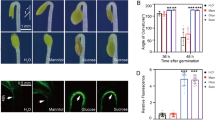
Abstract
Background
Branch angle is a pivotal component of tea plant architecture. Tea plant architecture not only affects tea quality and yield but also influences the efficiency of automatic tea plant pruning. However, the molecular mechanism controlling the branch angle, which is an important aspect of plant architecture, is poorly understood in tea plants.
Results
In the present study, three CsLAZY genes were identified from tea plant genome data through sequence homology analysis. Phylogenetic tree displayed that the CsLAZY genes had high sequence similarity with LAZY genes from other plant species, especially those in woody plants. The expression patterns of the three CsLAZYs were surveyed in eight tissues. We further verified the expression levels of the key CsLAZY1 transcript in different tissues among eight tea cultivars and found that CsLAZY1 was highly expressed in stem. Subcellular localization analysis showed that the CsLAZY1 protein was localized in the plasma membrane. CsLAZY1 was transferred into Arabidopsis thaliana to investigate its potential role in regulating shoot development. Remarkably, the CsLAZY1 overexpressed plants responded more effectively than the wild-type plants to a gravity inversion treatment under light and dark conditions. The results indicate that CsLAZY1 plays an important role in regulating shoot gravitropism in tea plants.
Conclusions
The results provide important evidence for understanding the functions of CsLAZY1 in regulating shoot gravitropism and influencing the stem branch angle in tea plants. This report identifies CsLAZY1 as a promising gene resource for the improvement of tea plant architecture.
Similar content being viewed by others
Background
The tea plant (Camellia sinensis) is an economic crop of great importance. Its leaves can be used to produce most traditional caffeinated teas, which are the second most popular beverage worldwide [1, 2]. The productivity of tea plants is greatly affected by the architecture of tea plants. A well-designed tree architecture should minimize competition with adjacent crops for environmental resources, such as light. In densely planted stands, a relatively wide branch angle may help the plant escape some diseases by decreasing humidity, but it makes the plant occupy more space and increases the extent of shade [3]; therefore, optimal tree architecture may contribute to increasing the yield and yield stability of crops [4]. Tea plant architecture is affected by geometric and environmental factors as well as the tea estate plantation elevation; besides, plucking patterns including manual plucking, shear harvesting and monsoon seasons also affect tea plant architecture [5]. According to their branch angle degree, tea plants are classified into three plant architecture types: open, half open, and erect.
Plant architecture is significantly associated with plant hormones, including gibberellic acid (GA), auxin, cytokinin, and strigolactones (SLs). GA is thought to promote upward growth and inhibit bending, and it is highly likely to be responsible for the weeping trait [6, 7]. Previous studies also suggested that genes associated with auxin and ethylene probably play crucial roles in shoot elongation [8, 9]. Cytokinin was identified as an important phytohormone that regulates plant shoot branching, it is synthesized in the roots and then transported throughout the plant for the development of the entire plant [10]. SLs are a group of newly identified plant hormones that are essential for regulating the shoot branch/tiller angle; they can inhibit auxin biosynthesis and attenuate rice shoot gravitropism, mainly by decreasing the local indoleacetic acid (IAA) content [11].
Many environmental signals, including light and gravity, can influence plant architecture [6, 12]. The branch angle is an important factor in determining plant structure and is regulated by specific genes. To date, many genes and transcription factors associated with branch angle have been identified. For instance, overexpression of OsPIN2 leads to increased tiller numbers, and altering OsPIN2 expression through genetic transformation can be directly used to modify rice architecture [13]. OsTAC1 controls tiller angle in rice [3], and changes in TAC1 have since been linked to upright tiller or branch angles in other plant species, including Arabidopsis [14], rice [15], poplar [4], peach [16, 17], and apple [18]. OsTAC4 participates in the regulation of rice tiller angle, and influences the endogenous auxin content, ultimately leading to reduced gravitropism and a tiller-spreading phenotype [19, 20].
In many plant species, LAZY1 plays an important role in regulating the plant branch angle. For example, the rice lazy1 mutant displays a tiller-spreading phenotype because gravitropism is reduced [21]. In Arabidopsis, a total of six LAZY genes have been identified, and mutating AtLAZY1 caused a large change in branch angle while the primary inflorescence stem remained vertical [22]. The other lazy mutations reversed the growth angle of lateral branches and roots, indicating that LAZY genes regulate the direction of polar auxin transport in response to gravity through the control of asymmetric PIN3 expression in the root cap columella [23]. In apple and poplar, evidence has shown that LAZY genes affected the vascular tissues of transgenic plants, thus modifying the branch angle [4, 16].
Although LAZY genes have been indicated to play an important role in modifying branch angle in a variety of plant species, the potential function of homologous genes in tea plants (Camellia sinensis) is still unknown. Branch angle is an important trait of tea plants that can influence the plant architecture as well as the mechanical harvesting of tea leaves. In this study, three LAZY genes were identified in the tea plant, and their expression levels in distinct tissues were characterized. CsLAZY1 was expressed predominately in stem and was located in the plasma membrane. Plants that overexpressed CsLAZY1 responded more effectively than the wild-type plants to gravity processing. Our results identify new candidate genes that can be used to breed new tea varieties with ideal plant architecture.
