Abstract
Phenotypic differences among breeding lines that introduce the same superior gene allele can be a barrier to effective development of cultivars with desirable traits in some crop species. For example, a deficient mutation of the Protein Disulfide Isomerase Like 1–1 (PDIL1-1) gene can cause accumulation of glutelin seed storage protein precursors in rice endosperm, and improves rice flour characteristics and food processing properties. However, the gene must be expressed to be useful. A deficient mutant allele of PDIL1-1 was introduced into two rice cultivars with different genetic backgrounds (Koshihikari and Oonari). The grain components, agronomic traits, and rice flour and food processing properties of the resulting lines were evaluated. The two breeding lines had similar seed storage protein accumulation, amylose content, and low-molecular-weight metabolites. However, only the Koshihikari breeding line had high flour quality and was highly suitable for rice bread, noodles, and sponge cake, evidence of the formation of high-molecular-weight protein complexes in the endosperm. Transcriptome analysis revealed that mRNA levels of fourteen PDI, Ero1, and BiP genes were increased in the Koshihikari breeding line, whereas this change was not observed in the Oonari breeding line. We elucidated part of the molecular basis of the phenotypic differences between two breeding lines possessing the same mutant allele in different genetic backgrounds. The results suggest that certain genetic backgrounds can negate the beneficial effect of the PDIL1-1 mutant allele. Better understanding of the molecular basis for such interactions may accelerate future breeding of novel rice cultivars to meet the strong demand for gluten-free foods.
Similar content being viewed by others
Background
Rice (Oryza sativa L.) is a staple food for more than half of the world’s population. Consumer preference causes strong demand for high-quality grain in rice cultivars, although increasing crop yield has long been an important requirement in many rice breeding programs (Champagne et al. 1999; Fitzgerald et al. 2009; Hori and Yano 2013; Custodio et al. 2019). Rice is mainly cooked and eaten as polished (white) rice, but it is also made into flour for noodles, cakes, dumplings, and gluten-free bread. Global improvements in living standards and greater demand for gluten-free foods are increasing the use of rice flours for breads, noodles, and cakes throughout the world (Ashida et al. 2009; Yano 2012; Ashida 2014; Kwak et al. 2017; Fradinho et al. 2019; Paz et al. 2020). However, few of the rice cultivars in current use are suitable for making these products (Sasahara et al. 2013; Ohta et al. 2015; Kwak et al. 2017). Therefore, new rice cultivars with both good processing attributes and high eating quality for various food applications need to be developed to meet consumer preferences, including gluten-free foods.
Many genes affect grain components, and cooking and eating quality (Fiaz et al. 2019; Guo et al. 2020). Recent progress in the genetic analysis of grain quality traits has identified many quantitative trait loci (QTLs) and their underlying genes. Several genes involved in the control of grain quality have been identified so far, but the regulatory mechanisms that contribute to the grain quality traits remain unclear (Zhang et al. 2021; Liu et al. 2022). Genetic control of rice grain quality has been successfully elucidated by gene identification through map-based cloning and the development of mutant lines.
The main components of polished rice grain (endosperm) are starch (up to 95% of dry weight), protein (5–7%), lipids (0.5–1%), as well as minerals, vitamins, and other micronutrients (less than 1%). All of these components affect the cooking characteristics and eating quality (Champagne et al. 1999; Fitzgerald et al. 2009). As the main component of the grain, starch largely controls the cooking characteristics and eating quality, but the seed storage proteins have the second most important effect on these traits (Martin and Fitzgerald 2002; Hori 2018; Hori et al. 2021b). Wheat flour contains the seed storage protein glutenin, which is necessary for the swelling and rising of doughs in breads, noodles, and cakes. Glutenin is converted to high-molecular-weight glutens with three-dimensional reticulate protein structures by inter-molecular disulfide bond formation with other seed storage proteins (gliadins) during food processing (Hu et al. 2021). However, seed storage proteins of rice grain do not form the same three-dimensional reticulate structures as wheat glutenin. This is a serious problem for the development of rice cultivars for flour and gluten-free food production. If the grain proteins in a rice cultivar formed inter-molecular disulfide bonds as effectively as the glutenin and gliadin do in wheat, such a cultivar would be a novel and useful ingredient for making breads, noodles, and cakes.
Rice grain has three classes of seed storage proteins: acid-soluble glutelin, alcohol-soluble prolamin, and salt-soluble globulin (Padhye and Salunkhe 1979; Juliano and Houston 1985). The glutelin in rice is homologous to glutenin in wheat, barley, and rye and to the 11S globulins in soybean and pea (Zhao et al. 1983; Takaiwa et al. 1987). Prolamin is typically found in cereal crops and is homologous to gliadin in wheat (Ogawa et al. 1987; Muench 1999). Globulin is accumulated in some cereal species including oats and rice (Padhye and Salunkhe 1979). Glutelin is initially synthesized on the endoplasmic reticulum (ER) as a 57-kDa precursor that is exported to the protein storage vacuole to form protein body II (PBII); this is converted to acid- and alkali-soluble subunits and post-translationally processed into two subunits linked by intra-molecular disulfide bonds (Yamagata et al. 1982).
Rice has at least 12 genes encoding protein disulfide isomerases (PDI), similar to Arabidopsis and maize (Houston et al. 2005). The endosperm storage protein 2 (esp2) gene was identified in a seed storage protein mutant line of rice that lacks the protein disulfide isomerase-like 1–1 (PDIL1-1) protein (Takemoto et al. 2002). The molecular function of PDIL1-1 is to create the intra-molecular disulfide bonds between two glutelin subunits to facilitate folding of the mature protein into its functional conformation and to enable ER export. Immunoblot analyses to investigate subcellular fractions show that the esp2 mutant line accumulates glutelin precursors as long undigested peptide chains containing both subunits, located inside large protein storage vacuoles in the endosperm (Takemoto et al. 2002; Satoh-Cruz et al. 2010; Fukuda and Kumamaru 2019). The PDIL1-1 protein has high enzyme activity for both formation and reduction of disulfide bonds, compared with other rice PDIL proteins (Onda et al. 2011; Onda and Kobori 2014). Transcriptomics and proteomics approaches have shown that PDIL1-1 also controls the expression of other seed storage protein and starch biosynthesis genes (Han et al. 2012; Kim et al. 2012). Recently, additional biochemical analysis showed that PDIL1-1 creates disulfide bonds mediated by microRNA5144, and it directly interacts with the cysteine protease OsCP1 protein (Kim et al. 2012; Xia et al. 2018). The importance of PDIL1-1 in the biosynthesis and accumulation of grain components such as seed storage proteins and starches in rice endosperm is clear.
