herbon LA. Unusual structure of geranium chloroplast DNA: A triplesized inverted repeat, extensive gene duplications, multiple inversions, and two repeat families. Proc Natl Acad Sci USA. 1987;84(3):769–73.
Wolfe KH, Morden CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci USA. 1992;89(22):10648–52.
Greiner S, Wang X, Rauwolf U, Silber MV, Mayer K, Meurer J, et al. The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. Sequence evaluation and plastome evolution. Nucleic Acids Res. 2008;36(7):2366–78.
Masood MS, Nishikawa T, Fukuoka SI, Njenga PK, Tsudzuki T, Kadowaki KI. The complete nucleotide sequence of wild rice (Oryza nivara) chloroplast genome: first genome wide comparative sequence analysis of wild and cultivated rice. Gene. 2004;340(1):133–9.
Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S. Plant DNA barcoding: from gene to genome. Biol Rev. 2015;90(1):157–66.
Cabras AA, Cruz RD. DNA barcoding of selected Pachyrhynchus species (Coleoptera: Curculionidae) from Mt. Apo natural park Philippines. Acta Biol Univ Daugavp. 2016;16:111–8.
Hansen S, Addison P, Benoit L, Haran JM. Barcoding pest species in a biodiversity hot-spot: The South African polyphagous broad-nosed weevils (Coleoptera, Curculionidae, Entiminae). Biodivers Data J. 2021;9: e66452.
Coissac E, Hollingsworth PM, Lavergne S, Taberlet P. From barcodes to genomes: extending the concept of DNA barcoding. Mol Ecol. 2016;25:1423–8.
Hollingsworth PM, Li DZ, van der Bank M, Twyford AD. Telling plant species apart with DNA: from barcodes to genomes. Philos Trans R Soc B. 2016;371(1702):20150338.
Liu C, Yang Z, Yang L, Yang J, Ji Y. The complete plastome of Panax stipuleanatus: Comparative and phylogenetic analyses of the genus Panax (Araliaceae). Plant Divers. 2018;40(6):265–76.
Sobreiro MB, Vieira LD, Nunes R, Novaes E, Coissac E, Silva-Junior OB, et al. Chloroplast genome assembly of Handroanthus impetiginosus: comparative analysis and molecular evolution in Bignoniaceae. Planta. 2020;252(5):91.
Pimenov MG, Georgievich M. Sosudistye Rasteniia SSSR. Leningrad: Nauka, Leningradskoe Otd-nie; 1981.
Korovin EP, Ferula L. In: Schischkin BK, editor. Flora SSSR. Izdatelstvo Akademii Nauk SSSR: Moscow, Leningrad; 1951. p. 62–142.
Ma XG, Liao CY, Chen Y, Xu HW. Ferula paeoniifolia sp. nov. (Apiaceae) from Sichuan, China. Nordic J Bot. 2019;37(3):e01751.
Wang XW, Liu M, Ru J, Wang JR, Wang YT. Morphological and anatomical characteristics of the fruit of Chinese Ferula and related taxa in Apiaceae. Acta Pataculturae Sinica. 2016;25(6):81–93.
Yang Z, Wong WSW, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22(4):1107–18.
Weiss H, Friedrich T, Hofhaus G, Preis D. The respiratory-chain NADH dehydrogenase (complex I) of mitochondria. Eur J Biochem. 1991;3:563–76.
Cramer WA, Yamashita E, Baniulis D, Hasan SS. The Cytochrome b6f Complex of Oxygenic Photosynthesis. Hoboken, NJ: John Wiley Sons, Ltd; 2011.
Wang P, Duan W, Takabayashi A, Endo T, Shikanai T, Ye JY, et al. Chloroplastic NAD(P)H dehydrogenase in tobacco leaves functions in alleviation of oxidative damage caused by temperature stress. Plant Physiol. 2006;141(2):465–74.
Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, et al. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5(9):2043–9.
Cay SB, Cinar YU, Kuralay SC, Inal B, Zararsiz G, Ciftci A, et al. Genome skimming approach reveals the gene arrangements in the chloroplast genomes of the highly endangered Crocus L. species: Crocus istanbulensis (B. Mathew) Ruksˇāns. PLoS ONE. 2022;17(6):e0269747.
Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol. 2000;54:499–518.
Xie DF, Tan JB, Yu Y, Gui LJ, Su DM, Zhou SD, et al. Insights into phylogeny, age and evolution of Allium (Amaryllidaceae) based on the whole plastome sequences. Ann Bot. 2020;125:1039–55.
Gao LZ, Liu YL, Zhang D, Li W, Gao J, Liu Y, et al. Evolution of Oryza chloroplast genomes promoted adaptation to diverse ecological habitats. Commun Biol. 2019;2:278.
Hudson GS, Mason JG. The chloroplast genes encoding subunits of the H+-ATP synthase. Photosynth Res. 1988;18:205–22.
Yin KQ, Zhang Y, Li YJ, Du FK. Different natural selection pressures on the atpF gene in evergreen Sclerophyllous and deciduous oak species: evidence from comparative analysis of the complete chloroplast genome of Quercus aquifolioides with other oak species. Int J Mol Sci. 2018;9:1042.
He S, Tan DY. Advances in studies of Ferula L. J Xinjiang Agri Univ. 2022;25(2):1–7.
Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–90.
Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45(4): e18.
Qu XJ, Moor MJ, Li DZ, Yi TS. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 2019;15(1):50.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9.
Zheng S, Poczai P, Hyvönen J, Tang J, Amiryousefi A. Chloroplot: an online program for the versatile plotting of organelle genomes. Front Genet. 2020;11: 576124.
Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–42.
Peden JF. Analysis of codon usage. PhD thesis. Nottingham: University of Nottingham; 1999.
Amiryousefi A, Hyvönen J, Poczai P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34(17):3030–1.
Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32(2):273–9.
Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2.
Zhou J, Peng H, Downie SR, Liu ZW, Gong X. A molecular phylogeny of Chinese Apiaceae subfamily Apioideae inferred from nuclear ribosomal DNA internal transcribed spacer sequences. Taxon. 2008;57(2):402–16.
Zhang D, Gao FL, Jakovlic I, Zou H, Zhang J, Li WX, Wang GT. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Eco Resour. 2020;20(1):348–55.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80.
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3.
Ronquist F, Huelsenbeck JP. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 2012;19(2):1572–4.
Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–8.
Rambaut A, Drummond A. FigTree, version 1.4.2. 2015. http://tree.bio.ed.ac.uk/software/fgtree/. Accessed 4 Oct 2021.
Yang Z, Dos RM. Statistical properties of the branch-site test of positive selection. Mol Biol Evol. 2011;28(3):1217–28.
Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80.
Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–91.
Gao FL, Chen CJ, Arab DA, Du ZG, He YH, Ho SYW. EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol Evol. 2019;9(7):3891–8.
Lan Y, Sun J, Tian R, Bartlett DH, Li R, Wong YH, et al. Molecular adaptation in the world’s deepest-living animal: insights from transcriptome sequencing of the hadal amphipod Hirondellea gigas. Mol Ecol. 2017;26(14):3732–43.
Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–7.
Altinordu F, Peruzzi L, Yu Y, He XJ. A tool for the analysis of chromosomes: KaryoType. Taxon. 2016;65(3):586–92.
Kljuykov EV, Liu M, Ostroumova TA, Pimenov MG, Tilney PM, Wyk BV, Staden JV. Towards a standardised terminology for taxonomically important morphological characters in the Umbelliferae. S Afr J Bot. 2004;70(3):488–96.
Acknowledgements
We acknowledge Yan Yu, Xianlin Guo, and Jun Wen for their help in collecting samples. We are grateful to Dengfeng Xie and Chang Peng for their help in software use.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 32170209, 32070221, 31872647). The funders were not involved in the design of the study, collection, analysis and interpretation of data, and manuscript preparation.
Author information
Authors and Affiliations
Contributions
S-DZ and X-JH designed the work. H-HQ, JC, and R-XZ collected the data; H-HQ and C-KL analyzed the data; H-HQ wrote the manuscript; MP, S-DZ, and X-JH revised the manuscript. All authors gave final approval of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The collection of all samples completely complies with national and local legislation permission. Plant samples used in the study were not included in the list of national key protected plants and were not collected from the national park or nature reserve when we collected them. According to national and local legislation, no specific permission was required for collecting these plants when we collected them.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Fig. S1. Phylogenetic tree reconstruction of the 62 taxa inferred from Bayesian inference (BI) analyses and Maximum likelihood (ML) based on nuclear internaltranscribed spacer (ITS) sequences. Numbers indicate Bayesian posterior probabilities (PP) and mamxium likelihood bootstrap values (BS), and (*) indicates maximumsupport in both two analysis, and (-) indicates mamxium likelihood bootstrap values (BS) less than 50 in Maximum likelihood (ML) analyses.
Additional file 2:
Fig. S2. Morphological features of mericarps of twelve species. (A) Dorsal side views ofmericarps. (B) commissural side views of mericarps. (C) transverse sections. Scale bars:A=1.0 mm;B=1.0 mm; C=0.5 mm.
Additional file 3:
Fig. S3. Partial alignment of amino acids sequences in another nine positively selected genes. (A-J): atpB, ndhA, ndhC, ndhI, ndhJ, and psbK, rpl20, rpoB, and rpoC1. The red blocks indicate the amino acids in twenty-two species with a high BEB posterior probability.
Additional file 4:
Table S1. The list of gene content in the twenty-two plastomes.
Additional file 5:
Table S2. Simple sequence repeats (SSRs) distribution in the twenty-two plastomes.
Additional file 6:
Table S3. The distribution of repeat sequences in the 22 plastomes.
Additional file 7:
Table S4. Codon usage and relative synonymous codon usage (RSCU) values of protein-coding genes of the 22 plastomes.
Additional file 8:
Table S5. The nrITS Genbank accession numbers of all species used in phylogenetic analysis.
Additional file 9:
Table S6. Synopsis of the morphological information from the 22 species.
Additional file 10:
Table S7. The result of positive selection analysis based on the branch-site model.
Additional file 11:
Table S8. Information for sample collections and specimen vouchers of the 22 species.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Qin, HH., Cai, J., Liu, CK. et al. The plastid genome of twenty-two species from Ferula, Talassia, and Soranthus: comparative analysis, phylogenetic implications, and adaptive evolution. BMC Plant Biol 23, 9 (2023). https://doi.org/10.1186/s12870-022-04027-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-022-04027-4