Abstract
Background
Streptomycetes from the rhizospheric soils are a rich resource of novel secondary metabolites with various biological activities. However, there is still little information related to the isolation, antimicrobial activity and biosynthetic potential for polyketide and non-ribosomal peptide discovery associated with the rhizospheric streptomycetes of Panax notoginseng. Thus, the aims of the present study are to (i) identify culturable streptomycetes from the rhizospheric soil of P. notoginseng by 16S rRNA gene, (ii) evaluate the antimicrobial activities of isolates and analyze the biosynthetic gene encoding polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs) of isolates, (iii) detect the bioactive secondary metabolites from selected streptomycetes, (iv) study the influence of the selected isolate on the growth of P. notoginseng in the continuous cropping field. This study would provide a preliminary basis for the further discovery of the secondary metabolites from streptomycetes isolated from the rhizospheric soil of P. notoginseng and their further utilization for biocontrol of plants.
Results
A total of 42 strains representing 42 species of the genus Streptomyces were isolated from 12 rhizospheric soil samples in the cultivation field of P. notoginseng and were analyzed by 16S rRNA gene sequencing. Overall, 40 crude cell extracts out of 42 under two culture conditions showed antibacterial and antifungal activities. Also, the presence of biosynthesis genes encoding type I and II polyketide synthase (PKS I and PKS II) and nonribosomal peptide synthetases (NRPSs) in 42 strains were established. Based on characteristic chemical profiles screening by High Performance Liquid Chromatography-Diode Array Detector (HPLC-DAD), the secondary metabolite profiles of strain SYP-A7257 were evaluated by High Performance Liquid Chromatography-High Resolution Mass Spectrometry (HPLC-HRMS). Finally, four compounds actinomycin X2 (F1), fungichromin (F2), thailandin B (F7) and antifungalmycin (F8) were isolated from strain SYP-A7257 by using chromatography techniques, UV, HR-ESI-MS and NMR, and their antimicrobial activities against the test bacteria and fungus were also evaluated. In the farm experiments, Streptomyces sp. SYP-A7257 showed healthy growth promotion and survival rate improvement of P. notoginseng in the continuous cropping field.
Conclusions
We demonstrated the P. notoginseng rhizospheric soil-derived Streptomyces spp. distribution and diversity with respect to their metabolic potential for polyketides and non-ribosomal peptides, as well as the presence of biosynthesis genes PKS I, PKS II and NRPSs. Our results showed that cultivatable Streptomyces isolates from the rhizospheric soils of P. notoginseng have the ability to produce bioactive secondary metabolites. The farm experiments suggested that the rhizospheric soil Streptomyces sp. SYP-A7257 may be a potential biological control agent for healthy growth promotion and survival rate improvement of P. notoginseng in the continuous cropping field.
Similar content being viewed by others
Background
The rhizosphere is a habitat where a high abundance of bacteria, actinobacteria and fungi, is observed. Those microbes are recruited by plant exudates released through the roots [1]. In this environment, they are exposed to a permanent state of competition; the production of specific antibiotics may play a decisive role in helping them survive within the rhizosphere microbial community [2]. Among them, it has been revealed that streptomycetes could help to promote the growth of host plants as well as to improve disease symptoms caused by plant pathogens through diverse mechanisms, including the production of bioactive metabolites, which are applied in the direct antagonism against pests and diseases [3, 4], in the changes of host physiological function [5], in the induction of host systemic acquired resistances [6], and so on. Among these characteristics, a significant common function for Streptomyces is the antibiotics production [7], which suggests that streptomycetes play a key role in the plant defense and are widely recognized for their potentials in the biocontrol [2]. For example, Blasticidin S, produced by Streptomyces griseochromogenes, was the first commercial antibiotic used in the control of rice blast in Japan [8]. Streptomyces sp. IA1, producing an antimicrobial actinomycin D, was isolated from Saharan soil in Amenas, Algeria. Moreover, strain IA1 and actinomycin D effectively reduced two fungal diseases, i.e., the Fusarium wilt of flax and chocolate spot of field bean [9]. Streptomyces sp. Acta 3034 from the rhizospheric soil of Clitorea sp., produced five new angucycline antibiotics, langkocyclines A1-A3, B1 and B2, which exhibited biological activities against Gram-positive bacteria and moderate inhibitory activities against several human tumor cell lines [10]. Streptomycetes may also promote the growth of the plant hosts by producing auxins that improve the root growth [11]. Therefore, members of the genus Streptomyces from rhizospheric soils are still a rich resource of secondary metabolites with various biological activities.
A diverse array of bioactive molecules are synthesized by the type I polyketide synthases (PKS I), type II polyketide synthase (PKS II or aromatic polyketide synthase) and nonribosomal peptide synthetases (NRPS) [12]. Degenerate PCR primers targeting on KS domain of PKS I and PKS II and A domain of NRPS have been applied into the screening of PKS and NRPS systems from sea sediments [13], cultured microorganisms including actinobacteria [14,15,16] and cyanobacteria [17]. To the best of our knowledge, data related to the functional genes responsible for secondary metabolites production in the soil-associated streptomycetes from the rhizosphere of Panax notoginseng are limited.
Panax notoginseng F. H. Chen (Araliaceae), named as Sanqi or Tianqi in China, is a well-known traditional Chinese medicine, which has wide application in promotion of blood circulation, removal of blood stasis, cure of cardiovascular diseases [18, 19]. The high-quality roots of P. notoginseng are obtained after growth for at least three years [20]. The long growth period easily results in the infection of soil-borne pathogens [21]. Up to now, several previous studies have been performed to investigate the rhizospheric soil and root endogenous microbial diversity during P. notoginseng cropping [22, 23]. In our long-term study, many bioactive bacteria and fungi with the production of many bioactive metabolites have been isolated from the environments related to P. notoginseng [24,25,26,27]. However, very little is known about the isolation, antimicrobial activity and biosynthetic potential for polyketide and non-ribosomal peptide discovery associated with the rhizospheric streptomycetes of P. notoginseng. Thus, the aims of the present study are to (i) identify culturable Streptomyces isolates from the rhizospheric soil of P. notoginseng by 16S rRNA gene, (ii) evaluate the antimicrobial activities and analyze the biosynthetic gene encoding polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs) of isolates, (iii) detect the bioactive secondary metabolites from the selected Streptomyces isolate, (iv) study the influence of the selected strain on the growth of P. notoginseng in the continuous cropping field. This study will provide a basis for the further discovery of the secondary metabolites from streptomycetes from the rhizospheric soil of P. notoginseng and their further utilization for biocontrol of plants.
