Abstract
Background
Plant cell wall degrading enzymes (PCWDEs) are a subset of carbohydrate-active enzymes (CAZy) produced by plant pathogens to degrade plant cell walls. To counteract PCWDEs, plants release PCWDEs inhibitor proteins (PIPs) to reduce their impact. Several transgenic plants expressing exogenous PIPs that interact with fungal glycoside hydrolase (GH)11-type xylanases or GH28-type polygalacturonase (PG) have been shown to enhance disease resistance. However, many plant pathogenic Fusarium species were reported to escape PIPs inhibition. Fusarium virguliforme is a soilborne pathogen that causes soybean sudden death syndrome (SDS). Although the genome of F. virguliforme was sequenced, there were limited studies focused on the PCWDEs of F. virguliforme. Our goal was to understand the genomic CAZy structure of F. viguliforme, and determine if exogenous PIPs could be theoretically used in soybean to enhance resistance against F. virguliforme.
Results
F. virguliforme produces diverse CAZy to degrade cellulose and pectin, similar to other necrotorphic and hemibiotrophic plant pathogenic fungi. However, some common CAZy of plant pathogenic fungi that catalyze hemicellulose, such as GH29, GH30, GH44, GH54, GH62, and GH67, were deficient in F. virguliforme. While the absence of these CAZy families might be complemented by other hemicellulases, F. virguliforme contained unique families including GH131, polysaccharide lyase (PL) 9, PL20, and PL22 that were not reported in other plant pathogenic fungi or oomycetes. Sequence analysis revealed two GH11 xylanases of F. virguliforme, FvXyn11A and FvXyn11B, have conserved residues that allow xylanase inhibitor protein I (XIP-I) binding. Structural modeling suggested that FvXyn11A and FvXyn11B could be blocked by XIP-I that serves as good candidate for developing transgenic soybeans. In contrast, one GH28 PG, FvPG2, contains an amino acid substitution that is potentially incompatible with the bean polygalacturonase-inhibitor protein II (PvPGIP2).
Conclusions
Identification and annotation of CAZy provided advanced understanding of genomic composition of PCWDEs in F. virguliforme. Sequence and structural analyses of FvXyn11A and FvXyn11B suggested both xylanases were conserved in residues that allow XIP-I inhibition, and expression of both xylanases were detected during soybean roots infection. We postulate that a transgenic soybean expressing wheat XIP-I may be useful for developing root rot resistance to F. virguliforme.
Similar content being viewed by others
Background
Inactivation of pathogen plant cell wall degrading enzymes (PCWDEs) is one of the strategies that plants employ to prevent infection. Several plant-derived extracellular PCWDEs inhibitor proteins (PIPs) were reported to not only reduce PCWDEs activities but also trigger defense response upon recognition of PCWDEs [1–3]. The importance of PIPs in plant defense has been demonstrated in transgenic plants expressing exogenous PIPs that show enhanced biotic resistance. For example, wheat xylanase inhibitors, such as xylanase inhibitor protein I (XIP-I) and Triticum aestivum xylanase inhibitor III (TAXI-III), have been shown to inhibit fungal GH11 xylanases [3–5]. Similarly, transgenic wheat with TAXI-III increased resistance to necrosis and head blight caused by Fusarium graminearum [6–8]. Another well-studied example is the polygalacturonase (PG) inhibitor proteins (PGIP), a leucine-rich repeat protein of plants that interact with fungal GH28 PG [2, 9]. Several transgenic plants expressing exogenous PGIPs have been shown to increase resistance against a broad spectrum of pathogens [10–16]. The mechanism of enhanced resistance in PGIP-transgenic plants has been recently demonstrated. In vivo expression of chimeric PGIP-PG in Arabidopsis showed that PGIP-PG interaction induced the production of oligogalacturonides, which serves as a damage-associated molecular mechanism to stimulate resistance [17].
Soybean sudden death syndrome (SDS), which is caused by a soilborne fungus F. virguliforme, is responsible for annual losses around US$190 million [18]. Breeding for SDS resistance is difficult because the interaction between F. virguliforme and soybean is quantitative [19]. Instead, it has been suggested that transgenic approaches may be suitable to manage SDS, and transgenic soybeans expressing exogenous toxin-specific antibody has been shown to reduce SDS foliar symptoms [18, 20]. However, symptoms caused by F. virguliforme include not only foliar symptoms but also root rot and vascular discoloration [18]. Soybeans that exhibit partial root resistance have been shown to have up-regulated genes involved in plant cell wall enhancement upon root infection by F. virguliforme [21]. Differences in root susceptibility of soybean genotypes also showed different expression patterns of genes involved in plant cell wall synthesis [22]. These studies indicated that plant cell wall modification maybe involved in resistance against F. virguliforme, which highlights the possibility of using transgenic soybeans that express exogenous PIPs to prevent and/or slow fungal colonization of soybean roots. Therefore, an in silico study would be useful before embarking in a time-consuming transgenic project, as it would be important to know if F. virguliforme secrets compatible PCWDEs to the transgenic exogenous PIPs during infection.
