Abstract
Gastric cancer (GC) is one of the most common malignancies causing death worldwide, and Helicobacter pylori is a powerful inducer of precancerous lesions and GC. The oral microbiota is a complex ecosystem and is responsible for maintaining homeostasis, modulating the immune system, and resisting pathogens. It has been proposed that the gastric microbiota of oral origin is involved in the development and progression of GC. Nevertheless, the causal relationship between oral microbiota and GC and the role of H. pylori in this relationship is still controversial. This study was set to review the investigations done on oral microbiota and analyze various lines of evidence regarding the role of oral microbiota in GC, to date. Also, we discussed the interaction and relationship between H. pylori and oral microbiota in GC and the current understanding with regard to the underlying mechanisms of oral microbiota in carcinogenesis. More importantly, detecting the patterns of interaction between the oral cavity microbiota and H. pylori may render new clues for the diagnosis or screening of cancer. Integration of oral microbiota and H. pylori might manifest a potential method for the assessment of GC risk. Hence it needs to be specified the patterns of bacterial transmission from the oral cavity to the stomach and their interaction. Further evidence on the mechanisms underlying the oral microbiota communities and how they trigger GC may contribute to the identification of new prevention methods for GC. We may then modulate the oral microbiota by intervening with oral-gastric bacterial transmission or controlling certain bacteria in the oral cavity.
Similar content being viewed by others
Background
The oral microbiota is a complicated ecosystem in the body. More than 700 bacterial species live in the human oral cavity, which include 11 bacterial phyla and 70 genera [1]. The main phyla of oral bacteria include Fusobacteria, Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria [2]. The composition of the oral microorganisms can be associated with the carcinogenesis of distant organs, especially the gastrointestinal (GI) tract. Many studies have provided evidence that oral microbiota play major roles in GI cancers [3,4,5]. Species, such as Tannerella forsythia, Porphyromonas gingivalis, Prevotella intermedia, Parvimonas, and Leptotrichia were correlated with the risk of various kinds of GI cancers [6,7,8,9,10]. Gastric cancer (GC) is one of the most common malignancies causing death. The direct relationship between oral microbes and the GC risk has not been completely assessed [11]. Microbial communities are considered an important factor in the incidence and development of GC [12]. The GC microbiome has been characterized by the enrichment of numerous bacterial genera and species, which often colonize the oral cavity as opportunistic pathogens or commensals [13]. Streptococcus, Lactobacillus [14,15,16,17], and Lactococcus [15] species were more common in patients with GC [11]. Relative abundance of Streptococcaceae family was greater in patients with GC than in other patients [17,18,19]. Helicobacter pylori is a powerful inducer of precancerous lesions and GC [20,21,22,23,24,25]. Shifts in nutrient availability and gastric acidity and the innate immune response disrupt microbial ecological balance in GC patients, contributing to the overgrowth and colonization of non-H. pylori bacteria [26]. This study was set to review the investigations done on oral microbiota and analyze various lines of evidence regarding the role of oral microbiota in GC, to date. In this regard, the possible roles of oral microbiota in GC, the effects of oral microbiota on metabolic pathways and carcinogenic induction, the interaction and relationship between H. pylori and oral microbiota in GC, as well as the current understanding with regard to the underlying mechanisms of oral microbiota in carcinogenesis are discussed.
Main text
The relationship between oral microbiota and GC
Research studies have proved that oral pathogens are necessary in the GC development (Table 1). It has been shown that changes in the volume of oral microbiota may affect maintaining the local microenvironment that is linked with the progression or development of GC [12]. Applying 16S rRNA marker gene analysis, studies have indicated a significant enrichment of oral-related bacteria in GC [15, 19, 31]. It has been found that the microbial composition of GC patients was significantly different from that of control group [13]. The oral cavity bacterial species including Leptotrichia, Fusobacterium, Haemophilus, Veillonella, and Campylobacter have higher relative abundances in patients with GC from Singapore and Malaysia compared to others [15]. The most taxa abundant in GC are related to the opportunistic pathogens or commensals that often colonize the oral cavity, such as genera Aggregatibacter, Alloprevotella, and Neisseria; species Streptococcus mitis/oralis/pneumoniae; and also strain Porphyromonas endodontalis.t_GCF_000174815 [13]. At the phylum level, the relative frequency of Firmicutes was significantly higher while the relative frequency of Bacteroidetes was lower in the patients with GC compared to healthy individuals (Padj for BH = 0.005 and 3.6e-5, respectively). In genus level, Streptococcus and Abiotrophia had higher relative abundances in GC patients increasing its risk (P = 0.0045 and 0.0045 for BH correction, respectively). While genera such as Prevotella7, Neisseria, Prevotella, Porphyromonas, and Haemophilus reduced the risk of stomach cancer (P = 1.89e-04, 9.33e-04, 3.24e-05, 0.002, and 0.022, respectively) [11]. A considerable rise in the relative excess of lactic acid (Lactobacillus and Lactococcus [15]) was detected in GC patients. Furthermore, it was revealed that Lactococcus OTU0002 has powerful cooccurrence interactions with other OTUs related to GC (Bacillus OTU0046 and Aneurinibacillus OTU0038). Previous studies have similarly reported an increase in Lactobacillus species abundance in GC [14, 16, 17]. Bacterial taxa including Streptococcus anginosus_OTU68 (q = 0.033), Peptostreptococcus_OTU16 (q = 0.03), Gemella_OTU17 (q = 0.033), Fusobacterium_ OTU33 (q = 0.04), and Slackia_OTU174 (q = 0.033) were enriched in GC [19].
It has been shown that the composition of gastric microbiota varies among the residents of the two cities of Colombia (high-risk Túquerres and low-risk Tumaco). A Veillonella sp. and Leptotrichia wadei (OTUs: operational taxonomic units) in Túquerres, and Staphylococcus sp. in Tumaco were significantly more abundant [32]. In one study, the LEfSe analysis on OTUs revealed that high abundant OTUs such as Serratia marcescens, Flavobacterium, Stenotrophomnonas, Klebsiella, Pseudomonas, and Achromobacter were enriched in GC samples compared with other samples [33]. Although recent studies have examined the relationship between Lactobacillus, Fusobacterium, Peptostreptococcus, and Streptococcus in GC patients compared with the control group [15, 27], there is little information about the composition of the microbiota structure with oral origin in GC tissue samples compared to adjacent non-tumor tissues (ANTTs). A study from China showed that the bacterial taxa in the samples of cancer were predominantly represented via oral bacteria (e.g., Streptococcus, Peptostreptococcus, Fusobacterium, and Prevotella), but lactic acid-producing bacteria (e.g., Lactobacillus brevis and Lactococcus lactis) and Serratia were more plentiful in ANTTs [12]. The results of LEfSe analysis showed that 33 taxa were enriched in the cancer subjects, like the genera Prevotella, Prevotella_7, Peptostreptococcus, Streptococcus, Selenomonas, Acinetobacter, Sphingomonas, Bacillus, and Lachnoanaerobaculum, and the species Pseudomonas aeruginosa, Acinetobacter baumannii, Prevotella oris, and Prevotella denticola; most of them were oral microbiota. Sixteen taxa were also enriched in the non-cancer subjects, like genera Serratia, Lactococcus, Helicobacter, and Niveispirillum and the species L. brevis, S. marcescens, H. pylori, and L. lactis [12]. Using the DESeq 2 package, it was shown that the eight genera (Streptococcus, Peptostreptococcus, Acinetobacter, Sphingomonas, Bacteroides, Bacillus, and Prevotella_1/_7) were enriched in the cancer subjects. Fusobacterium was considerably profuse in cancerous tissues. Helicobacter and Lactobacillus manifested a significant increase in the ANTTs [12]. Another study showed that, tumor tissue, in comparison to the non-malignant tissues of the stomach, had lower Proteobacteria and higher Bacteriodetes, Fusobacteria, Firmicutes, and Spirochaetes in Chinese samples. No significant change was observed in phylum-level taxa in Mexican samples [31]. Another study from China showed that merely one bacterial taxa (Comamonadaceae_OTU85) overlapped the findings from GC vs. superficial gastritis (SG), depleted in GC lesions in comparison to ANTTs (q = 0.024) [19]. Such results highlighted the potential pathogenic impact of the GC-related oral microbiota [12]. Altered GC acidity can increase the chances of oral bacteria colonizing the gastrointestinal tract. Accordingly, the development and occurrence of GC disturb the endogenous bacterial community structure; H. pylori may only limitedly affect the progression and/or development of malignant tumors [12].
Negative link between gastric microbiota with oral origin and GC
Some studies suggest a reversal of oral microbiota in GC (Table 2); for instance, a study from China found that some bacterial taxa including Acinetobacter_ OTU369 (q = 0.045), Comamonadaceae_OTU85 (q = 0.033), Candidatus_Portiera_OTU1596 (q = 0.041), and Vogesella_OTU661 (q = 0.03) were depleted in GC [19]. Bacteria from the Sphingomonadaceae family [13, 32], especially Sphingobium yanoikuyae species [13], are negatively associated with GC. In the study by Hu et al., analyses at the phyla level showed that the relative abundance of Proteobacteria (especially Neisseria and Haemophilus) in GC subjects was meaningfully decreased in comparison to healthy subjects (P < 0.001). In patients with GC compared with healthy controls, it was also shown that the relative frequencies of Fusobacterium (P = 0.004), Porphyromonas (P = 0.002), Haemophilus (P = 0.007), and Neisseria (P = 0.008) were significantly reduced [34]. Several studies have shown the significant depletion of genera Neisseria in GC [14, 30]. In the study by Avies-Jimenez et al., the species Streptococcus sinensis was greatly abundant in NAG compared to MAG-IM and lower in GC [14]. In Korean population, the L. lactis’s mean relative abundance was greater in normal control subjects compared to patients with GC [35] (Table 2). Such differences in the relationship between oral microbiota and GC may be due to differences in the populations studied, the kind of samples, the kind of study, the materials and methods used, and the analysis methods.