Results
Diversity of streptomycetes from the rhizospheric soil of P. notoginseng
The method of Serial dilution spread plate was used to isolate actinobacteria from the rhizospheric soil of P. notoginseng. After 1–4 weeks, individual colonies were picked from the plates based on the morphological characteristics of aerial hyphae and substrate mycelia, into the fresh plates to obtain pure cultures. To further ascertain the taxonomic status of the isolates, the 16S rRNA gene was PCR-amplified and sequenced. Forty two representative isolates belonging to the genus of Streptomyces were selected based on 16S rRNA gene sequencing (Table S3). The percentages of 16S rRNA gene identity to the closest type strains were presented in Table S3. The 42 strains belonged to the genus of Streptomyces with the identities from 97.24 to 100% (Table S3). Among them, 34 isolates (81%) were from the rhizospheric soil of healthy P. notoginseng, whereas 4 isolates (9.5%) from the rhizospheric soil of root-rot P. notoginseng and 4 isolates (9.5%) from the rhizospheric soil with serious nematodes infection. The culturable Streptomyces strains showed more abundance in the healthy rhizospheric soil (81%) than in the sick soil (19%). On the other hand, the 16S rRNA gene sequences of strain SYP-A7053, SYP-A7096 and SYP-A8135 showed 97.24, 98.83 and 98.44% identity to S. viridosporus ATCC 27479 (DQ442556), S. mexicanus DSM 41796 (AB249966) and S. caniferus ATCC 43699 (AB184640), respectively. These strains are most likely to represent new species, and taxonomic studies are underway. These data indicated the considerable diversity of species grouped within the genus of Streptomyces isolated from the rhizospheric soil of P. notoginseng.
Antimicrobial activity of the fermented extracts
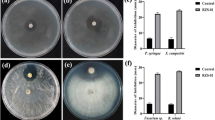
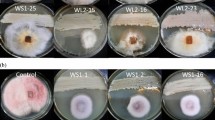
All 42 streptomycetes were cultivated in two fermentation media, GYM4 and SPM1. 40 (95%) of 42 fermented extracts exhibited strong and moderate activities against at least one of the tested pathogens (Table 1). In addition, the extract of each strain from different fermentation broth exhibited obviously different activity. For example, only 29 (69%) of 42 extracts from GYM4 fermentation showed activities against at least one of the test pathogens, whereas 40 (95%) of 42 extracts from SPM1 fermentation displayed activities against at least one of the tested pathogens. Furthermore, most extracts from SPM1 displayed stronger activities than those from GYM4. In terms of the inhibition activity against A. baumannii, 16 strains (38%) in SPM1 exhibited inhibition activities, while only 1 strain (2.3%) in GYM4 had the same antagonistic activity. Moreover, whichever media was used, three isolates, SYP-A7053, SYP-A7193 and SYP-A7257, appeared to have a broad spectrum of antimicrobial activities. The strong inhibitory activities of these strains against the tested pathogens suggested that these soil-derived streptomycetes may be potential candidates for the production of bioactive metabolites under the optimal cultivation condition. These results also revealed that Streptomyces isolates and their metabolites with antagonistic activity may play an important biological protective role on the healthy growth of P. notoginsen.
Distribution and phylogenetic analysis of biosynthetic genes
All 42 Streptomyces isolates were screened for the presence of KS domain related to PKS I and PKS II genes and A domain associated with NRPS genes. In total, 41 (98%) of 42 strains yielded sequence-verified genes related to at least one of three targeted genes (Table 1). More specifically, PKS I loci were detected in 30 strains (71%), PKS II loci in 38 strains (90%) and NRPS loci in 7 strains (17%). Meanwhile, 30 strains (71%) had at least two types of three genes, and 4 strains (9%), SYP-A7076, SYP-A7087, SYP-A7255 and SYP-A7750 possessed three types of the biosynthetic genes simultaneously (Table 1). These results suggested that the biosynthetic genes of PKS I and PKS II were widespread in the rhizospheric soil-derived streptomycetes of P. notoginseng.
Using BlastX search of GenBank, the PKS I sequence shared 93–100% sequence similarity with their closest matches (Table S4). Among them, KS sequences of 20 strains showed the high similarities at the amino acid level with that of Bacillus sp. from 98 to 100% (Table S4). PKS-I sequences retrieved from SYP-A7200, SYP-A7257 and SYP-A7283 clustered with a gene product (MerC) involved in the production of meridamycin [28] with the bootstrap value of 90% (Fig. 1). In addition, PKS-I sequences from Streptomyces strains SYP-A8195 showed the highest identity (96%) to the PKS-I gene responsible for biosynthesizing thiotetronate antibiotics [29], forming a separate clade with the bootstrap value of 100% (Fig. 1, Table S4).
Neighbor-joining tree of KS amino acid sequences of isolates from this study and reference sequences of known representative PKS I. The scale bar indicates 0.20 substitutions that occur per site. Percentage bootstrap values of neighbor-joining analysis from 1000 resamplings are indicated at the nodes
The vast majority (38 isolates, 90%) of the KS sequences are most closely related to modular PKS II genes according to amino acid identities and phylogenetic analyses (Fig. 2, Table S4). All of the 38 KS sequences had the amino acid identities ranging from 77 to 100% (Table S4). Among them, SYP-A7131 and SYP-A7750 shared 90 and 94% amino acid identities (Table S4) with the sequence involved in the production of actinorhodin [30], respectively, and also formed the separated clade supported by 80% of bootstrap values. The A domains related to NRPS were sequenced for analyzing their phylogenetic relationships (Table S4). Only 7 sequences had the correspondence BLAST matches to the A domain with the amino acid identities ranging from 92 to 97%.
Neighbor-joining tree of KS amino acid sequences of isolates from this study and reference sequences of known representative PKS II. The scale bar indicates 0.10 substitutions that occur per site. Percentage bootstrap values of neighbor-joining analysis from 1000 resamplings are indicated at the nodes
Secondary metabolite profiles of fermentation extracts of the isolates [44]
Based on the functional gene screening and antimicrobial activities, 8 strains were selected for small-scale fermentation and HPLC-DAD analysis, among which 6 strains (SYP-A7200, SYP-A7257, SYP-A7283, SYP-A7131, SYP-A7750, SYP-A8195) (marked with * in Table S3) grouped with the described functional chemical classes in phylogenetic relationship analysis and 2 strain, SYP-A7053 and SYP-A7193, had a broad spectrum of antimicrobial activities. Under investigating the spectra of culture extracts by HPLC–DAD, strain SYP-A7257 exhibited a characteristic chemical profile differing from another 5 strains with characteristic UV/Vis absorption at bands of 325 to 360 nm and 241 to 446 nm, indicating the chromophore of typical polyene macrolide and of typical actinomycin analogues, respectively (Figure S1). Therefore, the extract of strain SYP-A7257 was analyzed by HPLC-HRMS to predict the numbers and diversity of compounds with respect to the presence of PKS and NRPS biosynthetic genes. HPLC-HRMS analysis of the extract of strain SYP-A7257 (Figure S2, S3-S8) showed the total ion current (TIC) chromatogram with six main peaks in the positive mode at m/z 1269.6146 (F1), 693.3818 (F2), 1293.6149 (F3), 1277.6161 (F4), 665.3505 (F5) and 709.3771 (F6), respectively (Table 2). According to the UV/Vis, MS and restricting to known compounds produced by Streptomyces spp., these compounds could be preliminarily identified as follows: F1 as actinomycin X2, F2 as fungichromin, F3 as actinomycin X0β, F4 as actinomycin D, F5 as 1′,14-dihydroxyisochainin, F6 as hydrofungichromin, respectively (Fig. 3 and Table 2). Among them, compound F6 represents yet unknown derivative of fungichromin with [M + Na]+ at m/z 709.3771 (C35H58NaO13), indicating that compound F6 possessed an additional hydroxyl group compared to fungichromin (C35H58NaO12, [M + Na]+ at m/z 693.3818). This compound did not match with any published compounds in spite of having the same UV spectra with that of fungichromin (Figure S8). Thus, strain SYP-A7257 was selected as the target strain for further chemical and bioactive characterization.