Although the genome of F. virguliforme has been published [23], genomic structure of PCWDEs remains uncharacterized. In this study, we annotated PCWDEs in the F. virguliforme genome, and further focused on the orthologous GH11 xylanases and GH28 PGs of F. virguliforme. The goal was to understand the genomic PCWDEs structure of F. virguliforme and to evaluate if orthologous GH11 xylanases and GH28 PGs of F. virguliforme have potential to serve as targets for exogenous PIPs produced by transgenic soybeans.
Results and discussion
Identification of carbohydrate-active enzymes (CAZy) in the genome of F. virguliforme
CAZy are proteins with polysaccharide-degrading enzymatic activities on polysaccharides [24, 25]. We identified 629 putative genes that encode CAZy in the genome of F. virguliforme (Additional file 1: Table S1). Of the six CAZy classes, carbohydrate esterases (CE), glycoside hydrolases (GH), and polysaccharide lyases (PL) are PCWDEs. There were 66, 292, and 28 genes belonging to the CE, GH, and PL classes, respectively (Table 1). Three other classes with indirect roles on degrading carbohydrates are auxiliary activity (AA), carbohydrate-binding module (CBM), and glycosyl-transferase (GT). There were with 96, 31, and 116 genes identified in the AA, CBM, and GT classes, respectively (Table 2).
Identification of putative cellulose-degrading enzymes in the genome of F. virguliforme
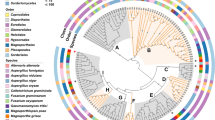
Cellulose is the most abundant component in plant cell walls, which results from the polymerization of glucose and the formation of a microfibril framework for other components to join [24, 26, 27]. Most cellulose-degrading enzymes are categorized within GH classes. GH1, GH3, and GH5 are prevalent PCWDEs that catalyze not only cellulose, but also hemicellulose and pectin (Table 1). Plant pathogenic oomycetes, and hemibiotrophic as well as necrotrophic fungi generally contain more GH1 degrading enzymes than biotrophic fungi. For example, the genome of F. virguliforme encodes five GH1 genes while most biotrophic fungi have none [28–30]. For enzymes in the GH3 family, F. virguliforme, hemibiotrophic and necrotrophic fungi, and Phytophthora species contain 8-38 genes compared to relatively fewer for biotrophic fungi and Pythium species (Fig. 1a). Endo- and exo-β-1,4-glucanases in the GH5 family are cellulose-degrading enzymes employed by both plant pathogenic fungi and oomycetes, and F. virguliforme has 15 GH family genes. In addition, F. virguliforme has one GH6 and three GH7 that not only have endo- and exo-β-1,4-glucanase but also cellobiohydrolase activity. GH12 encode cellulose/hemicellulose-degrading enzymes similar to GH3, which is common in F. virguliforme, plant pathogenic fungi and Phytophthora species but not in Pythium species. GH30 is dominant in oomycetes but not in plant pathogenic fungi, and none was found in F. virguliforme (Fig. 1a). On the other hand, GH45 and GH51 are fungi-specific degrading enzymes that have not been found in oomycetes [28, 30]. GH131 CAZy that encodes exo-β-1,3/1,6- and endo-1,4-glucanase was only found in F. virguliforme. In addition to GH families, some AA families, such as AA8 and AA9, have been reported to accelerate cellulose degradation. Instead of catalyzing carbohydrates, enzymes in the AA9 family (previously known as GH61) have copper-dependent lytic polysaccharide monooxygenase activity to assist degradation of lignocellulose [25, 31]. It has been suggested that plant pathogenic fungi have more AA9 genes than oomycetes [28–30], and 12 AA9 genes were found in F. virguliforme (Table 2).