Effects of oral microbiota on metabolic pathways and carcinogenic induction
It has been shown that the serological status of bacteria can significantly affect metabolic function. Metabolic contribution of bacteria correlates with carcinogenesis. It has been observed that bacterial metabolic pathways have been significantly increased in GC. The enrichment of carbohydrate absorption and digestion is found to be involved in generating short chain fatty acids (SCFAs) like butyrate, acetate, and propionate plus carbohydrate metabolism pathways in relation with the Lactococcus and Lactobacillus species enrichment in GC [15]. Castaño-Rodríguez et al., reported several bacterial metabolic pathways that were notably enriched in GC. In addition to carbohydrate metabolism pathways involved in the Lactococcus and Lactobacillus species enrichment in the GC, they detected the digestion enrichment and carbohydrates’ absorption affecting the SCFAs generation like butyrate, acetate, and propionate. Augmented bacterial SCFA rates may induce colonic cells’ hyperproliferation [36]. A significant rise in the relative lactic acid-producing bacteria’s abundance was seen in GC subjects [15]. Lactate can be a source of energy for the cells of tumor that induce glycolytic enzymes that increase the supply of ATP. This metabolite may potentiate inflammation and activate the angiogenesis of tumor [37,38,39].
The pathways’ enrichment related with SCFAs’ production in the subjects with GC has been detected by investigating the gastric samples’ microbiome by 16S rRNA marker gene assessment [15, 19]. Many metabolic pathways were significantly enriched in the samples of GC compared with adjacent non-cancerous samples, like those involved in carbohydrate metabolism (e.g., glycolysis and gluconeogenesis), energy metabolism (methane metabolism), and nucleotide metabolism (purine and pyrimidine metabolism) [12]. Purines can regulate immune cell responses and the cytokines release and are rich in the microenvironment of cancer [40]. It has been shown that the purine metabolism pathways are enriched in the cancer subjects [12]. Pathways related to the biosynthesis of L-ornithine, L-arginine, heme, biotin, and lipopolysaccharide (LPS) were enriched in GC group. The enrichment of LPS biosynthesis pathways in GC samples increased microbiota-induced inflammation [13]. LPS has been shown to increase inflammation in the tumor microenvironment and direct tumorigenesis [41, 42]. LPS and F. nucleatum cell extracts have been shown to raise inflammatory cytokines and chemokines and create a pro-inflammatory microenvironment that enhances the growth of cancer [43]. Pathways involved in pentose phosphate were predominantly abundant in GC [13]. S. anginosus—an oral bacterium—contains the enzyme alcohol dehydrogenase (ADH) that metabolizes alcohol to the carcinogenic acetaldehyde, causing cancer [44]. S. anginosus is responsible for inducing the nitric oxide synthesis and inflammatory cytokines causing carcinogenesis [45]. S. anginosus—a sulfate-reducing bacterium—affects colonic sulphur metabolism and induces inflammatory cytokines [46]. P. stomatis, P. micra, D. pneumosintes, and S. exigua also play a prominent role in progression of GC [19]. The nitrogen-containing compounds’ accumulation like nitrite and nitrate in the stomach may enhance gastric cells’ malignant transformation [47, 48]. Lactobacillus, and Nitrospirae are described as higher in GC and are involved in nitrate/nitrite metabolism [16]. N-nitroso compounds, which are formed in nitrate/nitrite metabolism, are important carcinogens. Bacteria such as Haemophilus, Staphylococcus, Clostridium, Neisseria, or Veillonella may be involved in the formation of these compounds, indicating that they may increase the risk of cancer [48, 49]. Metabolic enzymes associated with denitrification, including nitrous oxide reductase (COG4263) and nitrate reductase (COG1116) were enriched in cancer subjects’ gastric microbiota, compared to the non-cancer group [12].
Direct relationship between H. pylori and oral microbiota in GC
It appears that the H. pylori serological status has a notable effect on gastric microbiome α-diversity and composition. The gastric microbiome has been shown to be influenced by H. pylori serological status and changed in gastric carcinogenesis [15]. In fact, H. pylori affects the structure of the microbial community, and a meaningful increase in alpha diversity has been detected in H. pylori-positive samples in comparison with H. pylori-negative [12]. Bacterial load was risen considerably in H. pylori-positive patients in comparison to H. pylori-negative subjects. Infection with H. pylori showed a notable effect on bacterial load (P < 0.05). Therefore, infection with H. pylori might show the bacterial load of the gastric microbiota. This is probably due to variations in the gastric niche caused by H. pylori. Shannon’s diversity index in H. pylori-positive subjects (2.42 ± 0.58) was increased significantly compared to H. pylori-negative subjects (1.56 ± 0.39) (P < 0.05) [16]. However, there are studies that show H. pylori may be in the oral cavity and has interactions with oral microbes [50,51,52]. The ability of H. pylori to interact with the host and control the local environment was shown with this bacterium’s ability to activate the increased levels of MUC5B and MUC7. Increasing the amount of these oral H. pylori receptors may lead to retention and colonization in the oral cavity [53]. H. pylori has been observed to have a large capacity to accumulate with Fusobacterium spp. isolated from dental plaques (F. nucleatum and F. periodontium) [52]. In addition, P. gingivalis may affect such interactions. Therefore, H. pylori is related to the physiological function of F. nucleatum and P. gingivalis in dental plaque and vice versa [52]. Streptococci—a source of Streptococcus diffuse signal agents (SDSF)—may affect the morphological transformation of H. pylori into coccoid forms [54]. H. pylori has genes for the absorption and metabolic conversion of D- and L-lactose [55]. In supragingival plaques, the pH buffering process may be mediated in an ammonia-dependent way. H. pylori urease converts urea to CO2 and ammonia. Autoinducer-2 (AI-2) is a significant signaling material generated in dental plaque. It is a chemorepellent agent, promoting the H. pylori aggregates/biofilms dispersion and initiating negative chemotaxis against the signal source [56]. Therefore, this niche’s H. pylori colonization has to be prevented. The factors stimulating coccoid and the low AI-2 levels in supragingival plaque (early- to mid-stages) let dental H. pylori establish this niche as nonculturable forms. Subgingival plaque may prefer the mixed spiral and coccoid H. pylori populations [52].
It is not yet understood how H. pylori affects the structure and diversity of the oral microbiota. However, variations caused by H. pylori in the gastric niche affect the growth and colonization of microbes. CagA was associated with increased Gram-negative bacteria in the stomach, hence leading to LPS biosynthesis up-regulation. Through up-regulating LPS biosynthesis in the stomach and attenuating the oral microbiota defense against the microorganisms having a pathogenic potential, infection with H. pylori isolates possessing CagA can likely raise the risk of many illnesses [57]. The genera Actinomyces, Neisseria, Granulicatella, Helicobacter, Veillonella, Streptococcus, Fusobacterium, and Prevotella considerably vary between the H. pylori-positive and H. pylori-negative sample groups [58]. Haemophilus, Prevotella, Campylobacter, and Veillonella affect atrophic gastritis activated by H. pylori infection [59]. An altered microbial composition with the overgrowth of Prevotella, Veillonella, Streptococcus, and Lactobacillus was seen in the stomach of H. pylori-infected gastric adenocarcinoma and dyspeptic patients [18]. Neisseria, Haemophilus, Stenotrophomonas, and Serratia dominated the H. pylori-negative samples [33]. Significant changes of the gastric microbiota were detected in the H. pylori +/ CagA+ samples, and Helicobacter and Haemophilus genera abundances were increased [57]. The H. pylori +/ CagA+ group had greater Haemophilus and Helicobacter and lower Roseburia relative abundances in comparison with other subjects at the genus level [57].
No or inverse relationship between H. pylori and oral microbiome in GC
Infection by H. pylori is correlated with the reduced diversity of microbial alpha from H. pylori-negative to H. pylori-positive with CagA as a notable factor [58]. It has been recently investigated the H. pylori impacts on the richness, diversity, and interactions of microbes at the various phases of the disease (i.e. atrophic gastritis, GC, and intestinal metaplasia). Although a decrease in phyllotype richness, diversity, and evenness was reported in H. pylori-positive gastric biopsies compared to H. pylori-negative samples from chronic gastritis patients, no differences in classification diversity and evenness were seen [59]. This did not change even after controlling for the several stages of GC. However, at all stages the number of interactions between gastric microbes was significantly reduced. Moreover, H. pylori presence in superficial gastritis and intestinal metaplasia led to poorer GC-enriched and GC-depleted OTUs interactions, highlighting the potential role of H. pylori in alteration of microbial interactions [19].