Identification of secondary metabolites and their antimicrobial activities
To further confirm chemical structures of metabolic products, a serial of separation and purification were performed in the crude extract of strain SYP-A7257, and two compounds, corresponding to the HPLC-HRMS analysis, were obtained. Unfortunately, compound F6 was not obtained because of its low production in the extract. Compound F1 (Fig. 3) was isolated as a dark red amorphous powder with molecular weight of 1269.0 displaying maximal absorbance at 241 nm (shoulder) and 446 nm similar to those of known actinomycin derivates (Fig. S1). ESI-MS of this compound revealed molecular ion peak at m/z 1292.0 [M + Na]+ (Fig. S9), which was identical to that of actinomycin X2 further confirmed on the basis of 1H NMR and 13C NMR (Figure S10 and S11; Table S5) [33]. The other compound F2 (Fig. 3) exhibited a typical characteristic of polyene macrolides spectra (λmax 360, 330 and 325 nm) (Fig. S1). ESI-MS analysis suggested its molecular masses to be m/z 669.4 [M-H]− (Fig. S12) and revealed its molecular formula to be C35H58O12, according to the 1H and 13C NMR spectral data (Figure S13 and S14; Table S6). Thus, compound F2 was identified as a known polyene antibiotics fungichromin [32].
To attain more antimicrobial compounds from strain SYP-A7257, a larger scale (10 L) extraction from culture grown in SPM1 medium was performed and 30.0 g of the crude material was attained. Compounds F7 and F8 were obtained under a series of separation and purification procedures. The structures of F7 and F8 (Fig. 3) were determined by using data from MS, and NMR techniques (Table S7). Compound F7 was obtained as a pale yellowish powder. The molecular mass of F7 was determined by ESI-MS, which gave the mass of m/z 607.4 [M-H]− (Fig. S15) and revealed its molecular formula to be C33H52O10 in accord with the 1H and 13C NMR spectral data (Fig. S16 and S17; Table S7). Thus, compound F7 was identified as a known polyene macrolide thailandin B [34]. Compound F8 was identified as a polyene macrolide by a diode array detector (DAD)/UV spectra with three characteristic λmax at 289, 303, 319 nm. Positive ESI-MS of F8 showed the molecular ion [M + H]+ at m/z 705.5 (Fig. S18) and revealed its molecular formula C35H60O14 in accord with the 1H and 13C NMR spectral data (Figure S19 and S20; Table S7). Thus, compound F8 was identified as a known polyene macrolide antifungalmycin [35].
The results from the antimicrobial assays showed that compound F1 displayed strong activity against S. aureus with MIC value of 0.3 μg/ml and weak activities against other test strains (Table 3). Compounds F2, F7 and F8 exhibited weak activities against all of the test bacteria. However, they had strong activities against pathogenic fungi C. albicans with MIC values of 1.24, 1.25 and 1.10 μg/ml, respectively (Table 3).
The farm experiment results
The seedling survival and healthy growth of P. notoginseng in the continuous cropping field are very important for P. notoginseng planting. The farm experiment showed that the plants in SYP-A7257 treatment (T) group grew healthily with high stems and wide leaves of dark green, while plants in control (CK) group grew slowly and were lack of vigor. The dead seedlings observed usually presented as root-rot diseases. The average survival rates in T groups at every recorded time (30, 60, 90 and 120 days) were significantly higher than those in CK groups (P < 0.05). On 120 days after the first-spray, the average emergence rate in T plots was 83.0%, while 68.0% in CK plots (Table 4), indicating that strain SYP-A7257 may have protective effects on the healthy growth of P. notoginseng against root-rot disease in the continuous cropping field.
Discussion
Although members of Streptomyces have long been extensively screened for bioactive compounds, the emergence of drug-resistant bacteria and the relative ease of generating genome sequences have revived the importance of streptomycetes as producers of new natural products with various biological activities and resulted in renewed efforts in isolating these strains from the untapped environments [36]. Streptomycetes are Gram-positive bacteria ubiquitously living in soil, where they are the largest genus of Streptomycetaceae family (order Actinomycetales), consisting of nearly 600 formally described species [37], http://www.ezbiocloud.net). Many species are not pathogenic to human or the plants [36]. On the contrary, streptomycetes have ability to produce many enzymes and bioactive secondary metabolites [7]. All these substances give them potential for application in plant growth promotion and the biocontrol [36]. Thus, it has been assumed that high levels of antagonistic streptomycetes derived from naturally-occurring soils have a significant effect on the disease suppression [38]. Thus, antagonistic streptomycetes derived from rhizospheric soil are still an important source of novel natural products for chemical defense of plants. P. notoginseng as perennial plants is mainly cultivated for several years, which leads to infection by soil-derived pathogens including fungi, bacteria and nematodes [21]. There are many studies about exploring diversity of microorganisms from different environments and investigating the antagonistic activity of bacteria isolated from those environments, such as Bacillus spp. used as biocontrol agents [21,22,23]. However, the comprehensive investigation has been rarely performed on the isolation, antimicrobial activity and biosynthetic potential for polyketides and non-ribosomal peptide discovery associated to the Streptomyces strains, residing in the rhizospheric soil of Panax notoginseng.
In the present study, we have obtained 42 streptomycetes strains from 12 rhizospheric soil samples of P. notoginseng, and then these strains were subjected to the 16S rRNA gene analysis to ascertain their taxonomic relationships (Table S3). The culturable streptomycetes strains showed more abundance in the rhizospheric soil of healthy P. notoginseng (81%) than that of sick P. notoginseng (19%), which indirectly implied that the most species of the genus Streptomyces are beneficial to plant health [39]. To our knowledge, this is the first research of culturable Streptomyces strains from soil environments of P. notoginseng in Wenshan region of Yunnan province in China.
It is important to explore these streptomycetes having antibacterial and antifungal activities as producers of novel natural products or for direct use as biocontrol in soil environments of P. notoginseng [36]. Therefore, these isolates were evaluated for their antagonistic activities under two culture conditions. The antimicrobial experiments showed most of the Streptomyces strains gave high percentage of inhibition to all the tested pathogens. For example, isolate SYP-A7053 identified as Streptomyces viridosporus showed significant antifungal activity against F. solani (11.8 mm). Similarly, isolate SYP-A7193 isolated from the rhizospheric soil of the healthy P. notoginseng, showed significant antimicrobial activity against S. aureus, E. coli, E. faecium, P. mirabilis, A. baumannii and A. marplatensis. Interestingly, when employing medium SPM1 as fermentation broth, these isolates displayed higher inhibition activity against all the tested pathogens than those cultivated in medium GMY4. Similar finding was reported by Wu et al. (2018) [40], who used a cultivation-dependent procedure to change media type, resulting in the discovery of inthomycin B from a marine-derived Streptomyces sp. YB104. A recent study also revealed that a near-shore marine intertidal zone-associated Streptomyces strain (USC-633) was cultivated under a series of complex media to determine optimal parameters leading to the secretion of various array of secondary metabolites with antimicrobial activities against diversity drug-resistant bacteria [41]. Thus, our study indicates that these streptomycetes have potential ability to synthesize bioactive metabolites as defensive substances under different culture conditions. Their antagonistic activities to the plant pathogenic bacteria (A. marplatensis) and fungi (F. solani) also play a solid foundation for these soil-derived Streptomyces strains as biocontrol agents for P. notoginseng cultivation in the future. Thus, the selected strain SYP-A7257, which produced the antibiotics actinomycin and polyene macrolides derivates with the antibacterial and antifungal activity, respectively, was applied to study the biocontrol for P. notoginseng growth in the continuous cropping field. To the best of our knowledge, continuous cropping obstacles significantly affect P. notoginseng survival and cause severe mortality to plants [42]. In our farm experiments with continuous cropping soil, Streptomyces SYP-A7257 had showed desirable biocontrol potential for P. notoginseng healthy growth and improvement of the survival rates compared to CK plots without strain spray (Table 4), which suggested that Streptomyces sp. SYP-A7257 may be a promising biocontrol agent against root-rot disease to promote plants healthy growth because of the chemical defense substances secreted by this strain.