Rice with the esp2 mutant allele has good flour and food processing characteristics suitable for making bread, although the original esp2 mutant line has the disadvantage of quite low grain yield (Kawagoe 2010; Hori 2016). To develop novel rice cultivars showing good rice flour and food processing properties together with high grain yield, the esp2 mutant was crossed with a high-eating-quality cultivar and with a high yield cultivar, and the resulting lines were evaluated.
Materials and Methods
Plant Materials
The esp2 mutant EM747 was crossed with the high-eating-quality japonica rice cultivar, Koshihikari, to produce F1 plants. EM747 was induced by N-methyl-N-nitrosourea treatment of the japonica rice cultivar Taichung 65 (Takemoto et al. 2002). The EM747 plants have a nucleotide substitution at exon splicing sites in the PDIL1-1 gene (Os11g0199200, LOC_Os11g09280), resulting in the nonfunctional gene allele. One F6 line was selected as Koshihikari esp2, using single seed descent and marker assisted selections from the F2 generation to develop an esp2 mutant line with high agronomic performance. Next, Koshihikari esp2 was crossed with the high yielding indica cultivar Oonari and self-pollinated to the BC3F5 generation, from which Oonari esp2 was selected. Whole genome genotyping of Koshihikari esp2 and Oonari esp2 was carried out using the Fluidigm SNP genotyping platform (Fluidigm, San Francisco, CA, USA) with a set of 198 SNPs to detect polymorphisms between Koshihikari and EM747 and between Koshihikari and Oonari. Gene-specific DNA markers of 17 yield-related genes identified by Hori et al. (2021b) were used to genotype Koshihikari esp2 and Oonari esp2. Koshihikari is a cultivar with high eating and cooking quality, which has been the most widely grown type in Japan for over 40 years (Kobayashi et al. 2018; Hori et al. 2021a) while Oonari is cultivar with high grain yield (Kobayashi et al. 2016). In this study, we tried to develop novel breeding lines showing high quality for rice flour and food processing characteristics together with high grain yield.
Scoring Agronomic Traits
Rice cultivars and breeding lines were cultivated from 2013 to 2018, using six experimental fields in Japan. These were Tsukuba (36.02° N, 140.11° E) and Tsukubamirai (36.01° N, 140.02° E) at the Institute of Crop Science, NARO, Chikusei (36.30° N, 140.02° E) in the Oshima Nojo K.K., Fujieda (34.90° N, 138.28° E) at Shizuoka University, Kasai (34.88° N, 134.86° E) at Kobe University, and Chikugo (33.21° N, 130.49° E) at the Kyushu Okinawa Agricultural Research Center, NARO (Additional file 1: Fig. S2a). All experimental fields were within regions suitable for cultivation of Koshihikari and Oonari (Kobayashi et al. 2016, 2018). Koshihikari and Koshihikari esp2 were grown at Tsukuba in 2016, 2017, and 2018, at Tsukubamirai in 2015 and 2016, and at Fujieda and Kasai in 2017. Oonari and Oonari esp2 were cultivated at Tsukuba in 2016, and in all six experimental fields in 2017 and 2018. The planting density of each line was 20.0 individuals per m2 with two replications at all six experimental fields. Month-old seedlings were transplanted in mid-May and maturing plants were harvested in September at Tsukuba, Fujieda, and Kasai; seedlings were transplanted in early July and maturing plants were harvested in October at Tsukubamirai, Chikusei, and Chikugo. Agronomic traits were evaluated at each experimental field using standardized procedures.
Evaluation of Grain Components
Total protein and amylose contents in matured grains were evaluated in each year from 2013 to 2018 using the methods of Hori et al. (2021b). Apparent amylose content was determined using an Auto Analyzer II (Bran + Luebbe, Norderstedt, Germany). Crude protein content was determined by the combustion method with an induction furnace at 900 °C (American Association of Cereal Chemists International, Approved Method 46-30.01). Measurement of low-molecular-weight compounds was carried out in 2015 and 2016, using the methods of Human Metabolome Technologies (Tsuruoka, Japan). Briefly, rice flours from each line were homogenized in 600 µL methanol containing 10 µM internal standards, mixed with 600 µL chloroform and 240 µL water, then centrifuged at 2300 × g for 5 min. The aqueous supernatant fraction was filtered and recovered into 50 µL of MilliQ water prior to metabolite analysis using capillary electrophoresis time of flight-mass spectrometry (CE-TOFMS) (Agilent Technologies, CA, USA) (Soga et al. 2004). Peaks detected in the CE-TOFMS analysis were extracted using automatic integration software (MasterHands ver. 2.17.1.11) and annotated with putative metabolites based on their migration in CE and m/z values.
Evaluation of Grain Milling and Food Processing Properties
Grain milling characteristics and food processing properties were evaluated in Koshihikari and Koshihikari esp2 from 2013 to 2016 and in Oonari and Oonari esp2 in 2017 and 2018. Head brown rice grains of each line were polished to 90% of their original weight, to obtain polished (white) rice., and the polished rice yields for each line were recorded. The polished rice was milled by four methods: airflow wet grinding, airflow dry grinding, roll wet grinding, and roll dry grinding. Moisture, ash, damaged starch content, mean volume diameter (d50) of flour particles, and electricity consumption for milling were evaluated as rice flour characteristics, using the methods of Hayakawa et al. (2004) and Okunishi (2014). The mixing properties of rice flours were analyzed with a farinograph, using 500 BU as the standard. Rice breads were made from 20%, 30%, 50%, and 80% rice flours, with the balance wheat flour or appropriate gluten. The loaf volume, weight, and hardness after baking were evaluated. Sponge cakes were made with 100% rice flour, and their volume, weight, and center height after baking were evaluated. Rice noodles were made from 20 and 50% rice flours with the balance wheat flour or appropriate gluten, and their hardness, stickiness, and brittleness were evaluated by sensory eating tests after boiling noodles. Evaluations of food processing properties related to rice breads, cakes, and noodles were performed according to the methods of Hayakawa et al. (2004) and Okunishi (2014).