Comparison analysis for PCWDEs of Fusarium virguliforme that putatively target on different polysaccharides. Blue color indicates carbohydrate esterases (CE); red color indicates glycoside hydrolases (GH); and green color indicates polysaccharide lyases (PL). a CAZy with cellulase activity. GH1, GH3, and GH5 are universal PCWDEs that catalyze celluloses, hemicelluloses, and pectin. GH30 is common distributed in plant pathogenic fungi and abundant in oomycetes, but it was not found in the genome of F. virguliforme. Instead, GH131 was found only in the genome of F. virguliforme. b CAZy with hemicellulase activity. GH29, GH30, GH44, GH54, GH62, and GH67 are absent in the genome of F. virguliforme, but other functional redundant CAZy may complement the loss of these families. c CAZy with pectinase activity. F. virguliforme have most pectinases and unique PL9, PL20, and PL22 that only existed in F. virguliforme and close-related species Nectaria haematococca. In general, the genomic PCWDEs structure of F. virguliforme is similar to necrotrophic and hemibiotrophic pathogenic fungi
Identification of putative hemicellulose-degrading enzymes in the genome of F. virguliforme
Hemicellulose is composed of polymers such as xyloglucan, xylan and galactomannan, cross-links the cellulose microfibrils and provides strength to plant cell walls [24, 26, 27]. In addition to GH1, GH3, GH5, GH12, GH51, and GH131 that have both cellulose- and hemicellulose-degrading activities, GH2, GH10, and GH11 are important hemicellulose-degrading enzymes for plant pathogenic fungi including F. virguliforme (Table 1). However, these families are generally deficient in oomycetes, except GH10, which exists in Phytophthora species (Fig. 1b). GH29, GH30, GH44, GH54, GH62, and GH67 families are absent in the genome of F. virguliforme. A closely related species, Nectria haematococca (anamorph Fusaium solani), has no CAZy in the GH29 and GH30 either. Instead, F. oxysporum and F. verticillioides have at least two enzymes for each GH29 and GH30 [30]. Nevertheless, F. virguliforme contains two GH95 α-fucosidases that may have similar enzymatic activities to GH29 and GH30, which remove xyloses from xyloglucan [24]. F. virguliforme has no GH54 and GH62 that encode α-L-arabinofuranosidases, but N. haematococca, F. oxysporum, and F. verticillioides have at least one GH54 and one GH62 enzyme [30]. The function of GH54 and GH62 may be redundant to GH3, GH10, GH43, and GH51 [24], which could be found in the F. virguliforme genome (Table 2). Among these four families, GH43 is one of the largest CAZy that catalyzes both hemicellulose and pectin, and F. virguliforme has 26 genes. In addition, F. virguliforme has no GH44 or GH67 that are deficient in most plant pathogens. The loss of GH44 and GH67 may be complemented by GH74 and GH36, respectively, because both GH44 and GH 74 encode xyloglucanases while GH67 and GH36 both encode α-galactosidases (Table 2). Another group of CAZy active on hemicellulose is the CE class. CE1 is the most dominant hemicellulose-degrading family in plant pathogens, and in the case of F. virguliforme, 32 genes were found. CE families such as CE2, CE3, CE4, CE5, and CE12, were all identified in the genome of F. virguliforme as reported in other plant pathogens [28–30].
Identification of putative pectin-degrading enzymes in the genome of F. virguliforme
Pectin, a polymer of mainly D-galacturonic acids, is the most divergent part of plant cell walls because of the different modifications on the side chains. Based on these modifications, pectin is categorized into subgroups like homogalacturonan and rhamnogalacturonan. Pectin forms a matrix between microfibrils to control the porosity and cohesion [24, 26, 27, 32, 33]. Besides the universal plant cell wall degrading families (GH1, GH3, and GH5) and the most well studied GH28 PGs, GH53 and GH78 are common in most hemibitrophic and necrotrophic fungi as well as Phytophthora species while GH105 is more abundant in plant pathogenic fungi than oomycetes (Fig. 1c). Except for the lack in GH30 and GH54 that have been discussed in the hemicellulose section, F. virguliforme has all the GH families that catalyze pectin. Some CAZy in the CE class, such as CE8 and CE12, allow degradation of pectin by removing methyl and acetyl groups from galacturonic acids, respectively. Both families are common in all plant pathogens including F. virguliforme but not Pythium species [29]. The PL class specializes in pectin degradation. PL1 and PL3 are the most dominant and common pectin lyases of plant pathogens. Similar to hemibitrophic and necrotrophic fungi and oomycetes, F. virguliforme has eleven PL1 and ten PL3 that are more abundant than bitrophic fungi [28–30]. In addition, PL4, PL9, PL20, and PL22 families were identified in the F. virguliforme genome (Fig. 1c). While PL4 is commonly distributed in plant pathogens, PL9, PL20, and PL22 were found only in F. virguliforme and N. haematococca [30].
Evaluation of xylanases and PGs of F. virguliforme as PIPs targets
GH11 xylanases and GH28 PGs have been successfully used as targets for transgenic plants expressing exogenous PIPs. However, GH11 xylanases and GH28 PGs of some Fusarium species can escape PIPs inhibition by amino acid substitution [34, 35]. Two GH11 xylanases, XylA (FGSG_10999) and XylB (FGSG_03624) of F. graminearum, have amino acid substitutions at the thumb region that allowed them to escape XIP-I binding (Fig. 2a) [4]. On the other hand, site-directed mutagenesis of lysine to glutamine of position 97 increased affinity of F. verticillioides PG to PvPGIP2 [35]; more importantly, a single substitution at the 261 position of F. phyllophilum PG (FpPG) from alanine to threonine significantly reduced FpPG affinity to PvPGIP2 [36]. Amino acid substitutions in these studies supported the variable response of PGIPs to PGs of different Fusarium species [36, 37].