To the best of our knowledge, the majority of compounds from actinobacteria are demonstrated to be complex polyketides and non-ribosomal peptides [43]. It may be hypothesized that a genome with a vast number of biosynthetic gene clusters is more likely to lead to a positive hit under an PKS/NRPS gene screening procedure [43]. In our study, PCR screening of 42 soil-derived streptomycetes revealed that genes associated with PKS I and PKS II biosynthesis were widespread and generally distributed among different species of the genus Streptomyces, which was consistent with the viewpoint of Schneemann et al. (2010) [43]. Therefore, we firstly predicted that positive results in the gene screening of NRPS and PKS-II genes could provide the evidences of the corresponding metabolites. However, it is known that genome-sequencing projects on different actinobacteria have revealed a large number of biosynthetic gene clusters in each genome [44]. This viewpoint is proved by the genome information of Streptomyces peucetius ATCC 27952, possessing 68 biosynthetic gene clusters (BCGs) for various types of secondary metabolites, including non-ribosomal peptide synthase (NRPS), polyketide synthase (PKS I, II, and III), terpenes, and others [45]. Hence, the chosen primer system used in this work, in spite of favorable for the great majority of known PKS/NRPS genes, may not work in all cases of polyketides and nonribosomal peptides with uncommon molecular constructions. For example, strain SYP-A7257 exhibiting no NRPS amplicons produced actinomycin derivatives synthesized via a NRPS biosynthetic gene cluster [46, 47]. Meanwhile, the detected NRPS or PKS gene fragments may not clearly assure the presence of the corresponding metabolites. In the study of Schneemann et al. (2010) [43], 10 of the strains yielded PCR products with the PKS-specific primers, but no corresponding PKS products were detected under the laboratory conditions. Within our study, strain SYP-A7257 produced compounds F1 (fungichromin) and F2 (actinomycin derivates) synthesized via PKS and NRPS gene clusters [46, 47], respectively, which did not match the obtained PKS genes involved in biosynthesis of meridamycin (PKSI) [28] in the functional gene screening. It is possible that the detected biosynthetic gene clusters are cryptic under laboratory condition, which leads to a low efficiency in the discovery of their corresponding metabolites [44]. It is also possible that the detected biosynthetic gene clusters are nonfunctional. Nevertheless, the PCR-based prescreening of isolates with the primers targeting on genes involving in the biosynthesis of secondary metabolites is an effective approach for detecting useful bioactive compounds [43, 48, 49].
To our knowledge, the genes encoding PKS and NRPS systems from these Streptomyces isolates might both play a key role in producing secondary metabolites with antagonistic activities against plant pathogenic microorganisms [50]. This opinion was similar to the studies of Sharma et al. (2016) [51] and Passari et al. (2015) [52], which demonstrated that streptomycetes having antimicrobial activities were positive for the presence of the genes related to these two biosynthetic pathways in their genomes because genes encoding PKS-I and NRPS might be responsible for both of their involvement in the regulation of antimicrobial activity in microorganisms. Within the present study, the presence of the biosynthetic genes PKS and NRPS in the rhizospheric streptomycetes isolates related to P. notoginseng suggests that these beneficial strains may be the important producers of bioactive secondary metabolites. For example, strain SYP-A7257 having a positive hit in PKS gene screening produced eight antagonistic compounds consisting of one so far unidentified compound, among which four isolated compounds exhibited favorable activities against the test bacteria and fungi. Meanwhile, compounds F1 and F2 both displayed antagonistic activities to the plant pathogenic bacteria (A. marplatensis) with MIC values of 14.6 and 49.4 μg/ml, respectively. These results indicate that these rhizospheric streptomycetes may be good candidates for biocontrol agents of P. notoginseng because immense diversity of PKS genes detected could indicate a diversity of antimicrobial secondary metabolites.
Conclusion
In summary, this research demonstrates the distribution and diversity of the P. notoginseng rhizospheric soil-derived streptomycetes in respect to their metabolic potential for polyketides and non-ribosomal peptides. Many questions remain in respect to the ecological function of Streptomyces strains in the rhizospheric environment, their evolution and biogeographic distribution. However, a diversity of streptomycetes and their potential to produce bioactive secondary metabolites suggested that these Streptomyces isolates might represent a valuable resource of bioactive secondary metabolites with antimicrobial activities and genes encoding their biosynthesis. That potential should not be overlooked. Meanwhile, the farm experiments indicated that Streptomyces sp. SYP-A7257 can be used as a promising biocontrol agent for promoting the healthy growth of P. notoginseng due to the chemical defense substances produced from this strain.
Methods
Soil sample collection and isolation of actinobacteria strains
Twelve rhizospheric soil samples in the cultivation field of P. notoginseng were collected in the Wenshan region of Yunnan Province (Table S1). After removing the most amount of soil, the residue rhizospheric soil was gently stripped and collected around 1 cm from the main roots. Three parallel soil samples at each site were mixed to obtain one sample. All the soil samples were sieved (< 4 mm) to remove stones and plant residues and then were placed into sterile bags. Finally, they were kept at 4 °C before treatment.
Actinobacteria were isolated by using serial dilution spread plate method. Five grams of soil was mixed with 45 ml of sterile 0.9% NaCl and stirred for 1 h at 25 °C. The soil suspension stood for 10 min at room temperature to allow precipitation of large particles. Serial 10-fold dilutions (from 10− 3 to 10− 5) were made with 0.9% NaCl. 100 μl of diluents were transferred to Petri dishes with 38# agar medium (4.0 g yeast extract, 2.0 g malt flour, 4.0 g glucose, 1.0 ml trace elements solution, a few vitamins complex containing 0.001 g riboflavin, 0.001 g nicotinic acid, 0.001 g creatinol, 0.001 g calcium pantothenate, 0.001 g biotin, 0.001 g p-aminobenzoic acid, 0.001 g vitamin B1, 0.001 g vitamin B6, 20.0 g agar and 1.0 L distilled water, pH 7.2) supplemented with 100 mg/L nystatin. Trace elements solution contained 1.0 g FeSO4•7H2O, 1.0 g MnCl2•4H2O, 1.0 g ZnSO4•7H2O and 1.0 L distilled water. Then, the plates were incubated at 28 °C for 1–4 week, and all well separated actinobacterial colonies were picked from the original isolation plate and repeatedly sub-cultured on 38# agar medium until pure cultures were obtained as judged by uniform colony morphology. The purified strain was preserved as a spore suspension in 25% glycerol at − 80 °C. Totally 42 of purified isolates were obtained.