Quantitative RT-PCR Analysis of Gene Expression
The causes of phenotypic differences in rice flour characteristics and food processing properties in Koshihikari, Koshihikari esp2, Oonari, and Oonari esp2 were investigated by assessing the mRNA expression of 18 genes involved in seed storage protein synthesis, including the PDIL genes, the Luminal binding protein (BiP) gene family, and the ER membrane-localized oxidoreductase 1 (Ero1) gene in immature rice grains at the grain filling stage. Total RNA was extracted from grains and from leaves at the grain filling stage of growth in 2018, using the RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) and primed with the oligo(dT)12-18 primer with SuperScript II reverse transcriptase (Invitrogen, MA, USA). cDNA corresponding to 50 ng of total RNA was used as the template for each SYBR Green–based PCR reaction, using gene-specific primers (Additional file 2: Table S1). Transcription levels of PDI-like genes, BiP family genes, Ero1, Granule-bound starch synthase I (GBSSI), Starch synthase I (SSI), Starch synthase IIIa (SSIIIa), and Rice ubiquitin2 (UBQ) were determined according to the methods of Shibaya et al. (2016). Expression of the genes of interest was quantified relative to the expression of the UBQ gene. Gene expression data are presented as the means of at least three biological replicates, with three technical repeats for each biological replicate.
SDS-PAGE Analysis of Seed Storage Proteins
Storage proteins from mature grain harvested in 2018 were extracted in the following series of elution buffers: 0.1 M NaCl, 70% ethanol, 0.1 M acetic acid, 2% SDS, and 2% SDS with 0.01 M DTT. Each elution sample was vigorously shaken for 30 min at 4 °C and then centrifuged at 15,000 × g for 5 min at 4 °C. Supernatants were used as loading samples for gel electrophoresis, and the precipitates (pellets) were dissolved in the next buffer solution in the series. Reducing and non-reducing SDS-PAGE analyses (with or without 2-mercaptoethanol, respectively) were conducted on 12.5% polyacrylamide concentration gels. The total amount of protein was calculated as the sum of all proteins in each elution buffer, including supernatants and final precipitations in both reducing and non-reducing conditions, using the CN Coder MT700 mark2 (Yanako Technical Science, Tokyo, Japan).
Results
Development of the Koshihikari esp2 and Oonari esp2 Lines
From the cross of the esp2 mutant line EM747 with the high-eating-quality cultivar Koshihikari, 85 individual plants in the F2 generation showed a wide range of phenotypic variations for agronomic traits such as heading date and grain yield (Additional file 1: Fig. S1a–c). A DNA marker for the esp2 mutation was identified in the F2, F3, F4, F5, and F6 lines (e.g., Additional file 1: Fig. S1d). Koshihikari esp2 had good agronomic performance with early heading date and good grain yield (Fig. 1a, Additional file 1: Fig. S1b, c). Whole genome genotyping demonstrated that the Oonari esp2 line developed from Koshihikari esp2 has all 17 of the gene alleles from Oonari that are associated with high grain yield (Additional file 1: Fig. S1f). This confirmed that the marker-assisted selection successfully introduced the esp2 mutation into both breeding lines.
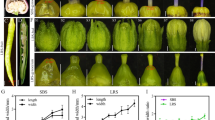
Phenotypic characteristics of rice from Koshihikari (K), Koshihikari esp2 (Ke), Oonari (O), and Oonari esp2 (Oe) lines. a Appearance of representative rice plants, matured grains and transections of grains in 2015. b Representative developed SDS-PAGE gel of seed storage proteins; black, white, and gray triangles indicate glutelin precursor, matured glutelin, and prolamin, respectively. c Glutelin precursor, d total protein, and e amylose content of mature rice grains; means of data from 2013 to 2018. f Principal component analysis, g hierarchical cluster analysis, and h relative area of low-molecular-weight metabolites in mature rice grains in 2015; black and gray triangles in g indicate increased and decreased levels of metabolites, respectively, in the esp2 lines as compared with the respective parental lines. Asterisks indicate significant difference from the Koshihikari parental line at P < 0.05 (*), < 0.01 (**), and < 0.001 (***)
In 2016, Koshihikari esp2 had earlier heading date (3 August vs. 19 August), increased unhulled grain weight (22.0 vs. 14.2 g/plant), and increased head brown grain weight (18.3 vs. 12.1 g/plant) compared with the original esp2 mutant line EM747 (Table 1). Interestingly, several agronomic traits in Koshihikari esp2 were different from those in the Koshihikari parent line (Table 1). For example, in 2016 the heading dates were similar in Koshihikari esp2 and Koshihikari (3 and 1 August, respectively), but significant decreases were observed in plant height (65.5 vs. 73.3 cm), number of tillers per plant (12.0 vs. 13.8), culm length (88.7 vs. 93.1 cm), unhulled grain weight (22.0 vs. 30.5 g/plant), and head brown rice grain weight (18.3 vs. 25.0 g/plant). Similar differences were observed in other years and at other field locations (Table 1, Additional file 1: Fig. S2, Additional file 2: Tables S2, S3). Oonari esp2 had high unhulled grain weight and head brown grain weight (e.g., 32.3 g/plant and 27.4 g/plant, respectively, in 2016; Table 1), but these were not significantly different from Oonari. Oonari esp2 and Oonari were also similar in other agronomic traits including heading date, plant height, culm length, and number of tillers (Table 1). In summary, incorporation of the esp2 mutant allele changed several agronomic traits in the Koshihikari genetic background, but not in Oonari.