As stated by Yu et al., oral-associated bacteria composition did not change by H. pylori colonization status, however, it changed between tumor gastric and paired non-malignant tissues in Mexican or Chinese samples [31]. Proteobacteria (e.g., Neisseria, Haemophilus, Stenotrophomonas, and Serratia) was the dominant species in the H. pylori -negative samples [33]. A study from Japan showed that proportion of Lactobacillus acidophilus was greater in H. pylori non-infected subjects than individuals with H. pylori infection, while the Lactobacillus salivarius proportion in H. pylori-infected people was high [60]. The relative Helicobacter abundance was associated inversely with the Firmicutes (r = − 0.49; P < 0.0001), non-Helicobacter Proteobacteria (r = − 0.59; P < 0.0001), Actinobacteria (r = − 0.54; P < 0.0001), and Bacteroidetes (r = − 0.43; P < 0.0001) abundances [30]. A work from Chile found that among the main phyla of gastric microbiota kept by children, children with H. pylori had a relatively lower Actinobacteria proportion than non-infected children. The frequency of five genera (i.e. Actinomyces, Streptococcus, Granulicatella, Rothia, and an undefined genus in family Neisseriaceae) in children with H. pylori was significantly reduced compared to non-infected children (P = 0.004–0.029). In contrast, the frequency of an unknown genus in the Comamonadaceae family was significantly risen in children infected with H. pylori versus non-infected children (P = 0.014). This reflects the fact that infection with H. pylori regenerates gastric microbiota, at least in infection, at several classification levels in children [61].
The relative genus Streptococcus abundance was declined markedly in H. pylori-positive (H. pylori+/CagA− and H. pylori+/CagA+) sample groups compared with the H. pylori-negative group (padj = 0.0216 and 0.0100, respectively) [58]. The relative abundance of Streptococcus showed no significant difference between the H. pylori+/CagA+ vs. H. pylori+/CagA− group (padj = 0.1716). Therefore, the expression of cagA gene did not affect the colonization of Streptococcus gastric [58]. In a study from Colombia, there was no significant association between the total gastric microbiota composition and carriage of the cagPAI or H. pylori population type. This shows that the changes in gastric microbial composition were highly independent of H. pylori colonizing strains. Streptococcus and Neisseria were genera seen more abundantly in people from the region with low GC risk [32].
Interaction between H. pylori and oral microbiome
Recent evidence suggests that commensal gastric microbes or their metabolites not only affect the ability of H. pylori to colonize the stomach but also modulate its pathogenicity potential directly [62, 63]. Many works have shown that infection with H. pylori is related with altered gastric microbiota and gastric dysbiosis is involved in some gastric diseases’ pathogenesis. It is not yet known whether H. pylori causes the growth of microorganisms or, conversely, the changed microbiota provides good conditions for the colonization of H. pylori. It is a two-way interaction; the H. pylori colonization prefers the growth of some bacteria, and vice versa, gastric dysbiosis can alter the gastric mucosa or lumen for the colonization of H. pylori [62].
It has been shown that H. pylori has the potential to alter the interactions between microbes [19]. Zhao et al., revealed that in the oral microbiota of the H. pylori-positive group, all interactions were significantly decreased, particularly for people infected with H. pylori+/CagA+ strains. Also, the oral microbiota of patients infected with H. pylori+/CagA+ was dominated by co-occurrence associations and showed one of the low network complexities because cooperation is destabilizing for the community. Therefore, the oral microbiota of people with H. pylori+/CagA+ strains might be more tolerant of alien species’ invasion [57]. H. pylori and taxa interactions were co-excluding in the samples of H. pylori +/CagA+. Some interactions were common between H. pylori − and H. pylori+/CagA− sample groups, including co-occurrence between OTU_68_Roseburia and OTU_10_Prevotella copri and between OTU_68_Roseburia and OTU_17_Propionibacterium, depleted in the H. pylori+/CagA+ group [57]. OTU_30_ Prevotella_histicola showed co-occurrence relations with OTU_28_Prevotella pallens and OTU_4_Veillonella dispar, which were ubiquitous in all subjects. The H. pylori+/CagA+ network group was dominated by cooperation associations; only one negative relationship was identified between OTU_11_Streptococcus and OTU_3_Prevotella. OTU_7_Roseburia interactions with OTU_30_ P. histicola and OTU_28_P. pallens that were detected in the groups representing the H. pylori − and H. pylori+/CagA−, depleted in the group representing H. pylori +/CagA+ [57].
The oral microbiome can possibly affect the bacteria that colonize the stomach. The close relationship between H. pylori and streptococci was confirmed by the fact that S. mitis and H. pylori were interacted upon co-cultivation via changed protein biosynthesis in H. pylori [64] though not validated under native and acidic conditions. The oral H. pylori physiology may potentially have modulated by Actinomyces spp. and Streptococcus spp.. These microorganisms may inhibit the growth of H. pylori in vitro [65]. The compounds secreted by Streptococcus mutans [66] and S. mitis [67] significantly reduce the durability of H. pylori. This effect is due to the H. pylori conversion to the nonculturable forms of coccoid. Streptococci SDSF may be involved in H. pylori morphological transformation into coccoid forms [54]. Some Streptococcaceae strains can have an impact on the final outcomes H. pylori infection. In coculture studies S. mitis caused the conversion of H. pylori to coccoid forms followed by growth inhibition [67]. The H. pylori cocooid form (vs. spiral form) shows not only a powerful impact on proliferation but also a poorer impact on apoptosis. The CagA and VacA expressions in the coccoid H. pylori were declined in comparison to the spiral form, while VacA was declined greater than that of CagA. The specific inhibitor of ERK1/2 notably blocked the increase in expression in Egr-1 and PCNA induced by the H. pylori cocooid form. Thus, the ERK1/2-Egr-1-PCNA pathway activation can affect cell proliferation triggered by cocooid H. pylori [68]. Furthermore, this coccoid form’s long latency in gastric mucosa was more associated with the development of GC than the spiral form [68, 69]. It also was shown that many Lactobacillus spp. including Lactobacillus casei, Lactobacillus murinus, L. salivarius and L. acidophilis inhibited H. pylori colonization [70,71,72,73]. Many Lactobacillus spp., as probiotics, can prevent H. pylori infection and improve H. pylori eradication in humans, although the mechanism is unknown [71]. L. salivarius WB 1004 may inhibit the binding of H. pylori to the gastric epithelial cells of murine and human and decrease IL-8 release in vitro [74]. L. salivarius, but L. casei or L. acidophilus generates abundant lactic acid as H. pylori inhibitor [75]. Lactobacillus gasseri OLL 2716 (LG21) has an ability to connect the gastric epithelium and withstand gastric acidity. It suppresses H. pylori and reduces gastric inflammation studied by the 13C-urea breath test and the serum pepsinogen levels [76]. Castaño-Rodríguez et al., found that the subject’s H. pylori serological status was related to a significant alteration in the predicted global microbial metabolic output. Using LEfSe, it was identified that KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways were enriched across the serological status of H. pylori; 20 predicted pathways (KEGG Level 3) were enriched in subjects with GC in comparison to controls. Additionally, carbohydrate absorption and digestion, which are somehow responsible for SCFAs production including propionate, butyrate, and acetate, were also enriched in GC [15].
Mechanisms underlying carcinogenic activity of oral microbiota
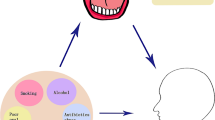
There are numerous potential mechanisms of action of oral microbiota that may cause carcinogenesis: I) Induction of chronic inflammation: Inflammatory mediators produced by oral bacteria, especially Fusobacterium, Porphyromonas, and Prevotella, cause oncogene activation, mutagenesis, DNA damage, cell cycle arrest, cell proliferation, tumor invasiveness, migration, metastasis, and angiogenesis [77, 78]. II) Inhibition of the host’s immune system: Oral microbiota such as P. gingivalis [79] and F. nucleatum [80,81,82] protect tumor cells by inhibiting immune responses. III) Anti-apoptotic activity: Oral bacteria such as F. nucleatum [83] and P. gingivalis [84] causes cancer growth by the activation of anti-apoptotic signaling pathways and inhibiting pro-apoptotic pathways that eventually lead to inhibition of cellular apoptosis. and IV) Carcinogenic substances: Oral bacteria produce some substances that play a role in chronic inflammation, genomic instability, accumulation of mutations, metastasis, and progression of GC [43, 85, 86] (Fig. 1).