DNA extraction, sequencing and analysis
The genomic DNA from the pure culture of each isolate was extracted by using a method of Muyzer et al (1993) [24]. The 16S rRNA genes were amplified by using the universal primer pairs (Table S2) [24], and then PCR products were subjected to gene sequencing by Sangon Biotech (Shanghai, China). The 16S rRNA gene sequences were compared with the EzTaxon-e databases by using the Basic Local Alignment Search Tool (nucleotide blast) (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Detection of PKSI, PKSII and NRPS genes
Three sets of degenerate primers targeting on genes encoding PKS I, PKS II and NRPS were performed as recommended in Table S2. Thermocycling conditions were similar to the method described by Li et al (2014) [14]. All correctly sized PCR products were immediately cloned by using the pEASY®-T5 Zero cloning kit (Transgen Co., China) based on the manufacturer’s suggested protocol. Plasmids were isolated and subjected to gene sequencing by Sangon Biotech (Shanghai, China). All sequences were compared with GenBank databases by using BlastX and were analyzed to construct the phylogenetic tree by the neighbor-joining method [53] using the MEGA software package, version 7.0. The stability of relationships was evaluated by performing bootstrap at 1000 resamplings [54].
Nucleotide sequence accession numbers
The 16S rRNA gene sequences of 42 isolates have been deposited in GenBank under the accession numbers given in Table S3. Accession numbers for PKSI, PKSII and NRPS gene sequences were assigned as follows: MH151921, MH151922, MH198457-MH198523, MH198525- MH198530 (Table S4).
Fermentation, extraction and HPLC-DAD analysis of culture extracts
Each isolate was grown in a 300-ml Erlenmeyer flask containing 100 ml fermentation broth at 28 °C with shaking at 250 rpm in the dark. Two media were employed to cultivate these strains, which included GYM4 [43] and SPM1 (40.0 g soluble starch, 5.0 g glucose, 25.0 g soybean meal, 5.0 g yeast extract, 0.5 g K2HPO4, 0.5 g MgSO4, 1.0 g CaCO3, 1.0 L distilled water, pH 7.5). After 5 to 7 days of cultivation, the fermentation broth was extracted with equal volume of ethyl acetate (EtOAc). Each EtOAc extract was then evaporated under reduced pressure to yield a crude extract. Subsequently, each EtOAc extract was dissolved in methanol (MeOH) for HPLC-DAD analysis on an Agilent 1260 series (Agilent Technologies, USA) with a Diode Array Detector (DAD) (200–600 nm) and a C18 RP-column (Platisil ODS-C18 5 μm, 4.6 × 250 mm), with a gradient from 10% acetonitrile in water to 100% acetonitrile over 60 min. The EtOAc extract of strain SYP-A7257 was further subjected to mass spectrometry by Agilent HPLC-HRMS system (Agilent 1260/6530 LC-QTOF/MS, USA) equipped with an Agilent ZORBAX SB-C18 column (4.6 × 50 mm, 1.8 μm) at 40 °C. Mass spectra data with the scanning range from 100 to 2000 amu were collected in the positive and negative modes. Compounds were identified by comparing molecular weights, UV spectra, and retention times with published chemical data from references and standard databases, such as Dictionary of Natural Products on DVD, version 22.2, and SciFinder 2007.
Isolation and identification of antimicrobial compounds
To obtain antimicrobial compounds, the fermentation study (3 L) was performed on targeting SYP-A7257, which were selected on the basis of functional gene screening, antimicrobial activity and chemical analysis. The fermentation of strain SYP-A7257 was carried out in SPM1 medium. Cultures were incubated at 28 °C for 5 days on rotary shakers at 300 rpm in the dark.
To obtain antimicrobial compounds of strain SYP-A7257, the mycelium and cultural liquid broth were separated by centrifugation at 6000 rpm. Metabolites were extracted from the supernatant with 3 L of ethyl acetate and from the mycelium with 500 ml MeOH. The extracts from mycelium and supernatant were combined and evaporated to obtain the crude sample (27 g), further fractioned by silica gel column chromatography using chloroform-methanol gradient elution to yield four fractions (A-D). Fraction A (0.8 g) was separated again by Sephadex LH-20 column chromatography using MeOH as eluant, and the resulting subfractions were combined and further purified by semi-preparative HPLC (CH3CN-H2O, 60%) to afford compound F1 (6.1 mg). Fraction D (0.6 g) was charged on ODS-C18 reverse chromatography column using MeOH-H2O gradient elution to give five subfractions (D1-D5). Subfraction D3 (60 mg) was further purified by semi-preparative HPLC (CH3CN-H2O, 35%) to afford compound F2 (9.4 mg).
Scale-up cultivation for strain SYP-A7257 was performed two times in a 5-L stirred fermentation tank in order to attain compounds F7 and F8 at a fermentation time of 96 h. For isolation, a 10-L aliquot of the culture filtrate were extracted with EtOAc to get the crude extract (30.0 g) and purified by chromatography on a silica gel column with chloroform-MeOH gradient elution, yielding five fractions (E-I). Fraction I (1.4 g) was subjected to chromatography on ODS-C18 reverse column using MeOH-H2O gradient to yield five subfractions (I1-I5). Fraction I3 (517.9 mg) was further purified by Sephadex LH-20 column chromatography using MeOH as eluant followed by preparative reversed-phase HPLC to afford compound F8 (2.0 mg). Fraction I5 (50mg) was subjected to preparative reversed-phase HPLC to give compound F7 (2.2 mg).
The NMR spectra were run on Bruker AVANCE-600 NMR spectrometers (Rheinstetten, Germany). The chemical shifts were expressed in d (ppm) using CDCl3, CD3OD and DMSO-d6 as solvent and TMS as internal reference. ESI-MS was recorded on an Aligent Quattro Premier Triple Quadrupole mass spectrometer. Semi-preparative HPLC was carried out on SHIMADZU 20A-DAD HPLC using Welch Materials Ultimate XB-C18 (5 μm) 10 × 250 mm column. Size exclusion chromatography was done on Sephadex LH-20 (Amersham, GE in USA).