Grain Components of the Koshihikari and Oonari esp2 Lines
Koshihikari esp2 and Oonari esp2 had translucent grains with some chalkiness (Fig. 1a; Hori 2018). Both esp2 lines accumulated more than twice the quantity of glutelin precursors in the matured grain as was observed in the respective parental lines (Fig. 1b, c). Accumulation of glutelin precursors is one of the representative phenotypes of the esp2 and other seed storage protein mutations associated with transport of proteins from ER to Golgi and protein bodies (PBI and PBII) (Takemoto et al. 2002; Fukuda and Kumamaru 2019). The total protein content of matured grain was similar in the two esp2 lines, but the amylose content was about 2% higher in the breeding lines than in the parental lines (Fig. 1d, e). Metabolome analysis with CE-TOFMS detected a total of 213 compounds in Koshihikari, Koshihikari esp2, Oonari, and Oonari esp2 in 2015 and 240 compounds in 2016 (Fig. 1g, Additional file 1: Fig. S3, Additional file 2: Table S4). Principal component hierarchical cluster analysis of the 2015 data clearly distinguished the different cultivars as the first principal component and the presence or absence of the esp2 mutation as the second principal component (Fig. 1f). Koshihikari esp2 and Oonari esp2 had increased content of several amino acids and organic acids such as asparagine, histidine, leucine, and allantoic acid compared with the respective parental lines, but decreased content of several fatty acids such as fumaric acid and 6-aminohexanoic acid in 2015 (Fig. 1h) and 2016 (Additional file 1: Fig. S3, Additional file 2: Table S4). Saccharide contents, including glucose, sucrose, and fructose, were not significantly different between Koshihikari esp2 and Koshihikari or between Oonari esp2 and Oonari (data not presented). These observations indicate that the alterations of grain components attributable to introduction of the esp2 mutation were similar in the Koshihikari and Oonari genetic backgrounds.
Grain Milling and Food Processing Properties in the Koshihikari and Oonari esp2 Lines
Several starch and seed storage protein mutants significantly decrease polished (white) rice yields due to high incidence of chalky and soft grains (Ashida et al. 2009; Mo and Jeung 2020). However, in the work reported here, the Koshihikari esp2 and Oonari esp2 lines showed the same proportion of polished rice (approximately 90.0%) as in their respective parental lines during three years of testing (Table 1). This suggests that the esp2 mutation maintained polished rice yield, unlike other reported starch and seed storage protein mutant lines.
Among the four milling methods used in all six years of the work reported here, the airflow wet grinding method produced the smallest mean diameter of rice flours with the lowest content of damaged starch (Fig. 2a, Additional file 2: Table S5). In 2013, Koshihikari esp2 had significantly smaller diameter of rice flour (38.6 µm) and decreased electricity consumption (1.0 kW) in the airflow wet grinding method and lower damaged starch content (5.4%) in the roll wet grinding method, as compared with Koshihikari (51.1 µm, 1.5 kW, and 6.4%, respectively). In contrast, Oonari esp2 scores for percent damaged starch, mean flour diameter, and electricity consumption were not significantly different from Oonari in all four milling methods.
Rice flour milling and food processing characteristics. a Damaged starch content, starch granule diameter, and electricity consumption; means of data from 2013 to 2018. b Bread-making properties; scale bars indicate 5 cm. c Sponge cake properties; scale bars indicate 1 cm. d Sensory testing of rice noodles. Asterisks indicate significant difference between the parental and the esp2 lines at P < 0.05 (*) and < 0.01 (**). Cultivar abbreviations as in Fig. 1
Rice flours milled by the airflow wet grinding method were chosen to evaluate in the making of bread, sponge cake, and noodles because this method produced the highest quality flour (i.e., lowest mean diameter and damaged starch content, Fig. 2a). Rice breads made with Koshihikari esp2 flour from 2013 to 2016 had larger volumes in all levels of rice:wheat content, compared to breads made with Koshihikari flour (Fig. 2b, Additional file 2: Table S5). For example, in 2013, volumes of rice breads made with 20% rice flour by roll wet grinding method were 1879.0 and 1854.6 mL for Koshihikari esp2 and Koshihikari, respectively. Rice breads made from Oonari esp2 and Oonari flours had similar volumes in all ratios of rice flour to wheat flour (e.g., volumes of 1900.1 and 1895.6 mL, respectively, at 20% rice flour in 2017), but these breads did not rise as high as breads made with the Koshihikari esp2 and Koshihikari flours (Fig. 2b, Additional file 2: Table S5). Sponge cakes made from Koshihikari esp2 rice flour had larger volume than those made of flour from the parental Koshihikari line (1346.7 vs. 1306.0 mL, respectively, in 2013; Fig. 2c, Additional file 2: Table S5). Sponge cakes made using the Oonari esp2 and Oonari flours had similar heights and volumes, (e.g., 1262.4 and 1271.1 mL, respectively, in 2017), but both were smaller than cakes made from the Koshihikari esp2 and Koshihikari flours (Fig. 2c, Additional file 2: Table S5). In sensory tests of rice noodles made from 50% rice flour, the Koshihikari esp2 noodles had softer texture and greater stickiness than Koshihikari noodles (Fig. 2d). Rice noodles made from Oonari esp2 and Oonari flours had similar texture, but they were more brittle and harder than the noodles made from Koshihikari and Koshihikari esp2 flours (Fig. 2d).
Differential Gene Expression of the esp2 Gene in Different Genetic Backgrounds
The PDIL and BiP genes code for the two main classes of chaperone involved in the folding of seed storage proteins in the ER lumen; the Ero1 gene product donates disulfide bonds to the PDIL gene products (Onda et al. 2009). In the work reported here, expression of the PDIL1-1 gene was absent from maturing endosperms in Koshihikari esp2 and Oonari esp2 (Fig. 3), which can be attributed to the deficient mutation of this gene. Interestingly, expression of mRNA in nine other PDIL genes, four of the BiP genes, and the Ero1 gene was significantly higher (P < 0.01 or better) in Koshihikari esp2 than in the Koshihikari parental line. Expression of the remaining genes was not significantly different in Koshihikari esp2 and Koshihikari. In contrast, in the Oonari esp2 line, expression of mRNA differed from the Oonari parental line only in PDIL5-2 (higher) and in PDIL1-1 and PDIL1-3 (lower; Fig. 3). Expression in Oonari esp2 of the remaining 15 genes was similar to both Oonari and Koshihikari.