Chronic inflammation
Chronic inflammation is known as the most prominent preventable cause of cancer [85,86,87,88]; some inflammatory cytokines may activate oncogenes [85]. Inflammation can also enhance progression of cancer and speed up invasion and metastasis processes [85, 86]. Oral microbiota, especially Fusobacterium, Porphyromonas, and Prevotella (anaerobic species), induce chronic inflammation. These bacteria incite the production of inflammatory mediators and adversely affect epithelial cells and extracellular matrix components. Oral pathogens associated with up-regulation of many cytokines such as interleukin-1β (IL-1β), IL-6, IL-17, IL-23, TNF-α, and other inflammatory mediators such as matrix metalloproteinases (MMPs) MMP-8 and MMP-9 are involved in carcinogenesis [77, 78]. P. gingivalis can promote local inflammation contributing to carcinogenesis [79]. Moreover, investigation of the anti-proliferative impact of the L. lactis cytoplasmic fraction on cancer cell line indicated a preventive influence on cell multiplication. L. lactis induced G0/G1 cell cycle arrest linked to an increase in expressions of p21 and p53, retinoblastoma protein phosphorylation, and a decrease in cyclin D1 expression, hence inducing apoptosis [89]. In recent studies, Fusobacterium species have attracted a lot of attention, with autophagy and TLR4 playing a crucial role in the inflammation they cause [90,91,92]. It has been shown that LPS and F. nucleatum cell extracts may raise chemokine and inflammatory cytokines and produce a proinflammatory microenvironment, promoting progression of cancer [43]. F. nucleatum can connect to cancerous and natural epithelial cells through FadA connection to E-cadherin [93]. This connection also stimulates β-catenin-regulated transcription, increasing the oncogenes cyclin D1 and c-Myc expression; Wnt signaling genes Wnt7a/Wnt7b/Wnt9a; and inflammatory genes nuclear factor-κB (NF-κB), IL-6, IL-8, and IL-18 which are responsible for carcinogenesis [93, 94]. IL-6 can induce oxidative stress and cause H2O2 transient accumulation in mitochondria and hence lead to mitochondrial damage [95, 96]. Most genes targeted by IL-6 contribute to the progression of the cell cycle and the suppression of the apoptosis. IL-6 may affect the development of cancer through influencing anti-apoptotic pathways [83]. IL-6 also influences the metastasis and invasion processes via increasing the expression of MMPs [97]. High content of IL-1β correlates with tumor migration, invasiveness, and higher aggressive tumor phenotype [98]. It was associated with lower E-cadherin expression, promoting cell migration [99]. TNF-α is also secreted responding to several factors, such as bacterial LPS. It triggers the generation of reactive oxygen compounds, prostaglandins, metalloproteinases, and leukotrienes [100]. For TNF-α-induced tumor growth, the activation of signaling pathways, such as Wnt and NF-κB, is crucial [101]. Furthermore, TNF-α may induce DNA damage through generation of reactive oxygen species [102]. It can affect invasion and motility processes via inducing expression of MMPs (Fig. 2) [103].
Oral bacteria increase the production of various types of inflammatory mediators such as interleukin-1β (IL-1β), IL-6, IL-17, IL-23, TNF-α, and MMP-8 and MMP-9, which are involved in DNA damage, tumor invasiveness, migration, metastasis and prevention of cell apoptosiss. High content of IL-1β correlates with tumor invasiveness and migration. IL-6 contributes to apoptosis suppression. IL-6 also influences the metastasis and invasion processes via increasing the expression of MMPs. TNF-α may induce DNA damage through generation of ROS. TNF-α can affect invasion and motility processes via inducing expression of MMPs. TNF-α: tumor necrosis factor-α, MMPs: matrix metalloproteinases, ROS: reactive oxygen species
Inhibition of the host’s immune system
Progression of cancer might be fueled by host immune system and microbiota interaction, particularly, in the gastrointestinal tract of the human in which there are plenty of bacteria; the immune system is very reactive [42]. Studies have suggested that P. gingivalis [79] and F. nucleatum [80, 81] could induce inhibition of the host’s immune response, and reducing these bacteria may lead to a decreased inhibition of immune responses. P. gingivalis can invade the eukaryotic cells through several virulence mechanisms, such as inhibiting the anti-oxidative and host’s immune systems and increasing the inflammation [104]. F. nucleatum has been reported to inhibit the proliferation and induction of T-cell apoptosis by expanding myeloid-derived immune cells [81]. F. nucleatum protects tumor cells from immune cell attack and natural killer (NK) -mediated killing [80]. Moreover, F. nucleatum can save the cells of the tumor from the immune cell attack and natural killer (NK)-mediated killing by interacting of its Fap2 protein with the inhibitory TIGIT (T cell immunoreceptor with Ig and ITIM domains) on the T and NK cells [80]. Various F. nucleatum strains can inhibit the NK cell killing of several tumors. It is mediated by the human TIGIT. The F. nucleatum Fap2 protein can directly interact with TIGIT, preventing the NK cell cytotoxicity [82].
Anti-apoptotic activity
Oral bacteria can affect the pathogenesis of cancers through influencing cytoskeletal rearrangements, inhibition of cellular apoptosis, cell proliferation, and activation of NF-ΚB [44]. F. nucleatum infection modulates numerous anti-apoptotic pathways. Toll-like receptor (TLR) activation causes bacteria stimulate NF-kB signaling [86]. FadA is a crucial pathogenic factor of F. nucleatum and changes methylation of the cyclin-dependent kinase inhibitor 2A (CDKN2A) promoter and infiltration of macrophage in cancer tissues [105]. F. nucleatum stimulates p38, which results in the MMP-13 and MMP-9 secretion and significantly affects cancer cell invasion and metastasis [106]. Also, F. nucleatum can induce signaling of β-catenin by its LPS. Enhancing the β-catenin expression and oncogenes C-myc and cyclin D1 is present in this process [90, 107]. The inflammatory cytokines activated by F. nucleatum LPS are IL-6, TNF-α, and IL-1β [44]. IL-6 may affect cancer development by influencing anti-apoptotic pathways [83]. P. gingivalis LPS may stimulate host response via TLRs, like TLR4 and TLR2 that may prevent apoptosis and enhance tumor proliferation; it therefore cooperates in the protection of tumor cells and the progression of cancer [84]. P. gingivalis functions anti-apoptotically through a lot of pathways’ modulation [108]. Intracellular P. gingivalis induces anti-apoptotic signaling of Jak1/Akt/Stat3 [109, 110]. This bacterium can discharge an anti-apoptotic enzyme nucleoside diphosphate kinase (NDK), breaking down ATP and inhibits the proapoptotic P2X7 receptor, thus modulating signaling of ATP/P2X7- [111]. It also accelerates progression via the cell cycle S-phase by manipulating cyclin/CDK activity; it decreases the p53 tumor suppressor level (Fig. 3) [112]. P. gingivalis results in significant pro-apoptotic Bad phosphorylation and inhibition, through enhancing the Bcl2 and Bax ratios [113].
Oral bacteria can affect the inhibition of cell apoptosis. F. nucleatum modulates numerous anti-apoptotic pathways. As a consequence of TLR activation, bacteria stimulate NF-kB signaling. F. nucleatum activates p38, which results in the MMP-9 and MMP-13 secretion and leads to cancer cell invasion and metastasis. Also, F. nucleatum may induce β-catenin signaling by its LPS and FadA. Stimulating the β-catenin expression and increasing the expression of oncogenes C-myc and cyclin D1 lead to cell proliferation. P. gingivalis LPS may stimulate host response via TLRs (TLR2 and TLR4) and enhance the growth of tumor. Also, P. gingivalis induces anti-apoptotic Jak1/Akt/Stat3 signaling. This bacterium can secrete a NDK enzyme, which cleaves ATP and prevents the proapoptotic P2X7 receptor activation, thus modulating ATP/P2X7-signaling. It also causes cell cycle arrest by manipulating cyclin/CDK activity and reduced levels of p53. TLR: Toll-like receptor, NF- kB: nuclear factor kappa B, p38: Mitogen-activated protein kinase p38, MMPs: matrix metalloproteinases, LPS: lipopolisaccharide, FadA: fusobacterial adhesin/invasin, Jak1: Janus kinase 1, Akt: protein kinase B, Stat3: Signal transducer and activator of transcription 3, Bad: Bcl-2-associated death promoter, CDK: cyclin-dependent kinase, p53: Tumor protein p53, NDK: nucleoside diphosphate kinase, ATP: Adenosine triphosphate, P2X7: Purinergic receptor
Carcinogenic substances
Substances which are generated by oral bacteria with a carcinogenic effect consist of organic acids, volatile sulfur compounds (VSC), reactive nitrogen species (RNS) and reactive oxygen species (ROS), and hydrogen peroxide (H2O2). The P. gingivalis NDK secretion may modulate the ATP-induced cytosolic and mitochondrial ROS and the antioxidant glutathione response (AGR) generated via the P2X7/NADPH-oxidase interactome [114]. ROS can markedly activate inflammation/cancer-associated transcription factors [115]. In this process, some species in the oral cavity produce H2O2. The peroxigenic oral bacteria consist of Streptococcus gordonii, S. oralis, Streptococcus sanguinis, S. mitis, Streptococcus oligofermentans [116], L. acidophilus, Lactobacillus fermentum, Lactobacillus minutus, Lactobacillus jensenii, and Bifidobacterium adolescentis [117]. These findings emphasize the relationship between free radicals and chronic inflammation and their effect in developing cancer [118].
The microorganisms that metabolize alcohol to acetaldehyde significantly affects the cancer development. Oral bacteria (e.g., Aggregatibacter actinomycetemcomitans, P. intermedia, P. gingivalis, and F. nucleatum) generate VSCs including methyl mercaptan (CH3SH), dimethyl disulfide (CH3SSCH3), hydrogen sulfide (H2S), and dimethyl sulfide ((CH3)2S) [44]. VSCs are toxic to tissues and may develop chronic inflammation [119]. H2S is a common genotoxic agent and causes cumulative mutations or genomic instability [120]. H2S has dichotomous influences on many gastrointestinal processes like cancer, inflammation, and apoptosis [121].
Oral microbiota are able to metabolize alcohol (ethanol) to acetaldehyde, due to possessing the enzyme alcohol dehydrogenase (ADH), which is involved in carcinogenesis [88, 122]. It has been shown that several species of oral bacteria such as S. mitis, S. gordonii, Streptococcus salivarius, S. sanguinis, and S. oralis [123] possess ADH, which metabolizes alcohol to acetaldehyde [124] with a potential for cancer development [44]. Genus Neisseria can produce the large amounts of the ADH enzyme, which generates the carcinogen acetaldehyde, and along with H. pylori with high generation of this enzyme, may affect alcohol-related gastric carcinogenesis [122].