Evaluation of antimicrobial activity
The extracts and compounds were dissolved in DMSO for antimicrobial activity assay using the 96-well flat-bottomed method [27] against human pathogenic bacteria (Staphylococcus aureus, Escherichia coli, Enterococcus faecium, Proteus mirabilis and Acinetobacter baumannii), human pathogenic fungi (Candida albicans) and P. notoginseng pathogenic bacteria (Achromobacter marplatensis). Disk diffusion method [55] was used to test the substance activity against the mycelial pathogenic fungus (Fusarium solani) of P. notoginseng. All these pathogens were stored in School of Life Science and Biopharmaceutics, Shenyang Pharmaceutical University, China. The bacterial suspension was diluted to the initial concentration (optical density of 0.3 at 600 nm; OD600 = 0.3). A 500 μl bacterial solution was added into 50 ml LB media. The bacterial solution was transferred into a 96-well plate (198 μl per well). Then, 2 μl of the extracts (50 g L− 1) or compounds (50 mg L− 1) dissolved in DMSO was added into a 96-well plate in order to reach the final volume of 200 μl per well. Here, untreated bacterial solution was considered as the negative control group. Bacterial solution treated with vancomycin served as and positive control group. The growth was measured by using a multiple photometric reader after 24 h of incubation at 37 °C. Every experiment was performed in triplicate. For inhibition test against mycelial pathogenic fungus, a 25 μl volume of the extract suspension was pipetted onto each disk (diameter, 7 mm) and incubated for 96 h at 28 °C in the dark before the inhibition zone measurement. A 25 μl volume DMSO was used as negative control. Geneticin (G418) served as positive control.
Actinomycin X2 (F1), fungichromin (F2), thailandin B (F7) and antifungalmycin (F8) were analyzed by the above same method to determine the MICs (Minimum Inhibitory Concentration) against all the tested microorganisms.
The farm experiments
The farm land is located at Bazi P. notoginseng plantation (104°6′12.9″E, 23°49′46.9″N) in Wenshan, Yunnan province, which is the main P. notoginseng producing region in China. The healthy P. notoginseng seedlings were harvested last winter. The field was covered with 2.5-m high shade shelter, which had a good ventilated condition. Chemical fumigants as described by Gao et al [56] was applied to treat the soil prior to replanting P. notoginseng. The land was divided into 6 plots on the basis of a completely random block design, three plots as treatment group and another three plots as control group. 400 of 1-year-old healthy P. notoginseng seedlings were planted in each plot (1.2 × 3.0 m) according to the method of Sun et al [57]. A 10-L of 1:200 diluent broth of strain SYP-A7257 fermented in SPM1 medium at 28 °C for 5 days was applied to the soil by root irrigation [58] in the treatment (T) group. Meanwhile, SPM medium dilution without strain cultivation was applied to the soil by root irrigation in the control (CK) group. The root irrigation experiments were continually carried out 3 times at 2, 3 and 7 months after the emergence of the P. notoginseng seedlings. Seedling emergence and dead seedling rate were recorded every 30 days after the first root irrigation. The survival rate (%) of each plot was calculated using the following equation: The survival rate (%) = (the number of survival seedlings/400) × 100.
Statistical analysis
The statistical analysis was performed by using Graph Pad Prism 5.01. All the data were present as means± SD of triplicate experiments. One-way analysis of variance (ANOVA) followed by Turkey’s test was used to express the statistical differences between groups. The value of P < 0.05 was presented as statistically significant.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Abbreviations
- PKS I:
-
Type I polyketide synthases
- PKS II:
-
Type II polyketide synthase
- NRPS:
-
Nonribosomal peptide synthetases
- HPLC-DAD:
-
High Performance Liquid Chromatography-Diode Array Detector
- HPLC-HRMS:
-
High Performance Liquid Chromatography-High Resolution Mass Spectrometry
- UV:
-
Ultraviolet
- NMR spectra:
-
Nuclear magnetic resonance spectra
- TIC:
-
Total ion current
- BCGs:
-
Biosynthetic gene clusters
- Blast:
-
Basic Local Alignment Search Tool
- EtOAc:
-
Ethyl acetate
- MeOH:
-
Methanol
- ANOVA:
-
One-way analysis of variance
References
Mendes LW, Raaijmakers JM, Hollander MD, Mendes R, Tsai SM. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 2017;12(1):212–24.
Wang C, Wang Y, Ma J, Hou Q, Liu K, Ding Y, Du B. Screening and whole-genome sequencing of two Streptomyces species from the Rhizosphere soil of Peony reveal their characteristics as plant growth-promoting Rhizobacteria. Biomed Res Int. 2018;2419686.
Nimaichand S, Devi AM, Tamreihao K, Ningthoujam DS, Li WJ. Actinobacterial diversity in limestone deposit sites in Hundung, Manipur (India) and their antimicrobial activities. Front Microbiol. 2015;6:413.
Viaene T, Langendries S, Beirinckx S, Maes M, Goormachtig S. Streptomyces as a plant's best friend? FEMS Microbiol Ecol. 2016;92(8):fiw119.
Ge BB, Cheng Y, Liu Y, Liu BH, Zhang KC. Biological control of Botrytis cinerea on tomato plants using Streptomyces ahygroscopicus strain CK-15. Lett Appl Microbiol. 2016;61(6):596–602.
Ceapă CD, Vázquez-Hernández M, Rodríguez-Luna SD, Cruz Vázquez AP, Jiménez SV, Rodríguez-Sanoja R, Alvarez-Buylla ER, Sánchez S. Genome mining of Streptomyces scabrisporus NF3 reveals symbiotic features including genes related to plant interactions. PLoS One. 2018;13(2):e192618.
Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, Klenk HP, Clément C, Ouhdouch Y, van Wezel GP. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol Mol Biol R. 2016;80(1):1–43.
Tapadar SA, Jha DK: Disease Management in Staple Crops: A Bacteriological Approach. Bacteria in Agrobiology: Disease Management. 2013:111–52. https://doi.org/10.1007/978-3-642-33639-3_5.
Toumatia O, Yekkour A, Goudjal Y, Riba A, Coppel Y, Mathieu F, Sabaou N, Zitouni A. Antifungal properties of an actinomycin D-producing strain, Streptomyces sp. IA1, isolated from a Saharan soil. J Basic Microb. 2015;55(2):221–8.
Kalyon B, Tan GY, Pinto JM, Foo CY, Wiese J, Imhoff JF, Süssmuth RD, Sabaratnam V, Fiedler HP. Langkocyclines: novel angucycline antibiotics from Streptomyces sp. Acta 3034(*). J Antibiot. 2013;66(10):609–16.
Han D, Wang L, Luo Y. Isolation, identification, and the growth promoting effects of two antagonistic actinomycete strains from the rhizosphere of Mikania micrantha Kunth. Mocrobiol Res. 2018;208:1–11.
Katz L, Baltz RH. Natural product discovery: past, present, and future. J Ind Microbiol Blot. 2016;43(2–3):155–76.
Wei Y, Zhang L, Zhou Z, Yan X. Diversity of gene clusters for Polyketide and nonribosomal peptide biosynthesis revealed by metagenomic analysis of the Yellow Sea sediment. Front Microbiol. 2018;9:295.
Li J, Dong JD, Yang J, Luo XM, Zhang S. Detection of polyketide synthase and nonribosomal peptide synthetase biosynthetic genes from antimicrobial coral-associated actinomycetes. Antonie Van Leeuwenhoek. 2014;106(4):623–35.
Marta M, Delphine A, Loïc M, Aymeric N, Magdalena C, Philippe D, Monique C, Barton HA, Marie-Pierre H, Nicolas S. A phenotypic and genotypic analysis of the antimicrobial potential of cultivable Streptomyces isolated from cave Moonmilk deposits. Front Microbiol. 2016;7:1455.
Yuan M, Yu Y, Li HR, Dong N, Zhang XH. Phylogenetic diversity and biological activity of Actinobacteria isolated from the Chukchi shelf marine sediments in the Arctic Ocean. Mar Drugs. 2014;12(3):1281–97.