In silico analysis of GH11 xylanases of Fusarium virguliforme. a F. graminearum contains amino acid substitutions that allow GH11 xylanases to escape XIP-I inhibition, including a substitution of threonine (T) to valine (V) for XylA (yellow blocks); and substitutions of asparagine (N) to cysteine (C), an insert of aspartic acid (D), and a substitution of T to C for XylB (yellow blocks). However, FvXyn11A and FvXyn11B are conserved in this region. The red block circles a string of 30 amino acids reported to induce necrosis [63]. The purple block and blue block indicate previously reported conserved residues. The name of necrosis-inducing xylanases were bold [8, 63, 64]. b Salmon color represents XIP-I. Golden color represents conserved thumb region of each xylanase. Control model of XIP-I inhibits Penicillium funiculosum GH11 xylanase XYNC. XIP-I perfectly fills into the catalyzing groove between two essential catalyzing residues glutamic acid (E) at position 85 (E85) and E176 that mimics substrates of XYNC. c The interaction between FvXyn11A and XIP-I, where the corresponding residues E114 and E205 were shown. d The interaction between FvXyn11B and XIP-I, where the corresponding residues E98 and E189 were shown
Two orthologous GH11 xylanases (FvXyn11A and FvXyn11B) and two orthologous PGs (FvPG1 and FvPG2) were identified for F. virguliforme (Table 3). Sequence analysis revealed that neither FvXyn11A nor FvXyn11B carry amino acid substitutions at the thumb region corresponding to XylA or XylB of F. graminearum (Fig. 2a). Protein-protein docking analysis was applied to further test the interaction between XIP-I and FvXyn11A as well as FvXyn11B. The results supported XIP-I forming inhibiting conformations with FvXyn11A and FvXyn11B in the same orientation to Penicillium funiculosum XYNC (Fig. 2b) [5]. In the case of FvPG1 and FvPG2, sequence alignment was uncertain at residue 97 because of the neighboring gaps (Fig. 3). However, because FvPG1 has an alanine at position 261 that is identical to Colletotrichum lupine PG (CluPG1) and Aspergillus niger PG (AnPGII), we speculated that the affinity strength of FvPG1 to PvPGIP2 would be similar to CluPG1 and AnPG II [2, 36]. The replacement of the nonpolar alanine to the polar threonine dramatically reduces FpPG affinity to PvPGIP2 [36], so we speculated FvPG2 would be less inhibited by PvPGIP2 because the corresponding position of FvPG2 is a larger, positively charged lysine.
Sequence alignment of GH28 polygalacturonase of Fusarium virguliforme with other fungi. The green blocks circle polymorphic residues [37]; the purple blocks indicate essential residues for binding substrates [65]; the orange blocks circle indispensible residues for catalyzing substrates [66]; and the red blocks circle residues (position 97 in top panel and 261 in bottom panel) that were reported to affect PvPGIP2 inhibition [35, 36]
FvXyn11A, FvXyn11B, FvPG1, and FvPG2 contained putative secretory peptides without trans-membrane domains. Moreover, their expressions were detectable during infection. Using an in vitro RNA-Seq dataset [38], we noticed FvXyn11B and FvPG2 were less active compared to FvXyn11A and FvPG1 in the in vitro condition (Fig. 4a). However, the expression of FvXyn11B and FvPG2 were significantly enhanced during root infection (Fig. 4b). It has been reported that PCWDEs of some Fusarium species displayed different expression patterns in different conditions. For example, two PGs of F. oxysporum, pg1 and pgx6, expressed actively during root infection, and the double knockout mutants of pg1 and pgx6 compromised virulence [39]. In addition, differential expression of GH11 xylanases was also reported that XylB had higher expression than XylA at 5 day-post-inoculation [40].
Expression comparison of FvXyn11B and FvPG2 in vitro and in planta. RNA of both conditions was extracted after 5 days post inoculation, from soybean dextrose broth and from soybean roots, respectively. a In vitro expression was indicated by counts per million (cpm) from a RNA-Seq data [38]. Asterisk indicated genes with raw counts below 1 cpm. b In planta expression indicated by log10(–ΔΔCt). Unlike in vitro condition, the expression of FvXyn11B increased and was significantly higher the FvXyn11A. The expression of FvPG2 was also increased to a level similar to FvPG1
Conclusion
In this study, we advanced the understanding of CAZy and PCWDEs in the genome of F. virguliforme and in silico analysis supported the possibility of developing transgenic soybeans with exogenous PIPs to enhance SDS resistance. As a soybean pathogen, F. virguliforme may have undergone selection pressure to PGIPs produced by soybean. Our analysis revealed a putative PvPGIP2-escaping FvPG2 had higher expression during root infection than in the in vitro condition. This indicated that the use of transgenic PVPGIP2 might not be a preferable option. Instead, F. virguliforme should rarely encounter XIP-I because xylanase inhibitor proteins are more dominant in graminaceous plants such as wheat. Xylanases play important roles in fungal virulence. The endo-β-1,4-xylanase Xyn11A was shown to required for virulence in Botrytis cinerea [41] and xylanases knock-down mutants of Magnaporthe oryzae also caused less lesions compared to wild type M. oryzae [42]. Our results revealed FvXyn11A and FvXyn11B lack amino acid substitutions that would avoid XIP-I inhibition. Because XIP-I has been reported to inhibit both GH10 and GH11 xylanases [3–5], we consider XIP-I a better candidate since multiple targets of XIP-I may extend the persistence of the transgenic soybeans. In addition to inhibition of GH11 xylanase, XIP-I was reported to reduce cell death induced by necrosis-inducing xylanases, such as XylA and XylB of F. graminearum [8], and an orthologous XIP-I from coffee has been shown to inhibit the germination of soybean rust urediniospores [43]. Developing a transgenic soybean that expresses an exogenous XIP-I might not only reduce soybean rust infection but also enhance resistance against SDS.