Results
Identification, conserved domain and sequence feature analysis of CsLAZYs
A total of six LAZY genes were identified in Arabidopsis thaliana [22]. Subsequently, these six AtLAZY genes were used as queries in Basic Local Alignment Search Tool (BLAST) analysis against the tea plant genome (http://tpia.teaplant.org/Blast.html) [24]. Initially, a total of 15 candidate unique genes were obtained for tea plants, and multiple sequence alignments of all LAZY genes were performed among tea plant, Arabidopsis and rice (data not shown). The results showed that only 3 unique genes contained regions of conserved sequence V that possess an ethylene-responsive element-binding factor-associated amphiphilic repression (EAR) motif (LxLxL) (Figure S1) and that this is an indispensable conserved domain of LAZY [22, 25]. Thereafter, the three obtained genes were referred to as CsLAZY1 (CSS025254), CsLAZY2 (CSS049138) and CsLAZY3 (CSS020288), and they were located in different scaffolds (Table 1). Their amino acid lengths were 399 aa (CsLAZY1), 367 aa (CsLAZY2) and 251 aa (CsLAZY3), respectively. Furthermore, the molecular weights (Mw) of CsLAZY1 to CsLAZY3 were 44.2, 41.2 and 29.0, and their isoelectric points (pI) were 6.55, 6.18 and 6.47, respectively (Table 1).
Evolution and phylogenetic analysis of LAZY genes
Previous studies and the existence of numerous fully sequenced plant genomes have made it possible to perform a comparative genomic analysis of LAZY genes across a broad range of plant species. LAZY genes have been identified as playing similar roles in many plant species, so we performed iterative BLAST searches to determine the phylogeny of LAZY1 genes. LAZY1 genes were identified from 21 distinct plant species, and a homology analysis of LAZY1 among algae, lowland species, monocots, and dicots provided further insight into the evolutionary processes of this gene family (Fig. 1A). Phylogenetic analyses showed that these LAZY1 genes were highly conserved among algae, monocots and dicots and that LAZY1 evolved from primitive organisms despite their overall relatively low sequence similarities. It was obvious that CsLAZY1 had higher sequence similarity with LAZY1 genes from other woody plants, including kiwifruit, grape, poplar and peach, indicating that CsLAZY1 was more highly conserved in the process of evolution within woody plants (Fig. 1A).
Phylogenetic relationships of LAZY genes from various plant species. A Phylogenetic tree of LAZY1 proteins identified from various plant species. Plant classifications are coded with different colours as shown in the legend, and the plant species and sequence accession IDs are listed. B Phylogenetic tree of the LAZY family protein sequences from eight plant species (Os: Oryza sativa, At: Arabidopsis thaliana, Sl: Solanum lycopersicon, Pt: Populus tomentosa, Vv: Vitis vinifera, Pp: Prunus persica, Ac: Actinidia chinensis, and Cs: Camellia sinensis). The phylogenetic trees were generated by MEGA 6.0 with the neighbor-joining algorithm with bootstrap analysis for 1000 replicates, and a p-distance model was used to ensure that the divergent domains could contribute to the topology of the NJ tree
To further understand their sequence homology and potential biological functions, we analyzed the evolutionary tree containing all LAZY gene family members from eight plant species, including Oryza sativa, Arabidopsis thaliana, Solanum lycopersicon, Populus tomentosa, Vitis vinifera, Prunus persica, Actinidia chinensis and Camellia sinensis. The complete LAZY gene families, including 32 members, were used for phylogenetic analysis. It was showed that these LAZY genes were mainly classified into three clades: class I, class II and class III (Fig. 1B). CsLAZY1 and CsLAZY2 were grouped into class I, and they both had high sequence similarities with the protein sequences of the AcLAZY1, AcLAZY2 and VvLAZY1 genes. CsLAZY3 was grouped into class II and showed high sequence similarity with the VvLAZY3, PpLAZY4, AtLAZY2 and AtLAZY4 genes.
Analysis of gene structures, cis-elements in promoters and tissue expression patterns of the three CsLAZY genes
To investigate the structural diversity of CsLAZY genes, we compared the exon/intron organization in the coding sequences of each CsLAZY gene, demonstrating that CsLAZY1 to CsLAZY3 contained 5, 5, and 3 exons, respectively (Fig. 2A). In terms of intron and exon length, CsLAZY1 was the longest while CsLAZY3 was the shortest. The coding sequence of CsLAZY1 was cloned and sequenced, demonstrating that the cloned cDNA was totally consistent with the genomic reference sequence.