Some species can generate acids more (e.g., aciduric Peptostreptococcus stomatis produces acetic, isocaproic, isobutyric, butyric, and isovaleric acids) [125]. Such acid production can affect the hypoxic and acidic microenvironment of the tumor, thus augmenting metastatic efficiency [126, 127]. Some oral bacteria of genera Lactobacillus, Streptococcus, Bifidobacterium, Leuconostoc, Lactococcus, and Pediococcus generate lactic acid [128]. These microorganisms are aciduric and acidogenic which may lower pH in the local environment by producing lactic acid [129]. Lactobacillus and Lactococcus species are known as probiotics and assumed good to the host. The production of lactic acid has immunomodulative, anti-inflammatory, and anti-cancer activities and contribute to H. pylori eradication [130,131,132]. Lactate also serves as energy source of the tumor, producing glycolytic enzymes to raise the supply of ATP. This metabolite may enhance inflammation and activate the angiogenesis of the tumor (Figs 4) [37,38,39, 133].
Oral bacteria produce some substances that play a role in chronic inflammation, genomic instability, tumor angiogenesis, and progression of gastric cancer. Some oral bacteria generate VSCs including CH3SH, H2S, CH3SSCH3, and (CH3)2S that may develop chronic inflammation. Oral bacteria are also involved in the production of ROS, RNS and H2O2, which may be involved in genotoxicity. Some species can generate organic acids (e.g., isobutyric, butyric, isocaproic, and isovaleric acids) that may contribute to cell metastasis. H2S may cause genomic instability, effects on inflammation, apoptosis, and many gastrointestinal processes like cancer. Other oral bacteria generate lactic acid, which is a source of energy for tumor cells and is involved in increasing ATP levels, which may exacerbate inflammation and angiogenesis. Some of them are able to metabolize alcohol to acetaldehyde by ADH enzyme, which is involved in carcinogenesis. VSCs: volatile sulfur compounds, CH3SH: including methyl mercaptan, H2S: hydrogen sulfide, CH3SSCH3: dimethyl disulfide, and (CH3)2S: dimethyl sulfide, ROS: reactive oxygen species, RNS: reactive nitrogen species, H2O2: hydrogen peroxide, ATP: Adenosine triphosphate, ADH: alcohol dehydrogenase
Conclusion
Several factors, including tooth flossing [134], poor oral hygiene [135,136,137,138], the metabolism of oral microbes [78], and tooth loss [136,137,138] have been found to affect the risk of gastric precancerous lesions and gastric non-cardia carcinoma. Nevertheless, the causal correlation between oral microbiota and GC was not obvious. It is proposed that identifying specific oral microbiota proteins can help detect early GC. Therefore, cancer may be prevented by targeting and inhibiting oral carcinogenic microbial proteins or by eradicating certain microbiome species. More importantly, detecting the patterns of interaction between the oral cavity microbiota and H. pylori may render new clues for the diagnosis or screening of cancer. Integration of oral microbiota and H. pylori might manifest a potential method for the assessment of GC risk. Hence it needs to be specified the patterns of bacterial transmission from the oral cavity to the stomach and their interaction. Further evidence on the mechanisms underlying the oral microbiota communities and how they trigger GC may contribute to the identification of new prevention methods for GC. We may then modulate the oral microbiota by intervening with oral-gastric bacterial transmission or controlling certain bacteria in the oral cavity.
Availability of data and materials
Not applicable.
Abbreviations
- GC:
-
Gastric cancer
- H. pylori :
-
Helicobacter pylori
- CagA:
-
Cytotoxin-associated gene A
- GI:
-
Gastrointestinal
- P. gingivalis :
-
Porphyromonas gingivalis
- P. intermedia :
-
Prevotella intermedia
- 16S rRNA:
-
16S ribosomal RNA
- S. mitis :
-
Streptococcus mitis
- S. oralis :
-
Streptococcus oralis
- S. anginosus :
-
Streptococcus anginosus
- S. marcescens :
-
Serratia marcescens
- ANTTs:
-
Adjacent non-tumor tissues
- L. lactis :
-
Lactococcus lactis
- L. brevis :
-
Lactobacillus brevis
- LEfSe analysis:
-
Linear discriminant analysis Effect Size
- SG:
-
Superficial gastritis
- SCFAs:
-
Short chain fatty acids
- LPS:
-
Lipopolysaccharide
- F. nucleatum :
-
Fusobacterium nucleatum
- ADH:
-
Alcohol dehydrogenase
- P. stomatis :
-
Peptostreptococcus stomatis
- P. micra :
-
Parvimonas micra
- D. pneumosintes :
-
Dialister pneumosintes
- S. exigua :
-
Salix exigua
- MUC5B:
-
Mucin 5B
- MUC7:
-
Mucin 5B
- F. periodontium :
-
Fusobacterium periodontium
- SDSF:
-
Streptococcus diffusible signal factor
- AI-2:
-
Autoinducer-2
- cagPAI:
-
cag pathogenicity island
- P. pallens :
-
Prevotella pallens
- P. histicola :
-
Prevotella_histicola
- ERK1/2:
-
Extracellular signal-regulated protein kinase 1/2
- PCNA:
-
Proliferating cell nuclear antigen
- Egr-1:
-
Early growth response protein 1
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- IL-1β:
-
Interleukin-1β
- IL-6:
-
Interleukin-6
- IL-17:
-
Interleukin-17
- IL-23:
-
Interleukin-23
- TNF-α:
-
Tumor necrosis factor alpha
- MMPs:
-
Matrix metalloproteinases
- MMP-8:
-
Matrix metalloproteinase-8
- MMP-9:
-
Matrix metalloproteinase-9
- p21:
-
A potent cyclin-dependent kinase inhibitor
- p53:
-
A tumor suppressor protein
- TLRs:
-
Toll-like receptors
- FadA:
-
Fusobacterium adhesin A
- NF- kB:
-
Nuclear factor kappa B
- p38:
-
Mitogen-activated protein kinase p38
- NK:
-
Natural killer
- TIGIT:
-
T cell immunoreceptor with Ig and ITIM domains
- Fap2:
-
Fusobacterium autotransporter protein 2
- CDKN2A:
-
Cyclin-dependent kinase inhibitor 2A
- C-MYC:
-
C-myelocytomatosis oncogene product
- Jak1:
-
Janus kinase 1
- Akt:
-
Protein kinase B, PKB
- Stat3:
-
Signal transducer and activator of transcription 3
- NDK:
-
Nucleoside diphosphate kinase
- Bad:
-
Bcl-2-associated death promoter
- CDK:
-
cyclin-dependent kinase
- NDK:
-
Nucleoside diphosphate kinase
- ATP:
-
Adenosine triphosphate
- P2X7:
-
Purinergic receptor
- Bcl-2:
-
B-cell lymphoma 2
- BAX:
-
BCL2-associated X protein
- VSCs:
-
Volatile sulfur compounds
- RNS:
-
Reactive nitrogen species
- ROS:
-
Reactive oxygen species
- H2O2 :
-
Hydrogen peroxide
- AGR:
-
Antioxidant glutathione response
- CH3SH:
-
Including methyl mercaptan
- H2S:
-
Hydrogen sulfide
- CH3SSCH3 :
-
Dimethyl disulfide
- (CH3)2S:
-
Dimethyl sulfide
- S. gordonii :
-
Streptococcus gordonii
References
Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43(11):5721–32.
Sampaio-Maia B, Caldas I, Pereira M, Pérez-Mongiovi D, Araujo R. The oral microbiome in health and its implication in oral and systemic diseases. In: Advances in applied microbiology, vol. 97: Elsevier; 2016. p. 171–210.
Chen X, Winckler B, Lu M, Cheng H, Yuan Z, Yang Y, Jin L, Ye W. Oral microbiota and risk for esophageal squamous cell carcinoma in a high-risk area of China. PLoS One. 2015;10(12):e0143603.
Han S, Yang X, Qi Q, Pan Y, Chen Y, Shen J, Liao H, Ji Z. Potential screening and early diagnosis method for cancer: tongue diagnosis. Int J Oncol. 2016;48(6):2257–64.
García-Castillo V, Sanhueza E, McNerney E, Onate SA, García A. Microbiota dysbiosis: a new piece in the understanding of the carcinogenesis puzzle. J Med Microbiol. 2016;65(12):1347–62.
Chen Y, Chen X, Yu H, Zhou H, Xu S. Oral microbiota as promising diagnostic biomarkers for gastrointestinal Cancer: a systematic review. OncoTargets and therapy. 2019;12:11131–44.
Mai X, Genco RJ, LaMonte MJ, Hovey KM, Freudenheim JL, Andrews CA, Wactawski-Wende J. Periodontal pathogens and risk of incident Cancer in postmenopausal females: the Buffalo OsteoPerio study. J Periodontol. 2016;87(3):257–67.
Yang Y, Cai Q, Shu X-O, Steinwandel MD, Blot WJ, Zheng W, Long J. Prospective study of oral microbiome and colorectal cancer risk in low-income and African American populations. Int J Cancer. 2019;144(10):2381–9.