Srivastava A, Singh VK, Patnaik S, Tripathi J, Singh P, Nath G, Asthana RK. Antimicrobial assay and genetic screening of selected freshwater cyanobacteria and identification of a biomolecule dihydro-2H-pyran-2-one derivative. J Appl Microbiol. 2016;122(4):881–92.
Park HJ, Kim DH, Park SJ, Kim JM, Ryu JH. Ginseng in traditional herbal prescriptions. J Ginseng Res. 2012;36(3):225–41.
Jong-Hoon K. Cardiovascular diseases and Panax ginseng: a review on molecular mechanisms and medical applications. J Ginseng Res. 2012;36(1):16–26.
Guo HB, Cui XM, An N, Cai GP. Sanchi ginseng (Panax notoginseng (Burkill) F. H. Chen) in China: distribution, cultivation and variations. Genet Resour Crop Ev. 2010;57(3):453–60.
Ma L, Cao YH, Cheng MH, Huang Y, Mo MH, Wang Y, Yang JZ, Yang FX. Phylogenetic diversity of bacterial endophytes of Panax notoginseng with antagonistic characteristics towards pathogens of root-rot disease complex. Anton Leeuw Int J G. 2013;103(2):299–312.
Fan ZY, Miao CP, Qiao XG, Zheng YK, Chen HH, Chen YW, Xu LH, Zhao LX, Guan HL. Diversity, distribution, and antagonistic activities of rhizobacteria of Panax notoginseng. J Ginseng Res. 2016;40(2):97–104.
Tan Y, Cui Y, Li H, Kuang A, Li X, Wei Y, Ji X. Diversity and composition of rhizospheric soil and root endogenous bacteria in Panax notoginseng during continuous cropping practices. J Basic Microb. 2017;57(4):337–44.
Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59(3):695–700.
Zhang MY, Xu H, Zhang TY, Xie J, Cheng J, Nimaichand S, Li SH, Li WJ, Zhang YX. Flavobacterium notoginsengisoli sp. nov., isolated from the rhizosphere of Panax notoginseng. Antonie Van Leeuwenhoek. 2015;108(3):545–52.
An YN, Zhang X, Zhang TY, Zhang MY, QianZhang DXY, Zhao F, Zhu LJ, Wang G, Zhang J. Penicimenolides A-F, Resorcylic Acid Lactones from Penicillium sp., isolated from the Rhizosphere Soil of Panax notoginseng. Sci Rep-UK. 2016;6:27396.
Xie J, Wu YY, Zhang TY, Zhang MY, Zhu WW, Gullen EA, Wang ZJ, Cheng YC, Zhang YX. New and bioactive natural products from an endophyte of Panax notoginseng. RSC Adv. 2017;7(60):38100–9.
Sun Y, Hong H, Samborskyy M, Mironenko T, Leadlay PF, Haydock SF. Organization of the biosynthetic gene cluster in Streptomyces sp. DSM 4137 for the novel neuroprotectant polyketide meridamycin. Microbiology. 2006;52(Pt 12):3507–15.
Tao W, Yurkovich ME, Wen S, Lebe KE, Samborskyy M, Liu Y, Yang A, Liu Y, Ju Y, Deng Z. A genomics-led approach to deciphering the mechanism of thiotetronate antibiotic biosynthesis. Chem Sci. 2015;7(1):376–85.
Susumu O, Takaaki T, Kozo O, Koji I. Biosynthesis of actinorhodin and related antibiotics: discovery of alternative routes for quinone formation encoded in the act gene cluster. Chem Biol. 2009;16(2):226–36.
Li Z, Rawlings BJ, Harrison PH, Vederas JC. Production of new polyene antibiotics by Streptomyces cellulosae after addition of ethyl (Z)-16-phenylhexadec-9-enoate. J Antibiot (Tokyo). 1989;42(4):577–84.
Song XQ, Jiang X, Sun JB, Zhang CY, Zhang Y, Lai-Chun LU, Jian-Hua JU. Antibacterial secondary metabolites produced by mangrove-derived Actinomycete Stremptmeces costaricanus SCSIO ZS0073. Nat Prod Res Dep. 2017;29:410–4.
Chen C, Song F, Wang Q, Abdel-Mageed WM, Guo H, Fu C, Hou W, Dai H, Liu X, Yang N. A marine-derived Streptomyces sp. MS449 produces high yield of actinomycin X 2 and actinomycin D with potent anti-tuberculosis activity. Appl Microbiol Biot. 2012;95(4):919–27.
Intra B, Greule A, Bechthold A, Euanorasetr J, Paululat T, Panbangred W. Thailandins a and B, new Polyene macrolactone compounds isolated from Actinokineospora bangkokensis strain 44EHWT, possessing antifungal activity against anthracnose Fungi and pathogenic yeasts. J Agr Food Chem. 2016;64(25):5171–9.
Wang YF, Wei SZ, Zhang ZP, Zhan TH, Tu GQ. Antifungalmycin, an antifungal macrolide from Streptomyces padanus 702. Natur Prods Biopros. 2012;2(1):41–5.
Labeda DP, Dunlap CA, Rong X, Huang Y, Doroghazi JR, Ju KS, Metcalf WW. Phylogenetic relationships in the family Streptomycetaceae using multi-locus sequence analysis. Antonie Van Leeuwenhoek. 2017;110(4):563–83.
Gallagher KA, Rauscher K, Ioca LP, Jensen PR. Phylogenetic and chemical diversity of a hybrid-Isoprenoid-producing Streptomycete lineage. Appl Environ Microb. 2013;79(22):6894–902.
Kinkel LL, Schlatter DC, Bakker MG, Arenz BE. Streptomyces competition and co-evolution in relation to plant disease suppression. Res Microbiol. 2012;163(8):490–9.
Chen X, Pizzatti C, Bonaldi M, Saracchi M, Erlacher A, Kunova A, Berg G, Cortesi P. Biological Control of Lettuce Drop and Host Plant Colonization by Rhizospheric and Endophytic Streptomycetes. Front Microbiol. 2016;7(714):1–12.
Wu Q, Zhang G, Wang B, Xin L, Yue S, Chen J, Zhang H, Hong W. Production and identification of Inthomycin B produced by a Deep-Sea sediment-derived Streptomyces sp. YB104 based on cultivation-dependent approach. Curr Microbiol. 2018;75(7):942–51.
English AL, Boufridi A, Quinn RJ, Kurtböke DI. Evaluation of fermentation conditions triggering increased antibacterial activity from a near-shore marine intertidal environment-associated Streptomyces species. Synthetic Syst Biotechnol. 2017;2(1):28–38.
Zhang Z, Wang W, Yang J, Cui X. Effects of continuous Panax notoginseng cropping soil on P notoginseng seed germination and seedling growth. Soils. 2010;42:1009–14.
Schneemann I, Nagel K, Kajahn I, Labes A, Wiese J, Imhoff JF. Comprehensive investigation of marine actinobacteria associated with the sponge halichondria panicea. Appl Environ Microbiol. 2010;76(11):3702–14.
Mao D, Okada BK, Wu Y, Xu F, Seyedsayamdost MR. Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr Opin Microbiol. 2018;45:156–63.