Analysis of gene structures, cis-elements in promoters and tissue expression patterns of the three CsLAZY genes. A Gene structures of the three CsLAZY genes from tea plants. B Prediction of cis-elements in the promoters of the three CsLAZY genes. C Expression patterns of the three CsLAZY genes in eight tissues
To explore the potential differences in non-coding regions of the CsLAZYs, a 2-kb flanking sequence upstream of the translation start codon was obtained, and many putative cis-regulatory elements in the promoter were identified using the PLACE and PlantCARE databases (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Light sensitive cis-elements, including Box 4, TCT-motif, ATC-motif, ATCT-motif, G-box, I-box, chs-CMA1a, MRE, and ACE, accounted for the largest proportion of all elements (Fig. 2B). CsLAZY1 contained two kinds of key light-sensitive cis-elements, including two G boxes and one ACE; CsLAZY2 contained seven kinds of light-sensitive cis-elements except for the ACE and TCT-motif elements. Seven kinds of hormone-sensitive cis-elements were obtained, including CGTCA-motif, GARE-motif, TCA-element, ERE, ABRE, TATC-box, and TGACG-motif. Notably, the promoter region of CsLAZY1 contained five kinds of hormone-sensitive cis-elements, including TGACG-motif, ABRE, TCA-element, GARE-motif, and CGTCA-motif (Fig. 2B). These cis-elements are MeJA-, GA-, SA- and ABA-responsive elements, implying that CsLAZY1 may play an important role in tea plant responses to hormones.
To understand the potential role of CsLAZYs in tea plants, we downloaded RNA-Seq data for eight tissues from the tea plant genome database. The data showed that the expression levels of the three CsLAZY genes were obviously specific to various tissues (Fig. 2C). For instance, CsLAZY1 had the highest expression level in stem, followed by in bud and leaf, while it was basically not expressed in fruit or root. In comparison, CsLAZY2 was expressed mainly in flower, and CsLAZY3 showed the highest expression level in leaf. Because stem bending is one of the main causes of branch angle development, CsLAZY1 probably plays a vital role in regulating the branch angle of tea plants.
Expression patterns of CsLAZY1 in tissues among different tea varieties
To further verify the tissue expression pattern of CsLAZY1, we examined the tissue expression level of CsLAZY1 in different tea varieties. A total of eight tea varieties with different branch angles, including four open-type varieties (Benshan, Foshou, Yaoshanxiulv, and Tieguanyin) and four erect-type varieties (Echa 5, Fuzao 2, Longjingchangye, and Zhenghedabaicha), were analysed. The expression level of the CsLAZY1 transcript varied significantly among the four tissues (leaf, bud, root and stem) (Fig. 3). It was showed that CsLAZY1 transcript was not detected in root of the eight tea varieties, and it had the highest expression level in stem, followed by that in leaf. Notably, no obvious difference in the tissue expression pattern was observed between the two different type tea plants. Unexpectedly, in the Tieguanyin cultivar, CsLAZY1 had the highest expression level in leaf, followed by that in stem.
Expression patterns of CsLAZY1 in four tissues of tea plants of two architecture types. A Tissue expression level of CsLAZY1 in four open-type tea cultivars (Benshan, Foshou, Yaoshanxiulv, and Tieguanyin). B Tissue expression level of CsLAZY1 in four erect-type tea cultivars (Echa 5, Fuzao 2, Longjingchangye, and Zhenghedabaicha). The expression levels of CsLAZY1 in eight cultivars were analyzed by qRT-PCR. Bars indicate the means ± SDs (n = 3) of three biological replicates. Different letters above the bars denote significantly different levels of expression determined by Duncan’s multiple range test (p < 0.05)
Subcellular localization of CsLAZY1 protein
In Arabidopsis, AtLAZY1, which contributes to the regulation of branch angles, is localized in the plasma membrane and nucleus [22]. To obtain insight into the molecular function of the CsLAZY1 protein, we constructed CsLAZY1-GFP and Pk7WGF2 35S-GFP fusion protein expression vectors to examine its subcellular localization. Transient expression in Arabidopsis protoplasts indicated that the CsLAZY1 protein was localized in the plasma membrane (Fig. 4A). In addition, the plasmid of CsLAZY1-GFP was transferred into Agrobacterium to infect Nicotiana benthamiana leaves and obtained identical results, namely, the CsLAZY1 protein was localized in the plasma membrane (Fig. 4B).
Overexpression of CsLAZY1 in Arabidopsis
To further investigate the role of CsLAZY1 in shoot gravitropism, we transferred CsLAZY1 into Arabidopsis thaliana. The expression of CsLAZY1 was detected using a real-time PCR assay in CsLAZY1-overexpression (OE) plants but not in wild-type (WT) plants, and the three OE lines were named OELAZY1-11, OELAZY1-20, and OELAZY1-24 (Fig. 5A). Subsequently, gravitropism assays recorded with time-lapse imaging were implemented to survey the responses of the WT and three OE lines to reorientation. All seedlings with a main stem of 5–10 cm were subjected to 90° inverted gravity processing. In the light, images were collected by computer-controlled cameras after 0, 30, 60, 90 and 120 min of inversion (Fig. 5B), and the angle of the hypocotyls was measured from the images. The OE plants clearly bent upward slightly at 30 min, while no bending was observed in the WT plants. After 90 min of inversion, the OE plants reached their maximum bending angles, while the WT plants bent upward slightly (Fig. 5B). After 30 min treatment, a significant difference in bending angle was observed between the OE and WT plants (Fig. 5C).