Methods
Identification of CAZy in the F. virguliforme genome
The Fusarium virguliforme genome sequence (accession AEYB01000000) was downloaded from NCBI and is available at http://fvgbrowse.agron.iastate.edu [23]. Augustus was used to predict putative proteins in the genome and transcriptome with F. graminearum as a model organism using default parameters except for the minexonintronprob (=0.1) and minmeanexonintronprob (=0.4) [44]. CAZy domains were identified in genomes with dbCAN and a cutoff of E value of 10-3 [45]. When a gene contained a CBM with other CAZy classes, the gene was classified in the later classes. When redundancies were detected, classification was determined based on the lowest E value (Additional file 1: Table S1). Protein annotation was based on the CAZy database [25, 46]. The genomic CAZy structure of F. virguliforme was compared to other plant pathogenic fungi and oomycetes [28–30].
In silico analyses of GH11 xylanases and GH28 PGs of F. virguliforme
Sequences of P. funiculosum GH11 xylanase XYNC [5] and F. phyllophilum GH28 FpPG [36] were used as queries to identify orthologous genes in F. virguliforme. Putative orthologous GH11 xylanases and GH28 PGs were determined at E value 10-50. MUSCLE in MEGA6 was used for protein sequence alignment [47]. SignalP 4.1 was used to detect secretory signal peptide [48]. SWISS-MODEL and QMEAN [49–52] were used to generate and evaluate a homology model for FvXyn11A (QMEAN6: 0.675) and FvXyn11B (QMEAN6: 0.708) based on Chaetomium thermophilum GH11 xylanase model 1h1a [53]. The protein-protein docking was performed by ZDOCK [54, 55]. The residue, E85 of P. funiculosum XYNC, E114 of FvXyn11A, and E98 of FvXyn11B, was set as indispensable interacting residuals with R179 of XIP-I and the modeling result was compare to interaction model 1te1 [5].
Expression analysis of GH11 xylanases and GH28 PGs in vitro and in planta
In vitro RNA-Seq transcriptome was downloaded from DDBJ/EMBL/GenBank accession GBJV00000000 and analyzed as previously described [38]. HTSeq (version 0.6.1) were applied to quantify mapped reads for each transcript [56]. Transcripts with less than 60 reads across six libraries were filtered out in R (version 3.0.1) [57]. A false discovery rate of 0.05 was used as significant cutoff in edgeR analysis (version 3.6.4) [57–60]. Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was used to measure gene expression during root infection. Soybean seeds were germinated for 5 days at 25 °C. Each radicle was inoculated with 15 μl of 1× 106 macroconidia per ml of F. virguliforme, and then incubated without light at 25 °C for 5 days before extracting total RNA by using TRIzol. Random primers were used to synthesize cDNA. Amplification efficiency of primers for qRT-PCR was determined based on four replicates and each replicate contained three concentration gradients (Table 3). Platinum® SYBR® Green qPCR SuperMix-UDG kit (Life Technologies) and Agilent Mx3005P qPCR System (Agilent Technologies) were used for qRT-PCR experiments. –ΔΔCt method was used to evaluate the expression of each gene [61] and gene expression was normalized to the translation elongation factor 1A of F. virguliforme (FvEF1A) [38, 62]. In planta gene expression analysis was repeated three times with three biological replicates for each. Statistics were conducted in R. Box-Cox power transformation was applied on raw data to fulfill the normal distribution of residuals. ANOVA and TukeyHSD were used to determine significance at p < 0.05.