Peters BA, Wu J, Pei Z, Yang L, Purdue MP, Freedman ND, Jacobs EJ, Gapstur SM, Hayes RB, Ahn J. Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. 2017;77(23):6777–87.
Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, Purdue MP, Abnet CC, Stolzenberg-Solomon R, Miller G, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2018;67(1):120–7.
Wu J, Xu S, Xiang C, Cao Q, Li Q, Huang J, Shi L, Zhang J, Zhan Z. Tongue coating microbiota community and risk effect on gastric Cancer. J Cancer. 2018;9(21):4039–48.
Chen X-H, Wang A, Chu A-N, Gong Y-H, Yuan Y. Mucosa-associated microbiota in gastric Cancer tissues compared with non-cancer tissues. Front Microbiol. 2019;10:1261.
Hu Y-L, Pang W, Huang Y, Zhang Y, Zhang C-J. The gastric microbiome is perturbed in advanced gastric adenocarcinoma identified through shotgun Metagenomics. Front Cell Infect Microbiol. 2018;8:433.
Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep. 2014;4:4202.
Castaño-Rodríguez N, Goh K-L, Fock KM, Mitchell HM, Kaakoush NO. Dysbiosis of the microbiome in gastric carcinogenesis. Sci Rep. 2017;7(1):15957.
Wang L, Zhou J, Xin Y, Geng C, Tian Z, Yu X, Dong Q. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol. 2016;28(3):261–6.
Eun CS, Kim BK, Han DS, Kim SY, Kim KM, Choi BY, Song KS, Kim YS, Kim JF. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter. 2014;19(6):407–16.
Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol. 2009;58(4):509–16.
Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, Wu WK, Wong SH, Chen Z, Sung JJ. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2018;67(6):1024–32.
Bakhti SZ, Latifi-Navid S, Zahri S, Yazdanbod A. Inverse association of helicobacter pylori cagPAI genotypes with risk of cardia and non-cardia gastric adenocarcinoma. Cancer medicine. 2019;8(10):4928–37.
Bakhti SZ, Latifi-Navid S, Safaralizadeh R. Helicobacter pylori-related risk predictors of gastric cancer: the latest models, challenges, and future prospects. Cancer Medicine. 2020.
Bakhti SZ, Latifi-Navid S, Zahri S. Unique constellations of five polymorphic sites of Helicobacter pylori vacA and cagA status associated with risk of gastric cancer. Infection, Genetics and Evolution. 2020;79:104167.
Abdi E, Latifi-Navid S, Latifi-Navid H, Safarnejad B. Helicobacter pylori vacuolating cytotoxin genotypes and preneoplastic lesions or gastric cancer risk: a meta-analysis. J Gastroenterol Hepatol. 2016;31(4):734–44.
Honarmand-Jahromy S, Siavoshi F, Malekzadeh R, Sattari TN, Latifi-Navid S. Multiple repeats of helicobacter pylori CagA EPIYA-C phosphorylation sites predict risk of gastric ulcer in Iran. Microb Pathog. 2015;89:87–92.
Safaralizadeh R, Dastmalchi N, Hosseinpourfeizi M, Latifi-Navid S. Helicobacter pylori virulence factors in relation to gastrointestinal diseases in Iran. Microb Pathog. 2017;105:211–7.
Brawner KM, Morrow CD, Smith PD. Gastric microbiome and gastric cancer. Cancer journal (Sudbury, Mass). 2014;20(3):211–6.
Hsieh Y-Y, Tung S-Y, Pan H-Y, Yen C-W, Xu H-W, Lin Y-J, Deng Y-F, Hsu W-T, Wu C-S, Li C. Increased abundance of Clostridium and Fusobacterium in gastric microbiota of patients with gastric Cancer in Taiwan. Sci Rep. 2018;8(1):158.
Sun JH, Li XL, Yin J, Li YH, Hou BX, Zhang Z. A screening method for gastric cancer by oral microbiome detection. Oncol Rep. 2018;39(5):2217–24.
Liu J, Xue Y, Zhou L. Detection of gastritis-associated pathogens by culturing of gastric juice and mucosa. Int J Clin Exp Pathol. 2018;11(4):2214.
Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, Figueiredo C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67(2):226–36.
Yu G, Torres J, Hu N, Medrano-Guzman R, Herrera-Goepfert R, Humphrys MS, Wang L, Wang C, Ding T, Ravel J. Molecular characterization of the human stomach microbiota in gastric cancer patients. Front Cell Infect Microbiol. 2017;7:302.
Yang I, Woltemate S, Piazuelo MB, Bravo LE, Yepez MC, Romero-Gallo J, Delgado AG, Wilson KT, Peek RM, Correa P. Different gastric microbiota compositions in two human populations with high and low gastric cancer risk in Colombia. Sci Rep. 2016;6:18594.
Li TH, Qin Y, Sham PC, Lau K, Chu K-M, Leung WK. Alterations in gastric microbiota after H pylori eradication and in different histological stages of gastric carcinogenesis. Scientific reports. 2017;7:44935.
Hu J, Han S, Chen Y, Ji Z. Variations of tongue coating microbiota in patients with gastric cancer. Biomed Res Int. 2015;2015.
Gunathilake MN, Lee J, Choi IJ, Kim Y-I, Ahn Y, Park C, Kim J. Association between the relative abundance of gastric microbiota and the risk of gastric cancer: a case-control study. Sci Rep. 2019;9(1):1–11.
Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, Moriyama EH, Copeland JK, Surendra A, Kumar S. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158(2):288–99.
Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest. 2013;123(9):3685–92.
Kennedy KM, Scarbrough PM, Ribeiro A, Richardson R, Yuan H, Sonveaux P, Landon CD, Chi J-T, Pizzo S, Schroeder T. Catabolism of exogenous lactate reveals it as a legitimate metabolic substrate in breast cancer. PLoS One. 2013;8(9):e75154.
Sonveaux P, Copetti T, De Saedeleer CJ, Végran F, Verrax J, Kennedy KM, Moon EJ, Dhup S, Danhier P, Frérart F. Targeting the lactate transporter MCT1 in endothelial cells inhibits lactate-induced HIF-1 activation and tumor angiogenesis. PLoS One. 2012;7(3):e33418.
Di Virgilio F. Purines, purinergic receptors, and cancer. Cancer Res. 2012;72(21):5441–7.
Rakoff-Nahoum S, Medzhitov R. Toll-like receptors and cancer. Nat Rev Cancer. 2009;9(1):57–63.
Gagliani N, Hu B, Huber S, Elinav E, Flavell RA. The fire within: microbes inflame tumors. Cell. 2014;157(4):776–83.
Gholizadeh P, Eslami H, Kafil HS. Carcinogenesis mechanisms of Fusobacterium nucleatum. Biomed Pharmacother. 2017;89:918–25.
Karpiński TM. Role of oral microbiota in cancer development. Microorganisms. 2019;7(1):20.
Chattopadhyay I, Verma M, Panda M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technology in cancer research & treatment. 2019;18:1533033819867354.
Morita E, Narikiyo M, Yano A, Nishimura E, Igaki H, Sasaki H, Terada M, Hanada N, Kawabe R. Different frequencies of Streptococcus anginosus infection in oral cancer and esophageal cancer. Cancer Sci. 2003;94(6):492–6.
Correa P. Human gastric carcinogenesis: a multistep and multifactorial process—first American Cancer Society award lecture on cancer epidemiology and prevention. Cancer Res. 1992;52(24):6735–40.
Alarcón T, Llorca L, Perez-Perez G. Impact of the microbiota and gastric disease development by helicobacter pylori. In: Molecular Pathogenesis and Signal Transduction by Helicobacter pylori: Springer; 2017. p. 253–75.
Hu Y, He L-H, Di Xiao G-DL, Gu Y-X, Tao X-X, Zhang J-Z. Bacterial flora concurrent with helicobacter pylori in the stomach of patients with upper gastrointestinal diseases. World J Gastroenterol: WJG. 2012;18(11):1257.
Al Sayed A, Anand PS, Kamath KP, Patil S, Preethanath R, Anil S. Oral cavity as an extragastric reservoir of helicobacter pylori. ISRN gastroenterology. 2014;2014.
Urban J. Helicobacter pylori–characteristics and pathogenic factors. Dental and Medical Problems. 2010;47(4):482–6.
Krzyżek P, Gościniak G. Oral helicobacter pylori: interactions with host and microbial flora of the oral cavity. Dental and medical problems. 2018;55(1):75–82.
Silva DG, Stevens RH, Macedo JM, Hirata R, Pinto AC, Alves LM, Veerman EC, Tinoco EM. Higher levels of salivary MUC5B and MUC7 in individuals with gastric diseases who harbor helicobacter pylori. Arch Oral Biol. 2009;54(1):86–90.
Krzyżek P, Gościniak G. A proposed role for diffusible signal factors in the biofilm formation and morphological transformation of helicobacter pylori. Turk J Gastroenterol. 2018;29(1):7.
Iwatani S, Nagashima H, Reddy R, Shiota S, Graham DY, Yamaoka Y. Identification of the genes that contribute to lactate utilization in helicobacter pylori. PLoS One. 2014;9(7):e103506.
Anderson JK, Huang JY, Wreden C, Sweeney EG, Goers J, Remington SJ, Guillemin K. Chemorepulsion from the quorum signal autoinducer-2 promotes Helicobacter pylori biofilm dispersal. MBio. 2015;6(4).