Gravitropic responses in wild-type (WT) and CsLAZY1 overexpression (OE) A.thaliana in the light. A Expression level of CsLAZY1 in WT and OE plants, the AtACTIN gene was used as the control. B WT and OE plants were treated at 90° inverted for 0, 30, 60, 90 and 120 min. Scale bar = 1 cm. C Bending angles of WT and OE plants. Bars indicate the means ± SDs (n = 3), and the asterisk above the bars denotes a significantly difference in bending angle determined by Duncan’s multiple range test (*P < 0.05, **P < 0.01)
In the dark, images were collected after 0, 30, 60, 90, 120, 150 and 180 min of inversion, and the angle of the hypocotyls was measured from the images. Both the WT and OE plants bent upward later in the dark than their corresponding plants in the light, implying that the light-sensitive cis-elements in the promoter may be associated with the function of the CsLAZY1 gene (Fig. 6A). In the dark, no bending angle was observed in the WT and OE plants after 30 min. The OE plants bent upward slightly after 60 min, while the WT plants bent upward slightly after 90 min (Fig. 6A). After 90 min, a significant difference in the bending angle was also observed between OE plants and WT plants (Fig. 6B). Consistent with the expression patterns, the evidence indicates that CsLAZY1 may play a vital role in the response to gravitropism in stem of tea plants.
Gravitropic responses in wild-type (WT) and CsLAZY1 overexpression (OE) A.thaliana in the dark. A WT and OE plants were subjected to 90°inversion for 0, 30, 60, 90, 120, 150 and 180 min. Scale bar = 1 cm. B Bending angles of WT and OE plants. Bars indicate the means ± SDs (n = 3), and the asterisk above the bars denotes a significantly difference in bending angle determined by Duncan’s multiple range test (*P < 0.05, **P < 0.01)
Discussion
Agricultural productivity is affected by various environmental factors that can result in lower crop yields. Plant architecture is one of the major constraints on crop yield, and branch angle plays a vital role in the formation of plant architecture [26]. Accumulated evidence indicates that LAZY1 plays a crucial role in plant responses to gravitropism and then regulates branch angles [22, 23, 27]. For tea plants, the branch angle is a critical factor that can greatly influence the productivity and efficiency of mechanical plucking. Nevertheless, the molecular mechanism controlling the branch angle of tea plants has scarcely been understood until now. In the present study, we identified three CsLAZY genes in tea plants and analyzed their phylogenetic relationships, gene structures and tissue-specific expression patterns. Subsequently, the biological function of the candidate gene CsLAZY1 was investigated, including its subcellular localization, tissue-specific expression patterns in different tea varieties, and heterologous overexpression analysis were performed, which revealed obvious differential responses to gravity.
The three CsLAZY genes exhibited different tissue expression patterns (Fig. 2C). CsLAZY3 was distinguished from the other two CsLAZY genes by clustering into a different subclade, which had a high expression level specifically in the second leaf. CsLAZY2 showed high sequence similarity with CsLAZY1 and had the highest expression level in flower, indicating that CsLAZY2 may play an important role in the development of flower. In comparison, CsLAZY1 had a far higher expression level in stem than in the other tissues (Fig. 2C), and similar tissue-specific expression patterns were observed in several other woody plants, such as poplar [4], peach [16] and apple [18]. To identify whether CsLAZY1 exhibits different tissue-specific expression patterns in different tea varieties, we examined its expression in two types of tea plants, open-type plants (Benshan, Foshou, Yaoshanxiulv and Tieguanyin) and erect-type plants (Echa 5, Fuzao 2, Longjingchangye, Zhenghedabaicha), categorized based on their branch angle. No obvious difference in the expression pattern was observed among the different tea varieties (Fig. 3). In poplar, the transcript level of PtLAZY1 showed similar expression profiles in different tissues, and no significant difference in tissue-specific expression profiles was observed between narrow-crown and broad-crown poplars [4]. In Arabidopsis, disruption of AtLAZY1 expression can cause the weakening of gravitropic response, and lead to branch angles to become larger [22, 28], thereby the results accounted for the similar expression patterns of CsLAZY1 in different tea varieties (Fig. 3). In addition, subcellular localization of CsLAZY1 was analyzed, demonstrating that the CsLAZY1 protein is located in the plasma membrane, which is consistent with previous studies in Arabidopsis [22, 23, 28]. Additionally, several other AtLAZY genes from Arabidopsis thaliana are localized in the plasma membrane [22, 28]. Unexpectedly, it has been confirmed that OsLAZY1 is located in the nucleus and that the nuclear localization of OsLAZY1 is essential for its function in rice [29, 30]. In fact, the OsBRXL4 protein interacts with OsLAZY1 at the plasma membrane, and their interaction determines the nuclear localization of OsLAZY1, thereby OsBRXL4 regulates shoot gravitropism and the rice tiller angle by affecting the nuclear localization of LAZY1 [30]. Thereby, the majority of LAZY1 from different plant species are localized in the plasma membrane, only a little difference is existed of OsLAZY1 localization in rice.