Abbreviations
AA, auxiliary activity; CAZy, carbohydrate-active enzymes; CBM, carbohydrate-binding module; CE, carbohydrate esterases; GH, glycoside hydrolases; GT, glycosyl-transferases; PCWDEs, plant cell wall degrading enzymes; PG, polygalacturonase; PGIP, polygalacturonase-inhibitor protein; PIPs, plant cell wall degrading enzyme inhibitor proteins; PL, polysaccharide lyases; SDS, sudden death syndrome; TAXI-III, Triticum aestivum xylanase inhibitor III; XIP-I, xylanase inhibitor protein I
References
Beliën T, Campenhout SV, Robben J, Volckaert G. Microbial endoxylanases: Effective weapons to breach the plant cell-wall barrier or, rather, triggers of plant defense systems? Mol Plant Microbe Interact. 2006;19:1072–81.
Federici L, Matteo AD, Fernandez-Recio J, Tsernoglou D, Cervone F. Polygalacturonase inhibiting proteins: players in plant innate immunity? Trends Plant Sci. 2006;11:65–70.
Misas-Villamil JC, Hoorn RA. Enzyme–inhibitor interactions at the plant–pathogen interface. Curr Opin Plant Biol. 2008;11:380–8.
Beliën T, Campenhout SV, Acker MV, Robben J, Courtin CM, Delcour JA, Volckaert G. Mutational analysis of endoxylanases XylA and XylB from the phytopathogen Fusarium graminearum reveals comprehensive insights into their inhibitor insensitivity. Appl Environ Microbiol. 2007;73:4602–8.
Payan F, Leone P, Porciero S, Furniss C, Tahir T, Williamson G, Durand A, Manzanares P, Gilbert HJ, Juge N, et al. The dual nature of the wheat xylanase protein inhitor XIP-I. J Biol Chem. 2004;279:36029–37.
Moscetti I, Tundo S, Janni M, Sella L, Gazzetti K, Tauzin A, Giardina T, Masci S, Favaron F, D’Ovidio R. Constitutive expression of the xylanase inhibitor TAXI-III delays Fusarium head blight symptoms in Durum wheat transgenic plants. Mol Plant Microbe Interact. 2013;26:1464–72.
Moscetti I, Faoro F, Moro S, Sabbadin D, Sella L, Favaron F, D’Ovidio R. The xylanase inhibitor TAXI-III counteracts the necrotic activity of a Fusarium graminearum xylanase in vitro and in durum wheat transgenic plants. Mol Plant Pathol. 2015;16:583–92.
Tundoa S, Moscettia I, Faorob F, Ml L, Giardinac T, Favarond F, Sellad L, D’Ovidioa R. Fusarium graminearum produces different xylanases causing host cell death that is prevented by the xylanase inhibitors XIP-I and TAXI-III in wheat. Plant Sci. 2015;240:161–9.
Di Matteo A, Bonivento D, Tsernoglou D, Federici L, Cervone F. Polygalacturonase-inhibiting protein (PGIP) in plant defence: a structural view. Phytochemistry. 2006;67:528–33.
Agüero CB, Uratsu SL, Greve C, Powell ALT, Labavitch JM, Meredith CP, Dandekar AM. Evaluation of tolerance to Pierce’s disease and Botrytis in transgenic plants of Vitis vinifera L. expressing the pear PGIP gene. Mol Plant Pathol. 2005;6:43–51.
Akhgari AB, Motallebi M, Zamani MR. Bean polygalacturonase-inhibiting protein expressed in transgenic Brassica napus inhibits polygalacturonase from its fungal pathogen Rhizoctonia solani. Plant Prot Sci. 2012;48:1–9.
Ferrari S, Sella L, Janni M, Lorenzo GD, Favaron F, D’Ovidio R. Transgenic expression of polygalacturonase-inhibiting proteins in Arabidopsis and wheat increases resistance to the flower pathogen Fusarium graminearum. Plant Biol. 2012;14:31–8.
Joubert DA, Slaughter AR, Kemp G, Becker JVW, Krooshof GH, Bergmann C, Benen J, Pretorius IS, Vivier MA. The grapevine polygalacturonase-inhibiting protein (VvPGIP1) reduces Botrytis cinerea susceptibility in transgenic tobacco and differentially inhibits fungal polygalacturonases. Transgenic Res. 2006;15:687–702.
Manfredini C, Sicilia F, Ferrari S, Pontiggia D, Salvi G, Caprari C, Lorito M, Lorenzo GD. Polygalacturonase-inhibiting protein 2 of Phaseolus vulgaris inhibits BcPG1, a polygalacturonase of Botrytis cinerea important for pathogenicity, and protects transgenic plants from infection. Physiol Mol Plant Path. 2005;67:108–15.
Powell ALT, Kan Jv, Have At, Visser J, Greve LC, Bennett AB, Labavitch JM. Transgenic expression of pear PGIP in tomato limits fungal colonization. Mol Plant Microbe Interact. 2000;13:942–50.
Wang X, Zhu X, Tooley P, Zhang X. Cloning and functional analysis of three genes encoding polygalacturonase-inhibiting proteins from Capsicum annuum and transgenic CaPGIP1 in tobacco in relation to increased resistance to two fungal pathogens. Plant Molec Biol. 2013;81:379–400.