Zhao Y, Gao X, Guo J, Yu D, Xiao Y, Wang H, Li Y. Helicobacter pylori infection alters gastric and tongue coating microbial communities. Helicobacter. 2019;24(2):e12567.
Klymiuk I, Bilgilier C, Stadlmann A, Thannesberger J, Kastner M-T, Högenauer C, Püspök A, Biowski-Frotz S, Schrutka-Kölbl C, Thallinger GG. The human gastric microbiome is predicated upon infection with helicobacter pylori. Front Microbiol. 2017;8:2508.
Schulz C, Schütte K, Koch N, Vilchez-Vargas R, Wos-Oxley ML, Oxley AP, Vital M, Malfertheiner P, Pieper DH. The active bacterial assemblages of the upper GI tract in individuals with and without helicobacter infection. Gut. 2018;67(2):216–25.
Iino C, Shimoyama T, Chinda D, Arai T, Chiba D, Nakaji S, Fukuda S. Infection of helicobacter pylori and atrophic gastritis influence Lactobacillus in gut microbiota in a Japanese population. Front Immunol. 2018;9:712.
Brawner K, Kumar R, Serrano C, Ptacek T, Lefkowitz E, Morrow C, Zhi D, Kyanam-Kabir-Baig K, Smythies L, Harris P. Helicobacter pylori infection is associated with an altered gastric microbiota in children. Mucosal Immunol. 2017;10(5):1169–77.
Espinoza JL, Matsumoto A, Tanaka H, Matsumura I. Gastric microbiota: an emerging player in helicobacter pylori-induced gastric malignancies. Cancer Lett. 2018;414:147–52.
Ianiro G, Molina-Infante J, Gasbarrini A. Gastric microbiota. Helicobacter. 2015;20:68–71.
Khosravi Y, Loke MF, Goh KL, Vadivelu J. Proteomics analysis revealed that crosstalk between helicobacter pylori and Streptococcus mitis may enhance bacterial survival and reduces carcinogenesis. Front Microbiol. 2016;7:1462.
Ishihara K, Miura T, Kimizuka R, Ebihara Y, Mizuno Y, Okuda K. Oral bacteria inhibit helicobacter pylori growth. FEMS Microbiol Lett. 1997;152(2):355–61.
Okuda K, Ishihara K, Miura T, Katakura A, Noma H, Ebihara Y. Helicobacter pylori may have only a transient presence in the oral cavity and on the surface of oral cancer. Microbiol Immunol. 2000;44(5):385–8.
Khosravi Y, Dieye Y, Loke MF, Goh KL, Vadivelu J. Streptococcus mitis induces conversion of helicobacter pylori to coccoid cells during co-culture in vitro. PLoS One. 2014;9(11):e112214.
Li N, Han L, Chen J, Lin X, Chen H, She F. Proliferative and apoptotic effects of gastric epithelial cells induced by coccoid helicobacter pylori. J Basic Microbiol. 2013;53(2):147–55.
Chan W-Y, Hui P-K, Leung K-M, Chow J, Kwok F, Ng C-S. Coccoid forms of helicobacter pylori in the human stomach. Am J Clin Pathol. 1994;102(4):503–7.
Franceschi F, Cazzato A, Nista EC, Scarpellini E, Roccarina D, Gigante G, Gasbarrini G, Gasbarrini A. Role of probiotics in patients with helicobacter pylori infection. Helicobacter. 2007;12:59–63.
Schmitz JM, Durham CG, Schoeb TR, Soltau TD, Wolf KJ, Tanner SM, McCracken VJ, Lorenz RG. Helicobacter felis–associated gastric disease in microbiota-restricted mice. Journal of Histochemistry & Cytochemistry. 2011;59(9):826–41.
Gotteland M, Brunser O, Cruchet S. Systematic review: are probiotics useful in controlling gastric colonization by helicobacter pylori? Aliment Pharmacol Ther. 2006;23(8):1077–86.
Sgouras D, Maragkoudakis P, Petraki K, Martinez-Gonzalez B, Eriotou E, Michopoulos S, Kalantzopoulos G, Tsakalidou E, Mentis Α. In vitro and in vivo inhibition of helicobacter pylori by Lactobacillus casei strain Shirota. Appl Environ Microbiol. 2004;70(1):518–26.
Kabir A, Aiba Y, Takagi A, Kamiya S, Miwa T, Koga Y. Prevention of helicobacter pylori infection by lactobacilli in a gnotobiotic murine model. Gut. 1997;41(1):49–55.
Aiba Y, Suzuki N, Kabir AM, Takagi A, Koga Y. Lactic acid-mediated suppression of helicobacter pylori by the oral administration of Lactobacillus salivarius as a probiotic in a gnotobiotic murine model. Am J Gastroenterol. 1998;93(11):2097–101.
Sakamoto I, Igarashi M, Kimura K, Takagi A, Miwa T, Koga Y. Suppressive effect of Lactobacillus gasseri OLL 2716 (LG21) on helicobacter pylori infection in humans. J Antimicrob Chemother. 2001;47(5):709–10.
Szkaradkiewicz AK, Karpinski T. Microbiology of chronic periodontitis. J Biol Earth Sci. 2013;3:14–20.
Ahn J, Chen CY, Hayes RB. Oral microbiome and oral and gastrointestinal cancer risk. Cancer Causes Control. 2012;23(3):399–404.
Le Bars P, Matamoros S, Montassier E, Le Vacon F, Potel G, Soueidan A, Jordana F, de La Cochetière M-F. The oral cavity microbiota: between health, oral disease, and cancers of the aerodigestive tract. Can J Microbiol. 2017;63(6):475–92.
Colucci F. An oral commensal associates with disease: chicken, egg, or red herring? Immunity. 2015;42(2):208–10.
Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, Kanno S, Yamamoto I, Ishigami K, Igarashi H, et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. 2016;22(2):557–66.
Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–55.
Haura EB, Turkson J, Jove R. Mechanisms of disease: insights into the emerging role of signal transducers and activators of transcription in cancer. Nat Clin Pract Oncol. 2005;2(6):315–24.
Michaud DS. Role of bacterial infections in pancreatic cancer. Carcinogenesis. 2013;34(10):2193–7.
Meng C, Bai C, Brown TD, Hood LE, Tian Q. Human gut microbiota and gastrointestinal cancer. Genomics, proteomics & bioinformatics. 2018;16(1):33–49.
Gallimidi AB, Fischman S, Revach B, Bulvik R, Maliutina A, Rubinstein AM, Nussbaum G, Elkin M. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget. 2015;6(26):22613.
Jia G, Zhi A, Lai P, Wang G, Xia Y, Xiong Z, Zhang H, Che N, Ai L. The oral microbiota–a mechanistic role for systemic diseases. Br Dent J. 2018;224(6):447–55.
Meurman JH. Infectious and dietary risk factors of oral cancer. Oral Oncol. 2010;46(6):411–3.
Kim SY, Kim J-E, Lee KW, Lee HJ. Lactococcus lactis ssp. lactis inhibits the proliferation of SNU-1 human stomach cancer cells through induction of G0/G1 cell cycle arrest and apoptosis via p53 and p21 expression. Ann New York Academy of Sciences. 2009;1171(1):270.
Chen Y, Peng Y, Yu J, Chen T, Wu Y, Shi L, Li Q, Wu J, Fu X. Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget. 2017;8(19):31802–14.
Tang B, Wang K, Jia Y-P, Zhu P, Fang Y, Zhang Z-J, Mao X-H, Li Q, Zeng D-Z. Fusobacterium nucleatum-induced impairment of Autophagic flux enhances the expression of Proinflammatory cytokines via ROS in Caco-2 cells. PLoS One. 2016;11(11):e0165701–1.
Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017;152(4):851–66 e824.
Han YW. Commentary: Oral bacteria as drivers for colorectal cancer. J Periodontol. 2014;85(9):1155–7.
Lam S, Yu J, Wong S, Peppelenbosch M, Fuhler G. The gastrointestinal microbiota and its role in oncogenesis. Best Pract Res Clin Gastroenterol. 2017;31(6):607–18.
Mathy-Hartert M, Hogge L, Sanchez C, Deby-Dupont G, Crielaard J-M, Henrotin Y. Interleukin-1β and interleukin-6 disturb the antioxidant enzyme system in bovine chondrocytes: a possible explanation for oxidative stress generation. Osteoarthr Cartil. 2008;16(7):756–63.
Murata M, Thanan R, Ma N, Kawanishi S. Role of nitrative and oxidative DNA damage in inflammation-related carcinogenesis. J Biomed Biotechnol. 2012;2012.
Kossakowska AE, Edwards DR, Prusinkiewicz C, Zhang MC, Guo D, Urbanski SJ, Grogan T, Marquez LA, Janowska-Wieczorek A. Interleukin-6 regulation of matrix metalloproteinase (MMP-2 and MMP-9) and tissue inhibitor of metalloproteinase (TIMP-1) expression in malignant non-Hodgkin’s lymphomas. Blood, The Journal of the American Society of Hematology. 1999;94(6):2080–9.
Voronov E, Shouval DS, Krelin Y, Cagnano E, Benharroch D, Iwakura Y, Dinarello CA, Apte RN. IL-1 is required for tumor invasiveness and angiogenesis. Proc Natl Acad Sci. 2003;100(5):2645–50.