Expression of seed storage protein biosynthesis genes in immature grain, for twelve PDIL, one Ero, and five Bip genes, relative to expression of UBQ. Asterisks indicate significant difference from the Koshihikari parental line at P < 0.01 (**) and < 0.001 (***); cultivar abbreviations as in Fig. 1
In the leaf tissue of rice plants at the grain filling stage, gene expression of PDIL1-1 was absent in the breeding lines of both cultivars (Additional file 1: Fig. S4), as was observed in immature rice grains. In addition, seven other genes had significantly different (P < 0.05) expression in the leaves of Koshihikari esp2 and Koshihikari, and two genes had different expression in Oonari esp2 and Oonari.
Expression of the GBSSI starch (amylose and amylopectin) biosynthesis genes increased, but expression of SSI and SSIIIa genes was unchanged in the endosperm of Koshihikari esp2 and Oonari esp2 in comparison with the Koshihikari parental line (Additional file 1: Fig. S5). GBSSI is associated with the biosynthesis of amylose and extra-long chain amylopectin, and SSI and SSIIIa are associated with biosynthesis of the side chains of amylopectin. These three genes are highly expressed in rice endosperm at the grain filling stage compared with other starch biosynthesis genes.
Composition of Seed Storage Proteins in the Two Genetic Backgrounds
Reducing and non-reducing gel electrophoresis of seed storage protein conformations and structures, including the disulfide bond content of rice endosperm, helped elucidate some of the differences in expression of the seed storage protein synthesis genes. In comparison with Koshihikari, several protein bands from 40 to 10 kDa were decreased in Koshihikari esp2 in the C2H5OH and CH3COOH fractions under reducing conditions (Fig. 4a). In non-reducing conditions in the C2H5OH, CH3COOH, and SDS fractions, several protein bands from 250 to 10 kDa were decreased in Koshihikari esp2 compared with the other three lines (Fig. 4b). Koshihikari esp2 had significantly more (P < 0.01) total protein than the Koshihikari parental line (Fig. 4c). In contrast, Oonari and Oonari esp2 had similar protein band patterns and total protein content.
SDS-PAGE analysis of seed storage proteins of mature rice grains, in centrifugation supernatants after serial treatment with NaCl, C2H5OH, CH3COOH, SDS, and SDS with DTT. a Under reducing conditions, and b non-reducing conditions (but no SDS-DTT step). c Total protein content, sum of all supernatants, and final precipitations. Cultivar abbreviations as in Fig. 1
Discussion
This study indicates that certain genetic backgrounds negated phenotypic and gene expression changes attributable to the mutant allele in another background. We developed two rice breeding lines by introducing the PDIL1-1 (esp2) mutation into two different genetic backgrounds, a high-eating-quality cultivar, Koshihikari, and a high-yielding cultivar, Oonari. A DNA marker for genotyping the PDIL1-1 mutation was useful for efficient marker-assisted selection. Both esp2 lines accumulated higher quantities of glutelin precursors and had higher amylose contents, with similar composition of low-molecular-weight metabolites such as amino acids, organic acids, and fatty acids in comparison with the respective parental lines. However, there were significant differences in agronomic traits, rice flour characteristics, and food processing properties between the two esp2 lines. The Koshihikari esp2 line had good rice flour characteristics and food processing properties, but decreased plant height and grain yield compared with Koshihikari. In the Oonari esp2 line, rice flour characteristics, food processing properties, and grain yield all remained similar to the parental Oonari line. These phenotypic differences are likely attributable to formation of high-molecular-weight protein complexes in the endosperm caused by observed increases in mRNA expression of the seed storage protein synthesis genes PDIL, BiP, and Ero1 in Koshihikari esp2, but not seen in Oonari esp2.
Previous studies regarding PDIL1-1 (esp2) have used knockout mutant lines and knockdown (RNAi) transgenic lines in the genetic backgrounds of japonica rice cultivars Kinmaze, Taichung 65, Nipponbare, Yukihikari, Dongjin, and Zhonghua11 (Takemoto et al. 2002; Onda et al. 2009; Satoh-Cruz et al. 2010; Onda et al. 2011; Han et al. 2012; Kim et al. 2012; Onda and Kobori 2014; Xia et al. 2018). These studies report that nonfunctional and decreased PDIL1-1 alleles result in similar phenotypes with altered stacking of seed storage proteins within the ER and decreases of enzymatic activity in the formation and reduction of disulfide bonds. Koshihikari is also a japonica rice cultivar and has been the most widely grown type in Japan for over 40 years (Kobayashi et al. 2018). Oonari is an indica rice cultivar derived from a cross between Milyang 42 and Milyang 25 (Kobayashi et al. 2016). According to the TASUKE database (Kumagai et al. 2019), both Koshihikari and Oonari have only one PDIL1-1 gene in their genome sequences. In the work reported here, the mutant PDIL1-1 gene changed the mRNA expression of several PDIL and BiP genes and the Ero1 gene in the immature endosperm of the Koshihikari esp2 line, but not in immature endosperm of Oonari esp2. This suggests that phenotypic expression of the PDIL1-1 (esp2) mutant allele may have been unattainable in Oonari due to a lack of expression of other PDIL and/or the BiP and Ero1 genes.
The rice genome contains 12 PDIL genes (Houston et al. 2005). Among these, seven of them (PDIL1-1, 1-2, 1-3, 1-4, 2-1, 2-2, and 2-3) contain two redox active cysteine pair domains. The redox active cysteine pair domain has a catalytic activity function for both formation and reduction of disulfide bonds. The PDIL1-1 protein has higher catalytic activity for disulfide bond formation than other PDIL proteins, such as PDIL1-4 and PDIL2-3, as shown by the oxidative RNase refolding assay (Onda and Kobori 2014). The PDIL1-1 and Ero1 genes function on the same electron transfer pathway for intra-molecular disulfide bond formation between acid- and alkali-soluble glutelin subunits (Onda et al. 2009). Expressions of several BiP genes are increased by ER stress and an unfolded protein response increase expression of BiP genes when deficient mutations of the PDIL1-1 and Ero1 genes are present (Onda et al. 2009, 2011; Satoh-Cruz et al. 2010; Han et al. 2012; Kim et al. 2012). In the study reported here, parts of the regulatory pathway via the PDIL1-1 gene were altered in the Koshihikari esp2 plants, as seen in those previous studies. Conversely, in the Oonari esp2 plants, no significant differences in mRNA expression were observed in many of the PDIL, BiP, and Ero1 genes. This suggests that there are unknown genes responsible for the expression of phenotypes attributable to the esp2 mutation. Further experimental studies are necessary to identify the unknown genes associated with these differences between Koshihikari and Oonari.