LAZY genes that share common domain sequences usually have a common origin and thus have similar functions [22, 25]. The homology analysis of LAZY-like genes in poplar and the functional investigation of PzLAZY suggested that PzLAZY may be involved in altering branch angle [4]. Among LAZY genes, an EAR motif is located in conserved region V, which plays a role in controlling hormonal systems and is related to the gravitropic response of plants. For instance, the binding of TOPLESS proteins to the EAR motif of AUX/IAA proteins can repress auxin-responsive genes [30, 31]. The three CsLAZY genes obtained from tea plant shared five limited sequence regions and were highly conserved in region V (Figure S1), indicating that they may play a role in the development of tea plants. Phylogenetic analysis showed that CsLAZY1 had high sequence similarity with LAZY1 from other woody plants, including those in grape, poplar and peach (Fig. 1A). This result indicates that LAZY1 experienced greater conservation in the process of its evolution within woody plants.
Furthermore, we obtained heterologous OE Arabidopsis plants, while no difference in phenotypes was observed between OE plants and wild type plants, which are also consistent with the results of a previous study [32]. Notably, the phenotype of the OE plants was obviously distinct from that of the wild types in response to gravity (Figs. 5 and 6). It was speculated that CsLAZY1 may play roles in altering branch angle by acting on the transportation of phytohormones. In Arabidopsis thaliana, six AtLAZY genes participate in early gravity signalling for shoot gravitropism [22, 23, 28]. AtLAZY1 leads to the asymmetric distribution of auxin, thus altering the rice tiller angle; AtLAZY1 also mediates gravity signalling in statocytes downstream of amyloplast displacement, leading to the development of asymmetric auxin distribution in gravity responsive organs [33,34,35]. In rice, OsLAZY1 controls the tiller angle by regulating shoot gravitropism through the inhibition of polar auxin transport [21, 29, 31]. We also analyzed the cis-elements in the promoters of CsLAZYs and found that MeJA, GA, SA and three ABA hormone-responsive elements existed in the promoter of CsLAZY1 (Fig. 2B). Collectively, CsLAZY1 may play roles in the response to gravitropism and alter the branch angle by acting on the transportation of phytohormones.
Conclusions
In this study, we identified three LAZY genes from tea plants and named them CsLAZY1 to CsLAZY3 based on their sequence similarity with the LAZY genes from Arabidopsis. The CsLAZY1 to CsLAZY3 genes showed distinct expression patterns in eight different tissues, and had the highest expression levels in stem, flower and leaf, respectively. Tissue-specific expression of CsLAZY1 was also identified in different tea varieties that exhibited distinct branch angles, confirming that CsLAZY1 had the highest expression level in the stem. The CsLAZY1 protein was localized in the plasma membrane based on a subcellular localization analysis. The overexpression of CsLAZY1 in Arabidopsis thaliana showed that overexpress plants responded more effectively than the wild type plants to gravity processing under light and dark conditions. The results indicated that CsLAZY1 plays an important role in regulating shoot gravitropism and affecting the branch angle in tea plants.
Materials and methods
Plant materials
A total of nine five-year-old tea plant cultivars (Camellia sinensis var. ‘Shuchazao’, ‘Benshan’, ‘Foshou’, ‘Yaoshanxiulv’, ‘Tieguanyin’, ‘Echa 5’, ‘Fuzao 2’, ‘Longjingchangye’, and ‘Zhenghedabaicha’) from the Tea Plant Cultivar and Germplasm Resource Garden in Guohe town (Anhui Agricultural University) were used for the collection of various tissues (the second leaf, apical bud, young root, and young stem). All tissues were sampled according to the demands of each experiment, and they were immediately frozen in liquid nitrogen and stored at -80 °C until utilization.
Identification and molecular cloning of CsLAZYs
The nucleotide and deduced amino acid sequences of 6 AtLAZY genes from Arabidopsis were obtained from TAIR (The Arabidopsis Information Resource) database (https://www.arabidopsis.org/). A genome-wide search of 6 AtLAZY genes was carried out using Basic Local Alignment Search Tool (BLAST) analysis with the 6 AtLAZY genes used as queries against the tea plant genome (http://tpia.teaplant.org/Blast.html) [36]. All nonredundant protein sequences were compared with AtLAZYs and OsLAZYs, and the genes possessing pivotal conserved domains were selected. To verify the coding regions of CsLAZYs, gene-specific primers were designed for the amplification of CsLAZY genes with cDNA templates from the young leaves of Camellia sinensis var. ‘Shuchazao’.
Phylogenetic analysis of LAZY genes
Gene sequences from rice, tomato, apple, peach, poplar, and grape were obtained from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html), and gene sequences from kiwifruit were obtained from the Kiwifruit Genome Database (http://kiwifruitgenome.org/). Multiple sequence alignment of LAZY protein sequences was performed using the ClustalW program. Phylogenetic trees were generated based on the full-length amino acid sequences by MEGA 6.0 with the neighbor-joining (NJ) algorithm. Bootstrap analysis with 1000 replicates was used to evaluate the significance of the nodes, and a p-distance model was used to ensure that the divergent domains could contribute to the topology of the NJ tree.