Wang F-m, H-q L, S-r L, S-p T. Yang L, Feng G-s: SHP-2 promoting migration and metastasis of MCF-7 with loss of E-cadherin, dephosphorylation of FAK and secretion of MMP-9 induced by IL-1 βin vivo andin vitro. Breast Cancer Res Treat. 2005;89(1):5–14.
Bradley J. TNF-mediated inflammatory disease. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland. 2008;214(2):149–60.
Rivas MA, Carnevale RP, Proietti CJ, Rosemblit C, Beguelin W, Salatino M, Charreau EH, Frahm I, Sapia S, Brouckaert P. TNFα acting on TNFR1 promotes breast cancer growth via p42/P44 MAPK, JNK, Akt and NF-κB-dependent pathways. Exp Cell Res. 2008;314(3):509–29.
Yan B, Wang H, Rabbani ZN, Zhao Y, Li W, Yuan Y, Li F, Dewhirst MW, Li C-Y. Tumor necrosis factor-α is a potent endogenous mutagen that promotes cellular transformation. Cancer Res. 2006;66(24):11565–70.
Leber T, Balkwill F. Regulation of monocyte MMP-9 production by TNF-α and a tumour-derived soluble factor (MMPSF). Br J Cancer. 1998;78(6):724–32.
Steffen M, Holt S, Ebersole JL. Porphyromonas gingivalis induction of mediator and cytokine secretion by human gingival fibroblasts. Oral Microbiol Immunol. 2000;15(3):172–80.
Park HE, Kim JH, Cho N-Y, Lee HS, Kang GH. Intratumoral Fusobacterium nucleatum abundance correlates with macrophage infiltration and CDKN2A methylation in microsatellite-unstable colorectal carcinoma. Virchows Arch. 2017;471(3):329–36.
Uitto V-J, Baillie D, Wu Q, Gendron R, Grenier D, Putnins EE, Kanervo A, Firth JD. Fusobacterium nucleatum increases collagenase 3 production and migration of epithelial cells. Infect Immun. 2005;73(2):1171–9.
Wu Y, Wu J, Chen T, Li Q, Peng W, Li H, Tang X, Fu X. Fusobacterium nucleatum potentiates intestinal tumorigenesis in mice via a toll-like receptor 4/p21-activated kinase 1 cascade. Dig Dis Sci. 2018;63(5):1210–8.
Gholizadeh P, Eslami H, Yousefi M, Asgharzadeh M, Aghazadeh M, Kafil HS. Role of oral microbiome on oral cancers, a review. Biomed Pharmacother. 2016;84:552–8.
Yilmaz Ö, Jungas T, Verbeke P, Ojcius DM. Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect Immun. 2004;72(7):3743–51.
Mao S, Park Y, Hasegawa Y, Tribble GD, James CE, Handfield M, Stavropoulos MF, Yilmaz Ö, Lamont RJ. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 2007;9(8):1997–2007.
Yilmaz Ö, Yao L, Maeda K, Rose TM, Lewis EL, Duman M, Lamont RJ, Ojcius DM. ATP scavenging by the intracellular pathogen Porphyromonas gingivalis inhibits P2X7-mediated host-cell apoptosis. Cell Microbiol. 2008;10(4):863–75.
Whitmore SE, Lamont RJ. Oral bacteria and cancer. PLoS Pathog. 2014;10(3):e1003933.
Yao Á, Jermanus C, Barbetta B, Choi C, Verbeke P, Ojcius DM, Yilmaz Ö. Porphyromonas gingivalis infection sequesters pro-apoptotic Bad through Akt in primary gingival epithelial cells. Molecular oral microbiology. 2010;25(2):89–101.
Choi CH, Spooner R, DeGuzman J, Koutouzis T, Ojcius DM, Yilmaz Ö. P orphyromonas gingivalis-nucleoside-diphosphate-kinase inhibits ATP-induced reactive-oxygen-species via P 2 X 7 receptor/NADPH-oxidase signalling and contributes to persistence. Cell Microbiol. 2013;15(6):961–76.
Spooner R, Yilmaz Ö. The role of reactive-oxygen-species in microbial persistence and inflammation. Int J Mol Sci. 2011;12(1):334–52.
Abranches J, Zeng L, Kajfasz JK, Palmer S, Chakraborty B, Wen Z, Richards VP, Brady LJ, Lemos JA. Biology of oral streptococci. Gram-Positive Pathogens. 2019:426–34.
Brauncajs M, Sakowska D, Krzeminski Z. Production of hydrogen peroxide by lactobacilli colonising the human oral cavity. Med Dosw Mikrobiol. 2001;53(4):331–6.
Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat Rev Cancer. 2003;3(4):276–85.
Milella L. The negative effects of volatile Sulphur compounds. J Vet Dent. 2015;32(2):99–102.
Attene-Ramos MS, Wagner ED, Plewa MJ, Gaskins HR. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res. 2006;4(1):9–14.
Singh SB, Lin HC. Hydrogen sulfide in physiology and diseases of the digestive tract. Microorganisms. 2015;3(4):866–89.
Muto M, Hitomi Y, Ohtsu A, Shimada H, Kashiwase Y, Sasaki H, Yoshida S, Esumi H. Acetaldehyde production by non-pathogenic Neisseria in human oral microflora: implications for carcinogenesis in upper aerodigestive tract. Int J Cancer. 2000;88(3):342–50.
Pavlova SI, Jin L, Gasparovich SR, Tao L. Multiple alcohol dehydrogenases but no functional acetaldehyde dehydrogenase causing excessive acetaldehyde production from ethanol by oral streptococci. Microbiology. 2013;159(Pt 7):1437.
Marttila E, Bowyer P, Sanglard D, Uittamo J, Kaihovaara P, Salaspuro M, Richardson M, Rautemaa R. Fermentative 2-carbon metabolism produces carcinogenic levels of acetaldehyde in C andida albicans. Mol Oral Microbiol. 2013;28(4):281–91.
Downes J, Wade WG. Peptostreptococcus stomatis sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol. 2006;56(4):751–4.
Lunt SJ, Chaudary N, Hill RP. The tumor microenvironment and metastatic disease. Clinical & experimental metastasis. 2009;26(1):19–34.
Mazzio EA, Smith B, Soliman KF. Evaluation of endogenous acidic metabolic products associated with carbohydrate metabolism in tumor cells. Cell Biol Toxicol. 2010;26(3):177–88.
Karpiński TM, Szkaradkiewicz AK. Characteristic of bacteriocines and their application. Pol J Microbiol. 2013;62(3):223–35.
Senneby A, Davies J, Svensäter G, Neilands J. Acid tolerance properties of dental biofilms in vivo. BMC Microbiol. 2017;17(1):165.
Kim J-E, Kim M-S, Yoon Y-S, Chung M-J, Yum D-Y. Use of selected lactic acid bacteria in the eradication of helicobacter pylori infection. J Microbiol. 2014;52(11):955–62.
Han KJ, Lee N-K, Park H, Paik H-D. Anticancer and anti-inflammatory activity of probiotic Lactococcus lactis NK34. J Microbiol Biotechnol. 2015;25:1697–701.
Kanayama M, Kato Y, Tsuji T, Konoeda Y, Hashimoto A, Kanauchi O, Fujii T, Fujiwara D. Enhancement of immunomodulative effect of lactic acid bacteria on plasmacytoid dendritic cells with sucrose palmitate. Sci Rep. 2018;8(1):1–12.
Sonveaux P, Végran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, De Saedeleer CJ, Kennedy KM, Diepart C, Jordan BF. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118(12):3930–42.
Salazar CR, Francois F, Li Y, Corby P, Hays R, Leung C, Bedi S, Segers S, Queiroz E, Sun J. Association between oral health and gastric precancerous lesions. Carcinogenesis. 2012;33(2):399–403.
Lockhart PB, Brennan MT, Sasser HC, Fox PC, Paster BJ, Bahrani-Mougeot FK. Bacteremia associated with tooth brushing and dental extraction. Circulation. 2008;117(24):3118.
Abnet CC, Kamangar F, Dawsey SM, Stolzenberg-Solomon RZ, Albanes D, Pietinen P, Virtamo J, Taylor PR. Tooth loss is associated with increased risk of gastric non-cardia adenocarcinoma in a cohort of Finnish smokers. Scand J Gastroenterol. 2005;40(6):681–7.
Al Asqah M, Al Hamoudi N, Anil S, Al-hamoudi WK. Is the presence of helicobacter pylori in the dental plaque of patients with chronic periodontitis a risk factor for gastric infection? Can J Gastroenterol. 2009;23.
Abnet CC, Qiao Y-L, Mark SD, Dong Z-W, Taylor PR, Dawsey SM. Prospective study of tooth loss and incident esophageal and gastric cancers in China. Cancer Causes Control. 2001;12(9):847–54.
Acknowledgements
Not applicable.
Funding
The National Institute for Medical Research Development (NIMAD) (grant number 958117), Tehran, Iran. The supporter had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
S.L-N. provided direction in the preparation of the manuscript. S.Z.B. performed primary literature search. S.Z.B. wrote the first draft of manuscript. S.L-N. discussed and revised the manuscript. S.Z.B. managed the references. S.L-N. approved the version to be published. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
No potential conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bakhti, S.Z., Latifi-Navid, S. Oral microbiota and Helicobacter pylori in gastric carcinogenesis: what do we know and where next?. BMC Microbiol 21, 71 (2021). https://doi.org/10.1186/s12866-021-02130-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-021-02130-4