In wheat cultivars, genotypes that code for high-molecular-weight glutenin subunits (HMW-GS) usually predict good bread- and dough-making quality based on the presence of individual glutenin subunits (Branlard et al. 2001). However, wheat cultivars having the same HMW-GS genotypes have also shown large phenotypic variations in bread-making scores. The PDI proteins are believed to be among several factors with the potential to change the bread-making properties of wheat (Demska et al. 2018). In fact, addition of recombinant PDI and Ero1 proteins to wheat flours increased disulfide bond formation in the bread dough, improving bread-making properties (Noguchi et al. 2015, 2016). In this study, Koshihikari esp2 had high bread-making scores even though this line has the nonfunctional PDIL1-1 gene allele. PDIL1-1 functions in the formation of intra-molecular disulfide bonds between two glutelin subunits. Improved bread-making properties in Koshihikari esp2 implies complementary effects of the PDIL1-1 gene with other PDIL and BiP genes to increase inter-molecular disulfide bonds among seed storage proteins, forming high-molecular-weight protein complexes.
High-yield rice cultivars are needed to produce large amounts of food for a growing global population. At the same time, income growth and urbanization will lead to increased consumer demands for high eating quality in various cereal foods including rice (Ito et al. 1989; Hori and Yano 2013; Hori 2018; Sharma et al. 2018; Hori et al. 2021b). The wide prevalence of celiac disease and wheat allergies is also increasing the demand for gluten-free foods (Ashida et al. 2009; Yano 2012; Montemurro et al. 2021). Therefore, it is important to develop high-yield rice cultivars for making rice flour foods including breads, noodles, and cakes.
Conclusions
Koshihikari and Oonari breeding lines with the esp2 mutant allele accumulated glutelin precursors and several low-molecular-weight metabolites such as amino acids and organic acids in their endosperm. The Koshihikari esp2 line showed the good flour characteristics and food processing properties than the Koshihikari parental line, whereas the Oonari esp2 line did not show a significant improvement in these traits compared to Oonari. Compared with the parental line, the endosperm of Koshihikari esp2 had elevated mRNA expression of the PDI and BiP gene families and formed high-molecular-weight protein complexes, but these alterations were not observed in Oonari esp2 endosperm. These results suggest that different genetic backgrounds can alter the phenotypic expression of a gene for rice flour characteristics and food processing properties, even when the same mutant gene allele is introduced to both genotypes. The work reported here has elucidated a part of the molecular basis of these phenotypic differences between breeding lines with the same mutant alleles in different genetic backgrounds. This may provide novel insights for further breeding efforts to introduce mutant gene alleles.
Abbreviations
- BiP:
-
Luminal binding protein
- DTT:
-
Dithiothreitol
- ER:
-
Endoplasmic reticulum
- Ero1:
-
ER membrane-localized oxidoreductase 1
- PBII:
-
Protein body II
- PDI:
-
Protein disulfide isomerase
- SDS-PAGE:
-
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SNP:
-
Single nucleotide polymorphism
References
Ashida K (2014) Properties of floury rice mutant and its utilization for rice flour. JARQ 48:51–56
Ashida K, Iida S, Yasui T (2009) Morphological, physical, and chemical properties of grain and flour from chalky rice mutants. Cereal Chem 86:225–231
Branlard G, Dardevet M, Saccomano R, Lagoutte F, Gourdon J (2001) Genetic diversity of wheat storage proteins and bread wheat quality. Euphytica 119:59–67
Champagne ET, Bett KL, Vinyard BT, McClung AM, Barton FE, Moldenhauer K et al (1999) Correlation between cooked rice texture and Rapid Visco Analyser measurements. Cereal Chem 76:764–771
Custodio MC, Cuevas RP, Ynion J, Laborte AG, Velasco ML, Demont M (2019) Rice quality: How is it defined by consumers, industry, food scientists, and geneticists? Trends Food Sci Technol 92:122–137
Demska K, Filip E, Skuza L (2018) Expression of genes encoding protein disulfide isomerase (PDI) in cultivars and lines of common wheat with different baking quality of flour. BMC Plant Biol 18:294
Fiaz S, Ahmad S, Noor MA, Wang X, Younas A, Riaz A et al (2019) Applications of the CRISPR/Cas9 system for rice grain quality improvement: perspectives and opportunities. Int J Mol Sci 20:888
Fitzgerald MA, McCouch SR, Hall RD (2009) Not just a grain of rice: the quest for quality. Trends Plant Sci 14:133–139
Fradinho P, Sousa I, Raymundo A (2019) Functional and thermorheological properties of rice flour gels for gluten-free pasta applications. Int J Food Sci Technol 54:1109–1120
Fukuda M, Kumamaru T (2019) Regulation of intracellular transport of the glutelin in the rice endosperm cell. Plant Morphol 31:31–35
Guo L, Chen W, Tao L, Hu B, Qu G, Tu B et al (2020) GWC1 is essential for high grain quality in rice. Plant Sci 296:110497
Han X, Wang Y, Liu X, Jiang L, Ren Y, Liu F et al (2012) The failure to express a protein disulphide isomerase-like protein results in a floury endosperm and an endoplasmic reticulum stress response in rice. J Exp Bot 63:121–130
Hayakawa K, Tanaka K, Nakamura T, Endo S, Hoshino T (2004) End use quality of waxy wheat flour in various grain-based foods. Cereal Chem 81:666–672
Hori K (2016) Detection of genetic factors responsible for grain quality and cooking characteristics of Japanese rice cultivars. J Jpn Soc Food Sci 63:484–487
Hori K (2018) Genetic dissection and breeding for grain appearance quality in rice. In: Sasaki T, Ashikari M (eds) Rice genomics, genetics and breeding. Springer, Singapore