Gene structure and promoter structure analysis of CsLAZY genes
Alignment of amino acid sequences was performed using T-COFFEE (http://tcoffee.org/) [37]. Based on the gene structure display server (GSDS 2.0, http://gsds.cbi.pku.edu.cn/index.php) program, we determined the exon/intron organization of CsLAZYs by comparing the coding sequences to their corresponding genomic sequences. To investigate cis-elements in the promoter sequences of the CsLAZY family genes, a 2 kb flanking sequence upstream of the translation start codon were isolated, and the PLACE and PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) were used to identify cis-regulatory elements in the promoters.
RNA extraction and real-time quantitative PCR analysis
Total RNA was extracted from tea leaves using the RNAprep Pure Plant Kit (cat DP432, Tiangen, Beijing) according to the manufacturer’s protocol. The quality and quantity of each RNA extract were detected using agarose gel electrophoresis and a Nanodrop 2000 (Thermo Fisher Scientific, US). First-strand cDNA was synthesized from total RNA using the PrimeScript RT Reagent Kit (cat RR036A, Takara, Japan) following the manufacturer’s protocol. A 10 μl total reaction volume, including 5 μl TB Green Enzyme, 1.2 μl cDNA, 3.2 μl water and 0.6 μl primer, was used for qRT-PCR, and the process was performed as described previously in detail [38, 39]. The CsGAPDH gene was selected as the internal control, and the relative gene expression values were analyzed using the 2−△Ct method [40]. All reactions were run with triplicate technical replicates for each sample, and three biological replicates were performed. The relevant primers are listed in Additional file 1, and the full length nucleotide and protein sequences of CsLAZY1 are listed in Additional file 2.
Subcellular localization of CsLAZY1 protein
The CsLAZY1 plasmid fused with GFP was constructed by Gateway Technology, and the ORFs of CsLAZY1 with a 25 bp vector adapter were amplified by RT-PCR. PCR products were inserted into the pDONR207 vector by BP clone enzyme mix, and then transferred into PK7WGF2 through LR reactions. The resultant empty vector and pk7WGF2-LAZY1 plasmids were transformed into Arabidopsis protoplast cells, and the protoplasts were examined after transformation overnight. Besides, the resultant vectors were also transformed into Agrobacterium GV3101 competent cells, and the construct and empty vector were transiently introduced into Nicotiana benthamiana leaves by injection. The tobacco leaves were held for 48 h at 25 °C in the dark after transformation, and the tobacco leaves and protoplasts were examined using an Olympus FV1000 confocal microscope (Olympus, Japan).
Arabidopsis transformation and branch angle measurements
The full-length cDNA sequences were ligated into PBI121 driven by CaMV35S and then transferred into Agrobacterium strain GV3101. Arabidopsis (Col) was transformed using the floral dip method as described previously [41]. Transformed plants were selected on the basis of their resistance to kanamycin, and 4-week-old homozygous T3 plants were used for further experiments. Three transgenic lines were subjected to 90° inverted gravity processing for analysis of the bending angle.
The plant response to the gravity angle was determined as follows: the stem was initially positioned in a standard alignment to allow angle changes at each time point to be detected, a tangent line was drawn along the initial stem and along the curved stem, and the angle between the two tangent lines was measured using IMAGEJ software.
Availability of data and materials
The data sets supporting the results of this article are available at the NCBI SRA database (https://www.ncbi.nlm.nih.gov/) under project accession number MW848488.
Abbreviations
- OE:
-
Overexpression
- WT:
-
Wild type
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- IAA:
-
Indoleacetic acid
- ABA:
-
Abscisic acid
- SA:
-
Salicylic acid
- GA:
-
Gibberellin
- MeJA:
-
Methyl Jasmonate
- SLs:
-
Strigolactones
- BRXL4:
-
Brevis Radix-like 4
- PIN:
-
PIN-FORMED
References
Yang TY, Lu X, Wang Y, Xie YX, Ma JZ, Cheng XM, Xia EH, Wan XC, Zhang ZL. HAK/KUP/KT family potassium transporter genes are involved in potassium deficiency and stress responses in tea plants (Camellia sinensis L.): expression and functional analysis. BMC Genomics. 2020;21:556.
Gong AD, Lian SB, Wu NN, Zhou YJ, Zhao SQ, Zhang LM, Cheng L, Yuan HY. Integrated transcriptomics and metabolomics analysis of catechins, caffeine and theanine biosynthesis in tea plant (Camellia sinensis) over the course of seasons. BMC Plant Biol. 2020;20:294.
Yu BS, Lin ZW, Li HX, Li XJ, Li JY, Wang YH, Zhang X, Zhu ZF, Zhai WX, Wang XK, Xie DX, Sun CQ. TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 2007;52:891–8.
Xu D, Qi X, Li JH, Han XJ, Wang JN, Jiang YZ, Tian YT, Wang YW. PzTAC and PzLAZY from a narrow-crown poplar contribute to regulation of branch angles. Plant Physiol Biochem. 2017;118:571–8.