Benedetti M, Pontiggia D, Raggi S, Cheng Z, Scalon F, Ferrari S, Ausubel F, Cervone F, Lorenzo GD. Plant immunity triggered by engineered in vivo release of oligogalacturonides, damage-associated molecular patterns. Proc Natl Acad Sci U S A. 2015;12:5533–8.
Hartman GL, Chang H-X, Leandro LF. Research advances and management of soybean sudden death syndrome. Crop Prot. 2015;73:60–6.
Lightfoot D. Two decades of molecular marker-assisted breeding for resistance to soybean sudden death syndrome. Crop Sci. 2015;55:1460–84.
Brar H, Bhattacharyya MK. Expression of a single-chain variable-fragment antibody against a Fusarium virguliforme toxin peptide enhances tolerance to sudden death syndrome in transgenic soybean plants. Mol Plant Microbe Interact. 2012;6:817–24.
Iqbal MJ, Yaegashi S, Ahsan R, Shopinski KL, Lightfoot DA. Root response to Fusarium solani f. sp. glycines: temporal accumulation of transcripts in partially resistant and susceptible soybean. Theor Appl Genet. 2005;110:1429–38.
Radwan O, Liu Y, Clough SJ. Transcriptional analysis of soybean root response to Fusarium virguliforme, the causal agent of sudden death syndrome. Mol Plant Microbe Interact. 2011;24:958–72.
Srivastava SK, Huang X, Brar HK, Fakhoury AM, Bluhm BH, Bhattacharyya MK. The genome sequence of the fungal pathogen Fusarium virguliforme that causes sudden death syndrome in soybean. PLoS One. 2014;9:e81832.
Glass NL, Schmoll M, Cate JHD, Coradetti S. Plant cell wall deconstruction by ascomycete fungi. Annu Rev Microbiol. 2013;67:477–98.
Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2013;42:D490–D5.
Lagaert S, Beliën T, Volckaert G. Plant cell walls: Protecting the barrier from degradation by microbial enzymes. Semin Cell Dev Biol. 2009;20:1064–73.
van den Brink J, de Vries R. Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol. 2011;91:1477–92.
Blackman L, Cullerne D, Hardham A. Bioinformatic characterisation of genes encoding cell wall degrading enzymes in the Phytophthora parasitica genome. BMC Genomics. 2014;15:785.
Zerillo MM, Adhikari BN, Hamilton JP, Buell CR, Vesque CAL, Tisserat N. Carbohydrate-active enzymes in Pythium and their role in plant cell wall and storage polysaccharide degradation. PLoS One. 2013;8(9):E72572.
Zhao Z, Liu H, Wang C, Xu R-J. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics. 2013;14:274.
Levasseur A, Drula E, Lombard V, Coutinho P, Henrissat B. Expansion of the enzymatic repertoire of the CAZy databse to integrate auxiliary redox enzymes. Biotechnol Biofuels. 2013;6:41.
Caffal KH, Mohnen D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res. 2009;344:1879–900.
Willats WG, McCartney L, Mackie W, Knox JP. Pectin: cell biology and prospects for functional analysis. Plant Mol Biol. 2001;47:9–27.
Beliën T, Campenhout SV, Acker MV, Volckaert G. Cloning and characterization of two endoxylanases from the cereal phytopathogen Fusarium graminearum and their inhibition profile against endoxylanase inhibitors from wheat. Biochem Biophys Res Commun. 2005;327:407–14.
Raiola A, Sella L, Castiglioni C, Balmas V, Favaron F. A single amino acid substitution in highly similar endo-PGs from Fusarium verticillioides and related Fusarium species affects PGIP inhibition. Fungal Genet Biol. 2008;45:776–89.
Benedetti M, Andreani F, Leggio C, Galantini L, Matteo AD, Pavel NV, Lorenzo GD, Cervone F, Federici L, Sicilia F. A single amino-acid substitution allows endo-polygalacturonase of Fusarium verticillioides to acquire recognition by PGIP2 from Phaseolus vulgaris. PLoS One. 2013;8:e80610.
Sella L, Castiglioni C, Roberti S, D'Ovidio R, Favaron F. An endo-polygalacturonase (PG) of Fusarium moniliforme escaping inhibition by plant polygalacturonase-inhibiting proteins (PGIPs) provides new insights into the PG–PGIP interaction. FEMS Microbiol Let. 2004;240:117–24.
Chang H-X, Domier LL, Radwan O, Yendrek C, Hudson M. Identification of multiple phytotoxins produced by Fusarium virguliforme including a phytotoxic effector (FvNIS1) associated with sudden death syndrome foliar symptoms. Mol Plant Microbe Interact. 2016;29:96–108.