Hori K, Yano M (2013) Genetic improvement of grain quality in japonica rice. In: Varshney RK, Tuberosa R (eds) Translational genomics for crop breeding. Abiotic stress, yield and quality, vol II. Wiley Blackwell, Ames
Hori K, Saisho D, Nagata K, Nonoue Y, Uehara-Yamaguchi Y, Kanatani A et al (2021a) Genetic elucidation for response of flowering time to ambient temperatures in Asian rice cultivars. Int J Mol Sci 22:1024
Hori K, Suzuki K, Ishikawa H, Nonoue Y, Nagata K, Fukuoka S et al (2021b) Genomic regions involved in differences in eating and cooking quality other than Wx and Alk genes between indica and japonica rice cultivars. Rice 14:8
Houston NL, Fan C, Xiang QY, Schulze JM, Jung R, Boston RS (2005) Phylogenetic analyses identify 10 classes of the protein disulfide isomerase family in plants, including single-domain protein disulfide isomerase-related proteins. Plant Physiol 137:762–778
Hu X, Cheng L, Hong Y, Li Z, Li C, Gu Z (2021) An extensive review: how starch and gluten impact dough machinability and resultant bread qualities. Crit Rev Food Sci Nutr 62:1–12
Ito S, Peterson EWF, Grant WR (1989) Rice in Asia: is it becoming an inferior good? Am J Agric Econ 71:32–42
Juliano BO, Houston DF (1985) Rice: chemistry and technology, 2nd edn. American Association of Cereal Chemistry, St. Paul
Kawagoe Y (2010) The rice esp2 mutant accumulates protein aggregates in the endosperm and has better qualities for rice bread. Gamma Field Symp 49:27–29
Kim YJ, Yeu SY, Park BS, Koh HJ, Song JT, Seo HS (2012) Protein disulfide isomerase-like protein 1–1 controls endosperm development through regulation of the amount and composition of seed proteins in rice. PLoS ONE 7:e44493
Kobayashi N, Ishii T, Yamaguchi M, Hirabayashi H, Takeuchi Y, Kuroki M et al (2016) Breeding of rice high-yielding cultivar “Oonari” by improving shattering habit of “Takanari.” Breed Res 18:143
Kobayashi A, Hori K, Yamamoto T, Yano M (2018) Koshihikari: a premium short-grain rice cultivar: its expansion and breeding in Japan. Rice 11:15
Kumagai M, Nishikawa D, Kawahara Y, Wakimoto H, Itoh R, Tabei N et al (2019) TASUKE+: a web-based platform for exploring GWAS results and large-scale resequencing data. DNA Res 26:445–452
Kwak J, Yoon MR, Lee JS, Lee JH, Ko S, Tai TH et al (2017) Morphological and starch characteristics of the japonica rice mutant variety Seolgaeng for dry-milled flour. Food Sci Biotechnol 26:43–48
Liu X, Ding Q, Wang W, Pan Y, Tan C, Qiu Y et al (2022) Targeted deletion of the first intron of the Wxb allele via CRISPR/Cas9 significantly increases grain amylose content in rice. Rice 15:1
Martin M, Fitzgerald MA (2002) Proteins in rice grains influence cooking properties. J Cereal Sci 36:285–294
Mo Y, Jeung JU (2020) The use of floury endosperm mutants to develop rice cultivars suitable for dry milling. Plant Biotechnol Rep 14:185–191
Montemurro M, Pontonio E, Rizzello CG (2021) Design of a “clean-label” gluten-free bread to meet consumers demand. Foods 10:462
Muench DG, Ogawa M, Okita TW (1999) The prolamins of rice. In: Shewry PR, Casey R (eds) Seed proteins, 2nd edn. Springer; Heidelberg
Noguchi T, Nishibori F, Shiono K, Oka D, Noguchi H, Takano K (2015) Influence of disulfide bond formation via recombinant PDI-Ero1 processing of proteins and baking quality. Food Preserv Sci 41:267–272
Noguchi T, Shiono K, Oka D, Noguchi H, Takano K (2016) The action of the SS bond formation due to the PDI-ERO1 to wheat gliadin (Glia A) on the dough formation and baking quality. Food Preserv Sci 42:9–14
Ogawa M, Kumamaru T, Satoh H, Iwata N, Omura T, Kasai Z et al (1987) Purification of protein body-I of rice seed and its polypeptide composition. Plant Cell Physiol 28:1517–1527
Ohta H, Yamaguchi M, Fukushima A, Kaji R, Tsuda N, Nakagomi K et al (2015) “Yumefuwari”, a new rice cultivar for rice flour bread. Bull Natl Agric Res Cent Tohoku Reg 117:15–27
Okunishi T (2014) Future on rice flour bread. J Cook Sci Jpn 48:385–391
Onda Y, Kobori Y (2014) Differential activity of rice protein disulfide isomerase family members for disulfide bond formation and reduction. FEBS Open Bio 4:730–734
Onda Y, Kumamaru T, Kawagoe Y (2009) ER membrane-localized oxidoreductase Ero1 is required for disulfide bond formation in the rice endosperm. Proc Natl Acad Sci USA 106:14156–14161
Onda Y, Nagamine A, Sakurai M, Kumamaru T, Ogawa M, Kawagoe Y (2011) Distinct roles of protein disulfide isomerase and P5 sulfhydryl oxidoreductases in multiple pathways for oxidation of structurally diverse storage proteins in rice. Plant Cell 23:210–223
Padhye VW, Salunkhe DK (1979) Extraction and characterization of rice proteins. Cereal Chem 56:389–393
Paz GM, King JM, Prinyawiwatkul W, Tyus CMO, Aleman RJS (2020) High-protein rice flour in the development of gluten-free muffins. J Food Sci 85:1397–1402
Sasahara H, Miura K, Shimizu H, Goto A, Shigemune A, Nagaoka I et al (2013) “Koshinokaori”, a new rice noodle cultivar. Bull NARO Agric Res Cent 19:15–29
Satoh-Cruz M, Crofts AJ, Takemoto-Kuno Y, Sugino A, Washida H, Crofts N et al (2010) Protein disulfide isomerase like 1–1 participates in the maturation of proglutelin within the endoplasmic reticulum in rice endosperm. Plant Cell Physiol 51:1581–1593
Sharma R, Nguyen TT, Grote U (2018) Changing consumption patterns-drivers and the environmental impact. Sustain 10:4190
Shibaya T, Hori K, Ogiso-Tanaka E, Yamanouchi U, Shu K, Kitazawa N et al (2016) Hd18, encoding histone acetylase related to Arabidopsis FLOWERING LOCUS D, is involved in the control of flowering time in rice. Plant Cell Physiol 57:1828–1838
Soga T, Kakazu Y, Robert M, Tomita M, Nishioka T (2004) Qualitative and quantitative analysis of amino acids by capillary electrophoresis-electrospray ionization-tandem mass spectrometry. Electrophoresis 25:1964–1972