Ponmurugan P, Gnanamangai BM, Manjukarunambika K. Architectural effect of different tea clones on the development of blister blight disease. J Appl Bot Food Qual. 2019;92:7–14.
Li SZ, Zheng TC, Zhuo XK, Li ZJ, Li LL, Li P, Qiu LK, Pan HT, Wang J, Cheng TR, Zhang QX. Transcriptome profiles reveal that gibberellin-related genes regulate weeping traits in crape myrtle. Hortic Res. 2020;7:54.
Yoshida M, Yamamoto H, Okuyama T, Nakamura T. Negative gravitropism and growth stress in GA3-treated branches of Prunus spachiana Kitamura f. spachiana cv. Plenarosea. J Wood Sci. 1999;45:368–72.
Liu JJ, Zeng YF, Yan PC, He CY, Zhang JG. Transcriptional and hormonal regulation of weeping trait in Salix matsudana. Genes. 2017;8:359.
Dun EA, Ferguson BJ, Beveridge CA. Apical dominance and shoot branching. Divergent opinions or divergent mechanisms? Plant Physiol. 2006;142:812–9.
Domagalska MA, Leyser O. Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol. 2011;12:211–21.
Sang DJ, Chen DQ, Liu GF, Liang Y, Huang LZ, Meng XB, Chu JF, Sun XH, Dong GJ, Yuan YD, Qian Q, Li JY, Wang YH. Strigolactones regulate rice tiller angle by attenuating shoot gravitropism through inhibiting auxin biosynthesis. Proc Natl Acad Sci U S A. 2014;111:11199–204.
Busov VB, Brunner AM, Strauss SH. Genes for control of plant stature and form. New Phytol. 2008;177:589–607.
Chen YN, Fan XR, Song WJ, Zhang YL, Xu GH. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol J. 2012;10:139–49.
Guseman JM, Webb K, Srinivasan C, Dardick C. DRO1 influences root system architecture in Arabidopsis and Prunus species. Plant J. 2017;89:1093–105.
Jiang JH, Tan LB, Zhu ZF, Fu YC, Liu FX, Cai HW, Sun CQ. Molecular evolution of the TAC1 gene from rice (Oryza sativa L.). J Genet Genomics. 2012;39:551–60.
Dardick C, Callahan A, Horn R, Ruiz KB, Zhebentyayeva T, Hollender C, Whitaker M, Abbott A, Scorza R. PpeTAC1 promotes the horizontal growth of branches in peach trees and is a member of a functionally conserved gene family found in diverse plants species. Plant J. 2013;75:618–30.
Hollender CA, Waite JM, Tabb A, Raines D, Chinnithambi S, Dardick C. Alteration of TAC1 expression in Prunus species leads to pleiotropic shoot phenotypes. Hortic Res. 2018;5:26.
Wang LM, Cai WB, Du CH, Fu Y, Xie X, Zhu YD. The isolation of the IGT family genes in Malus × domestica and their expressions in four idiotype apple cultivars. Tree Genet Genomes. 2018;14:46.
Li H, Sun HY, Jiang JH, Sun XY, Tan LB, Sun CQ. TAC4 controls tiller angle by regulating the endogenous auxin content and distribution in rice. Plant Biotechnol J. 2021;19:64–73.
Zeng YX, Chen Y, Ji ZJ, Liang Y, Yang CD. Twenty-four alleles at twelve quantitative trait loci act additively to control tiller angle in cultivated rice. Plant Growth Regul. 2019;88:195–203.
Li PJ, Wang YH, Qian Q, Fu ZM, Wang M, Zeng DL, Li BH, Wang XJ, Li JY. LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 2007;17:402–10.
Yoshihara T, Spalding EP, Iino M. AtLAZY1 is a signaling component required for gravitropism of the Arabidopsis thaliana inflorescence. Plant J. 2013;74:267–79.
Yoshihara T, Spalding EP. LAZY genes mediate the effects of gravity on auxin gradients and plant architecture. Plant Physiol. 2017;175:959–69.
Wei CL, Yang H, Wang SB, Zhao J, Liu C, Gao LP, Xia EH, Lu Y, Tai YL, She GB, Sun J, Cao HS, Tong W, Gao Q, Li YY, Deng WW, Jiang XL, Wang WZ, Chen Q, Zhang SH, Li HJ, Wu JL, Wang P, Li PH, Shi CY, Zheng FY, Jian JB, Huang B, Shan D, Shi MM, Fang CB, Yue Y, Li FD, Li DX, Wei S, Han B, Jiang CJ, Yin Y, Xia T, Zhang ZZ, Bennetzen JL, Zhao SC, Wan XC. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc Natl Acad Sci U S A. 2018;115:E4151–8.
Kagale S, Links MG, Rozwadowski K. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 2010;152:1109–34.
Bai F, Reinheimer R, Durantini D, Kellogg EA, Schmidt RJ. TCP transcription factor, BRANCH ANGLE DEFECTIVE 1 (BAD1), is required for normal tassel branch angle formation in maize. Proc Natl Acad Sci U S A. 2012;109:12225–30.