Thuan NH, Dhakal D, Pokhrel AR, Luan LC, Pham TTV, Shrestha A, Sohng JK. Genome-guided exploration of metabolic features of Streptomyces peucetius ATCC 27952: past, current, and prospect. Appl Microbiol Biot. 2018;2:1–16.
Kong D, Lee MJ, Lin S, Kim ES. Biosynthesis and pathway engineering of antifungal polyene macrolides in actinomycetes. J Ind Microbiol Biot. 2013;40(6):529.
Wang X, Tabudravu J, Rateb ME, Annand KJ, Qin Z, Jaspars M, Deng Z, Yu Y, Deng H. Identification and characterization of the actinomycin G gene cluster in Streptomyces iakyrus. Mol BioSyst. 2013;9(6):1286–9.
Qin S, Li J, Chen H, Zhao G, Zhu W, Jiang C, Xu L, Li W. Isolation, diversity, and antimicrobial activity of rare Actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl Environ Microbiol. 2009;75(19):6176–86.
Gontang EA, Gaudêncio SP, William F, Jensen PR. Sequence-based analysis of secondary-metabolite biosynthesis in marine actinobacteria. Appl Environ Microbiol. 2010;76(8):2487–99.
Bibi F, Strobel GA, Naseer MI, Yasir M, Alghamdi AAK, Azhar EI. Microbial Flora associated with the halophyte–Salsola imbricate and its biotechnical potential. Front Microbiol. 2018;9:65.
Sharma P, Kalita MC, Thakur D. Broad Spectrum Antimicrobial Activity of Forest-Derived Soil Actinomycete, Nocardia sp. PB-52. Front Microbiol. 2016;7(856):347.
Passari AK, Mishra VK, Saikia R, Gupta VK, Singh BP. Isolation, abundance and phylogenetic affiliation of endophytic actinomycetes associated with medicinal plants and screening for their in vitro antimicrobial biosynthetic potential. Front Microbiol. 2015;6(273):273.
Saitou NNM, Nei MC, Saitou N, Nei M. The neighbor-joining method-a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–25.
Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39(4):783–91.
Mearns-Spragg A, Bregu M, Boyd KG, Burgess JG. Cross-species induction and enhancement of antimicrobial activity produced by epibiotic bacteria from marine algae and invertebrates, after exposure to terrestrial bacteria. Lett Appl Microbiol. 1998;27(3):142–6.
Gao S, Sosnoskie LM, Cabrera JA, Qin R, Hanson BD, Gerik JS, Wang D, Browne GT, Thomas JE. Fumigation efficacy and emission reduction using low-permeability film in orchard soil fumigation. Pest Manag Sci. 2015;72(2):306–14.
Sun Y, Yang L, Wei M, Huang T. Effects of different treatments and GA3 concentration on induction seedling of Panax notoginseng. Spec Wild Econ Anim Plant Res. 2013;04:47–9.
Chen JL, Sun SZ, Miao CP, Wu K, Chen YW, Xu LH, Guan HL, Zhao LX. Endophytic Trichoderma gamsii YIM PH30019: a promising biocontrol agent with hyperosmolar, mycoparasitism, and antagonistic activities of induced volatile organic compounds on root-rot pathogenic fungi of Panax notoginseng. J Ginseng Res. 2016;40(4):315–24.
Acknowledgements
Not applicable.
Funding
This research program is financially supported by the Science and Technique Programs of Yunnan Province (2016ZF001–001, 2017IB038), Liao Ning Revitalization Talents Program (XLYC1902072), Liaoning Nature Foundation (2019ZD0464, 2019MS298), Basic Scientific Research Program in Higher Education of Liaoning Province (2017LQN17).
Author information
Authors and Affiliations
Contributions
PF conceived and designed the study. PF, ZM and HS performed the experiments. ZM and CJ collected and provided the soil samples. WY and ZY reviewed and edited the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have no conflicts of interest with any parties or individuals.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1 Figure S1
. HPLC-DAD analysis of crude extracts of SYP-A7257 cultured in SYM1 medium. Extracts were analyzed using Agilent 1260 series (Agilent Technologies, USA) with a Diode Array Detector (DAD) (200–600 nm) and a C18 RP-column (Platisil ODS-C18 5 μm, 4.6 × 250 mm), with a gradient from 10% acetonitrile in water to 100% acetonitrile over 60 min., 1.0 ml/min as the flow rate. Figure S2. TIC chromatogram of the extract of strain SYP-A 7257 and MS spectra of peaks F1–F6 obtained by HPLC–HRESIMS in positive mode. Figure S3. HR-ESI-MS in positive-ion mode and UV/vis characteristics of compound F1. Figure S4. HR-ESI-MS in positive-ion mode and UV/vis characteristics of compound F2. Figure S5. HR-ESI-MS in positive-ion mode and UV/vis characteristics of compound F3. Figure S6. HR-ESI-MS in positive-ion mode and UV/vis characteristics of compound F4. Figure S7. HR-ESI-MS in positive-ion mode and UV/vis characteristics of compound F5. Figure S8. HR-ESI-MS in positive-ion mode and UV/vis characteristics of compound F6. Figure S9. ESI-MS in positive and negative-ion mode of compound F1. Figure S10.1H NMR spectrum of compound F1 in CDCl3 recorded at 600 MHz. Figure S11.13C NMR spectrum of compound F1 in CDCl3 recorded at 150 MHz. Figure S12. ESI-MS in positive and negative-ion mode of compound F2. Figure S13.1H NMR spectrum of compound F2 in CD3OD recorded at 600 MHz. Figure S14.13C NMR spectrum of compound F2 in CD3OD recorded at 150 MHz. Figure S15. ESI-MS in negative-ion mode of compound F7. Figure S16.1H NMR spectrum of compound F7 in CD3OD recorded at 600 MHz. Figure S17.13C NMR spectrum of compound F7 in CD3OD recorded at 150 MHz. Figure S18. ESI-MS in positive-ion mode of compound F8. Figure S19.1H NMR spectrum of compound F8 in CD3OD recorded at 600 MHz. Figure S20.13C NMR spectrum of compound F8 in CD3OD recorded at 150 MHz. Table S1. Characteristics of rhizospheric soil samples of P. notoginseng in the Wenshan region of Yunnan Province, China. Table S2. PCR primers used in this study. Table S3.Streptomyces isolated from different soil of P. notoginseng with similarity values of 16S rRNA gene sequences to the closest cultivated species. Table S4. KS domain and NRPS amino acid sequences of the soil-derived Streptomyces isolates from the rhizospheric soil of P. notoginseng. Table S5. NMR Data for compound F1 (600 MHz) in CDCl3 (δ in ppm, J in Hz). Table S6. NMR Data for compound F2 (600 MHz) in CD3OD (δ in ppm, J in Hz). Table S7. NMR Data for compound F7and F8 (600 MHz) in CD3OD (δ in ppm, J in Hz).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Peng, F., Zhang, MY., Hou, SY. et al. Insights into Streptomyces spp. isolated from the rhizospheric soil of Panax notoginseng: isolation, antimicrobial activity and biosynthetic potential for polyketides and non-ribosomal peptides. BMC Microbiol 20, 143 (2020). https://doi.org/10.1186/s12866-020-01832-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-020-01832-5