Ruiz G, Di Pietro A, Roncero I. Combined action of the major secreted exo- and endopolygalacturonases is required for full virulence of Fusarium oxysporum. Mol Plant Pathol. 2015;DOI: 10.1111/mpp.12283.
Sella L, Gazzetti K, Faoro F, Odorizzi S, D’Ovidio R, Schäfer W, Favaron F. A Fusarium graminearum xylanase expressed during wheat infection is a necrotizing factor but is not essential for virulence. Plant Physiol Biochem. 2013;64:1–10.
Nl B, Espino JJ, González C. The endo-β-1,4-xylanase Xyn11A is required for virulence in Botrytis cinerea. Mol Plant Microbe Interact. 2006;19:25–32.
Nguyen QB, Itoh K, Vu BV, Tosa Y, Nakayashiki H. Simultaneous silencing of endo-b-1,4 xylanase genes reveals their roles in the virulence of Magnaporthe oryzae. Mol Microbiol. 2011;81:1008–19.
Vasconcelos EA, Santana CG, Godoy CV, Seixas CD, Silva MS, Moreira LR, Oliveira-Neto OB, Price D, Fitches E, Filho EX, et al. A new chitinase-like xylanase inhibitor protein (XIP) from coffee (Coffea arabica) affects Soybean Asian rust (Phakopsora pachyrhizi) spore germination. BMC Biotechnol. 2011;11:14.
Keller O, Kollmar M, Stanke M, Waack S. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics. 2011;27:757–63.
Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W51.
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–8.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;12:2725–9.
Petersen TN, Brunak S, Heijne G, Nielsend H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6.
Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201.
Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–50.
Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;doi: 10.1093/nar/gku340.
Guex N, Peitsch M, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss- PdbViewer: A historical perspective. Eletrophoresis. 2009;30:S162–S73.
Hakulinen N, Turunen O, Janis J, Leisola M, Rouvinen J. Three-dimensional structures of thermophilic b-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa. Eur J Biochem. 2003;270:1399–412.
Moreira IS, Fernandes PA, Ramos MJ. Protein-protein docking dealing with the unknown. J Comp Chem. 2010;31:317–42.
Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS One. 2011;6:e24657.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106.
Yendrek C, Ainsworth E, Thimmapuram J. The bench scientist’s guide to statistical analysis of RNA-Seq data. BMC Res Notes. 2012;5:506.
MacDonald J. affycoretools: Functions useful for those doing repetitive analyses with Affymetrix GeneChips. R package version 1321; 2008.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. p. 397–420.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–8.
Kim H-K, Yun S-H. Evaluation of potential reference genes for quantitative RT-PCR analysis in Fusarium graminearum under different culture conditions. J Plant Pathol. 2011;27:301–9.
Noda J, Brito N, González C. The Botrytis cinerea xylanase Xyn11A contributes to virulence with its necrotizing activity, not with its catalytic activity. BMC Plant Biol. 2010;10:38.
Rotblat B, Enshell-Seijffers D, Gershoni JM, Schuster S, Avni A. Identification of an essential component of the elicitation active site of the EIX protein elicitor. Plant J. 2002;32:1049–55.
Federici L, Caprari C, Mattei B, Savino C, Matteo AD, Lorenzo GD, Cervone F, Tsernoglou D. Structural requirements of endopolygalacturonase for the interaction with PGIP (polygalacturonase-inhibiting protein). Proc Natl Acad Sci USA. 2001;98:13425–30.
van Santen Y, Benen JAE, Schröter K-H, Kalk KH, Armand S, Visser J, Dijkstra BW. 1.68-Å crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis. J Biol Chem. 1999;274:30474–80.
Acknowledgements
We thank Mr. Ian Hu from the Department of Biochemistry, University of Cambridge, for instructive suggestions on protein structural modeling.
Funding
The project was partially supported by the United States Department of Agriculture-Agricultural Research Service and the University of Illinois.
Availability of data and materials
Genome sequence of F. virguliforme is available in NCBI accession AEYB01000000 and in vitro RNA-Seq data is available in DDBJ/EMBL/GenBank accession GBJV00000000. Predicted coding sequences for 629 CAZy are available in the Additional file 1: Table S1.
Authors’ contributions
HXC designed the study, analyzed the data, and wrote the manuscript; CRY assisted with data analyses and wrote the manuscript; GCA assisted with understanding of the data and wrote the manuscript; GLH initiated and supervised the study and wrote the manuscript. All authors read and approved the final version.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article is available at http://dx.doi.org/10.1186/s12866-017-1020-8.
Additional file
Additional file 1: Table S1.
CAZy of Fusarium virguliforme. (XLSX 480 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Chang, HX., Yendrek, C.R., Caetano-Anolles, G. et al. Genomic characterization of plant cell wall degrading enzymes and in silico analysis of xylanses and polygalacturonases of Fusarium virguliforme . BMC Microbiol 16, 147 (2016). https://doi.org/10.1186/s12866-016-0761-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-016-0761-0