Takaiwa F, Kikuchi S, Oono K (1987) A rice glutelin gene family - a major type of glutelin mRNAs can be divided into two classes. Mol Gen Genet 208:15–22
Takemoto Y, Coughlan SJ, Okita TW, Satoh H, Ogawa M, Kumamaru T (2002) The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol 128:1212–1222
Xia K, Zeng X, Jiao Z, Li M, Xu W, Nong Q et al (2018) Formation of protein disulfide bonds catalyzed by OsPDIL1;1 is mediated by microRNA5144-3p in rice. Plant Cell Physiol 59:331–342
Yamagata H, Sugimoto T, Tanaka K, Kasai Z (1982) Biosynthesis of storage proteins in developing rice seeds. Plant Physiol 70:1094–1100
Yano H (2012) Comparison of oxidized and reduced glutathione in the bread-making qualities of rice batter. J Food Sci 77:182–188
Zhang H, Xu H, Jiang Y, Zhang H, Wang S, Wang F et al (2021) Genetic control and high temperature effects on starch biosynthesis and grain quality in rice. Front Plant Sci 12:757997
Zhao WM, Gatehouse JA, Boulter D (1983) The purification and partial amino acid sequence of a polypeptide from the glutelin fraction of rice grains; homology to pea legumin. FEBS Lett 162:96–102
Acknowledgements
We are grateful to the technical staff of the Field Management Division at NARO and to K. Oshima at the Oshima Nojo, K.K. for their assistances in field management. We gratefully acknowledge Y. Nakamura, M. Iizumi, T. Takahashi, and C. Yazawa at NARO and K Ozawa at Tokyo University of Agriculture for their kind assistance in all experiments. We acknowledge Dr. Matthew Shenton at NARO for English correction of the manuscript.
Funding
This work was supported by the Project of the NARO Bio-oriented Technology Research Advancement Institution (25035B, 28014B) and by the Strategic Innovation Promotion Program “Technologies for Smart Bio-industry and Agriculture” (DDB2007).
Author information
Authors and Affiliations
Contributions
KH, TO, KN, YK, and TY designed the experiments. KH, TO, KN, KI, MH, KH, KS, TI, MY, YT, SK, YT, TI, HY, TY, TK, and YK developed plant materials, genotyped by DNA markers, evaluated grain component contents, agronomic traits, grain milling characteristics, food processing properties, transcription analysis and SDS-PAGEs in the developed rice lines. KH, OT, KN, TY, and TK analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Fig. S1.
Development of rice breeding lines introducing the esp2 mutation. a Pedigree of Koshihikari esp2 and Oonari esp2. b Days to heading in 85 F2 individuals derived from a cross between EM747 and Koshihikari. c Diversity of panicle size phenotypes in 85 F2 individuals from the cross of EM747 and Koshihikari. d DNA marker for detecting the esp2 mutant allele. e Whole genome genotype of Koshihikari esp2. f Whole genome genotype and positions of PDIL and 17 yield-related genes in Oonari esp2. K, Koshihikari; Ke, Koshihikari esp2; O, Oonari; Oe, Oonari esp2. Fig. S2. Grain yield of Koshihikari esp2 and Oonari esp2 in 2017 and 2018. a Locations of the six experimental fields in Japan. b Head brown rice weight of Koshihikari and Koshihikari esp2 at three locations in 2017. c Head brown rice weight in Oonari and Oonari esp2 at six locations in 2017 and 2018. Asterisks indicate significant difference from the Koshihikari parental line at P < 0.001. Fig. S3. Results of metabolome analysis of matured grains in 2016. a Principal component analysis of metabolites (n = 3). b Hierarchical cluster analysis of metabolites. Black and gray triangles indicate increased and decreased compounds, respectively, in the esp2 lines in comparison with the respective parental lines. c Quantities of low-molecular-weight metabolites. Asterisks indicate significant difference from the Koshihikari parental line at P < 0.05 (*), < 0.01 (**) and < 0.001 (***). Cultivar abbreviations as in Additional file 1: Fig. S1. Fig. S4. Expression of seed storage protein biosynthesis genes in rice leaves at the grain filling stage. The expression of twelve PDIL, one Ero, and five Bip family genes relative to the expression of UBQ. Asterisks indicate significant difference from the Koshihikari parental line at P < 0.05 (*), < 0.01 (**), and < 0.001 (***). Cultivar abbreviations as in Additional file 1: Fig. S1. Fig. S5. Expression of starch biosynthesis genes GBSSI, SSI, and SSIIIa in rice immature grain and in leaves at the grain filling stage. Gene expression is relative to expression of the UBQ. Asterisks indicate significant difference from Koshihikari at P < 0.05 (*), < 0.01 (**), and < 0.001 (***). Cultivar abbreviations as in Additional file 1: Fig. S1.
Additional file 2: Table S1.
Primer sequences used in this study. Table S2. Agronomic characteristics of Koshihikari, Koshihikari esp2, Oonari, and Oonari esp2 lines grown in the Chikusei, Fujieda, Kasai, and Chikugo fields in 2017 and 2018. Table S3. Agronomic characteristics of Koshihikari, Koshihikari esp2, Oonari, and Oonari esp2 lines grown in the Tsukubamirai field in 2015, 2016, 2017, and 2018. Table S4. Low-molecular-weight metabolites in Koshihikari, Koshihikari esp2, Oonari, and Oonari esp2 lines in 2015 and 2016. Table S5. Rice flour milling and food processing characteristics of rice from the Koshihikari, Koshihikari esp2, Oonari, and Oonari esp2 lines from 2013 to 2018.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hori, K., Okunishi, T., Nakamura, K. et al. Genetic Background Negates Improvements in Rice Flour Characteristics and Food Processing Properties Caused by a Mutant Allele of the PDIL1-1 Seed Storage Protein Gene. Rice 15, 13 (2022). https://doi.org/10.1186/s12284-022-00560-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-022-00560-w