Zhu M, Hu YJ, Tong AZ, Yan BW, Lv YP, Wang SY, Ma WH, Cui ZB, Wang XX. LAZY1 controls tiller angle and shoot gravitropism by regulating the expression of auxin transporters and signaling factors in rice. Plant Cell Physiol. 2020;61:2111–25.
Taniguchi M, Furutani M, Nishimura T, Nakamura M, Fushita T, Iijima K, Baba K, Tanaka H, Toyota M, Tasaka M, et al. The Arabidopsis LAZY1 family plays a key role in gravity signaling within statocytes and in branch angle control of roots and shoots. Plant Cell. 2017;29:1984–99.
Zhang N, Yu H, Yu H, Cai YY, Huang LZ, Xu C, Xiong GS, Meng XB, Wang JY, Chen HF, Liu GF, Jing YH, Yuan YD, Liang Y, Li SJ, Smith SM, Li JY, Wang YH. A core regulatory pathway controlling rice tiller angle mediated by the LAZY1-dependent asymmetric distribution of auxin. Plant Cell. 2018;30:1461–75.
Li Z, Liang Y, Yuan YD, Wang L, Meng XB, Xiong GS, Zhou J, Cai YY, Han NP, Hua LK, Liu GF, Li JY, Wang YH. OsBRXL4 Regulates Shoot Gravitropism and Rice Tiller Angle through Affecting LAZY1 Nuclear Localization. Mol Plant. 2019;12:1143–56.
Yoshihara T, Iino M. Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and -independent gravity signaling pathways. Plant Cell Physiol. 2007;48:678–88.
Hollender CA, Hill JL Jr, Waite J, Dardick C. Opposing influences of TAC1 and LAZY1 on lateral shoot orientation in Arabidopsis. Sci Rep. 2020;10:6051.
Yoshihara T, Spalding EP. Switching the direction of stem gravitropism by altering two amino acids in AtLAZY1. Plant Physiol. 2020;182:1039–51.
Furutani M, Hirano Y, Nishimura T, Nakamura M, Taniguchi M, Suzuki K, Oshida R, Kondo C, Sun S, Kato K, Fukao Y, Hakoshima T, Morita MT. Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control. Nat Commun. 2020;11:76.
Ge LF, Chen RJ. Negative gravitropic response of roots directs auxin flow to control root gravitropism. Plant Cell Environ. 2019;42:2372–83.
Xia EH, Li FD, Tong W, Li PH, Wu Q, Zhao HJ, Ge RH, Li RP, Li YY, Zhang ZZ, Wei CL, Wan CH. Tea Plant Information Archive: a comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnol J. 2019;17:1938–53.
Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–17.
Li YY, Mi XZ, Zhao SQ, Zhu JY, Guo R, Xia XB, Liu L, Liu SR, Wei CL. Comprehensive profiling of alternative splicing landscape during cold acclimation in tea plant. BMC Genomics. 2020;21:65.
Liu SR, Mi XZ, Zhang R, An YL, Zhou QY, Yang TY, Xia XB, Guo R, Wang XW, Wei CL. Integrated analysis of miRNAs and their targets reveals that miR319c/TCP2 regulates apical bud burst in tea plant (Camellia sinensis). Planta. 2019;250:1111–29.
Zhao SQ, Wang XW, Yan XM, Guo LX, Mi XZ, Xu QS, Zhu JY, Wu AL, Liu LL, Wei CL. Revealing of MicroRNA involved regulatory gene networks on terpenoid biosynthesis in Camellia sinensis in different growing time points. J Agric Food Chem. 2018;66:12604–16.
Zhang XR, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc. 2006;1:641–6.
Acknowledgements
We thank the Tea Plant Cultivar and Germplasm Resource Garden (Guohe town, Lu Jiang County, Anhui Province, China) for providing tea plant samples.
Funding
This work was financially supported by the Anhui Provincial Natural Science Foundation (1808085QC92), the China Postdoctoral Science Foundation (2017 M621991), and the National Key Research and Development Program of China (2019YFD1001601).
Author information
Authors and Affiliations
Contributions
XBX performed the experiments and wrote the manuscript. XZM, LJ, RG, JYZ, HX, LL, YLA, and CZ were involved in the experiments and data analysis. SRL and CLW conceived the project, designed the research and revised the paper. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. The relevant primers
Additional file 2
. Gene sequences of CsLAZY1
Additional file 3: Figure S1
. Sequence alignment of five conserved regions of all LAZY family genes from rice, Arabidopsis and tea plants. All the LAZY genes were finally confirmed based on the EAR motif.
Additional file 4: Figure S2
. Expression analysis of AtLAZY1 and CsLAZY1 in the WT and overexpression Arabidopsis plants.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xia, X., Mi, X., Jin, L. et al. CsLAZY1 mediates shoot gravitropism and branch angle in tea plants (Camellia sinensis). BMC Plant Biol 21, 243 (2021). https://doi.org/10.1186/s12870-021-03044-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-021-03044-z