Abstract
Zinc (Zn) is an essential micronutrient for living organisms, and understanding the molecular mechanisms of Zn deficiency may help to develop strategies to mitigate this problem. Microarray analysis of Zn deficient rice revealed the up-regulation of several genes involved in Zn transport. Moreover many genes involved in starch synthesis/transport were up-regulated by Zn deficiency in rice roots and shoots. Furthermore, starch granules were detected mainly in the cortical cells of these tissues. The gene encoding inactive RNase was much more highly transcribed than those encoding active RNases. Although the level of RNA degradation in a crude extract of Zn-deficient shoots was higher than that of Zn-sufficient shoots, addition of Zn significantly reduced the level of degradation. These results indicate that RNA degradation could be regulated by the amount of Zn in the cell, and that the tolerance of rice plants to low levels of Zn is promoted by the accumulation of starch and inactive RNase.
Similar content being viewed by others
Background
Zinc (Zn) is an essential micronutrient for almost all organisms, and its deficiency represents a serious nutritional problem in humans and plants. Zn is a non redox active element and serves as a cofactor for large number of enzymes involved in DNA transcription, protein, nucleic acid, carbohydrate, and lipid metabolism (Ishimaru et al. [2011]; Broadley et al. [2007]; Marschner [1995]). For example, DNA and RNA polymerases require Zn as a cofactor, and Zn is also essential for cell division. Indeed, Zn concentration in plant meristems is much higher than in other tissues (Kitagishi & Obata [1986]). Zn also plays a role in the structural stability of certain proteins, such as those containing Zn-finger domains, a dominant feature of many transcription factors. Genomic research in Arabidopsis revealed that roughly 22% of transcription factors contain a Zn-finger domain (Riechmann et al. [2000]); therefore, Zn may be important in regulating gene expression.
Many regions of the world, particularly those with calcareous soils, lack sufficient Zn, resulting in poor plant growth. Therefore, the mechanism for tolerance to Zn deficiency should be elucidated to mitigate Zn deficiency. Several mechanisms have been investigated to clarify the physiological basis of differential Zn efficiency among wheat genotypes. For example, cultivars tolerant to Zn deficiency secrete higher amounts of mugineic acid family phytosiderophores (MAs) than intolerant cultivars (Cakmak et al. [1994]; Walter et al. [1994]; Zhang et al. [1989]; Suzuki et al. [2006]; Cakmak et al. [1996]). MAs are synthesized from L-methionine (Mori & Nishizawa [1987]; Shojima et al. [1990]; Ma et al. [1995]; Ma et al. [1999]). Nicotianamine (NA) synthase (NAS) transforms three molecules of S-adenosyl-L-methionine to one molecule of NA, and NA aminotransferase (NAAT) catalyzes the amino transfer of NA. The keto form is subsequently reduced to 2′-deoxymugineic acid (DMA) by DMA synthase (DMAS) (Bashir et al. [2006]; Bashir & Nishizawa [2006]; Bashir et al. [2010]). Zn-DMA, is suggested to be preferred over Zn2+ for uptake in barley roots (Suzuki et al. [2006]), while rice roots absorb less Zn-DMA compared to Zn2+ (Suzuki et al. [2008]). Similarly the secretion of MAs increases in Zn deficient barley, while it decreases in rice (Suzuki et al. [2006]; Suzuki et al. [2008]). Despite this, Zn-DMA is suggested to be the preferred form for the long distance transport in rice (Suzuki et al. [2008]). Difference in DMA secretion is suggested to increase tolerance in rice (Widodo et al. [2010]). Moreover, a modelling study also proposed a strong correlation between DMA secretion and rooting density, and suggested a role of DMA for Zn absorption in rice (Ptashnyk et al. [2011]). The identification of Zn-NA complexes in rice phloem sap also suggests that NA performs a significant role in Zn transport (Nishiyama et al. [2012]). Zn uptake by roots, and translocation within the plant, are associated with Zn efficiency (Rengel & Graham [1996]), while the availability of Zn at the cellular level is also suggested to be related to Zn efficiency (Cakmak et al. [1997]). Moreover the expression level or activity of enzymes requiring Zn is shown to be different between Zn-efficient and Zn-inefficient cultivars (Hacisalihoglu et al. [2003]). Partitioning of carbohydrates also shows correlation with tolerance to Zn-deficiency stress (Pearson & Rengel [1997]). Rice cultivars differing in Zn efficiency have been used for physiological and genetic analyses of tolerance to Zn deficiency (Gao et al. [2005]; Hajiboland et al. [2005]; Hoffland et al. [2006]; Wissuwa et al. [2006]). Among graminaceous plants, rice is highly sensitive to Zn-deficient stress; thus, we investigated the physiological change and gene expression pattern in rice during Zn deficiency. Profiling of the genes involved in Zn-deficient stress is critical to elucidate the mechanisms of tolerance to or damage by Zn deficiency. We performed a microarray analysis with Zn-deficient and Zn-sufficient rice to identify genes whose expression increases in response to Zn deficiency. Based on our combined expression and phenotypic analyses, we demonstrated that besides up-regulation of genes involved in Zn uptake and transport, Zn deficiency enhances starch accumulation both in roots and shoots, and that Zn deficiency induces the expression of a gene encoding putative inactive RNase, which may function as vegetative storage protein. Further, the level of RNA degradation was increased by Zn deficiency.
Results
Up-regulation of genes involved in Zn transport
Microarray analysis revealed that the genes involved in Zn uptake and transport were up-regulated, both in roots and shoots of Zn deficient rice. The expression of OsNAS1 particularly increased in rice shoots (Table 1). OsNAS1 is reported to be up-regulated by iron deficiency in root and shoot tissue (Inoue et al. [2003]; Bashir et al. [2011]; Ishimaru et al. [2009]). Moreover, the expression of OsNAS3 was also upregulated in root and shoot tissue. The expression of OsNAS3 is not regulated by Fe deficiency and is reported to be up-regulated by Zn deficiency (Suzuki et al. [2008]). As NA was not detected in Zn deficient rice shoot, the NA catalyzed by OsNAS3 may be transformed to DMA (Suzuki et al. [2008]). The up-regulation of OsNAAT1 supports this hypothesis. Besides this the expression of Zn transporters, OsZIP4 OsZIP5 and OsZIP8 increased significantly in Zn deficient roots and shoots, while the expression of OsZIP7 increased in shoot tissue (Table 1). OsZIP4 and OsZIP5 are plasma membrane Zn transporters induced by Zn deficiency (Lee et al. [2010]; Yang et al. [2009]; Ishimaru et al. [2005]). Moreover, the expression of OsHMA1 was also induced by Zn deficiency in shoot tissue. OsHMA1 is supposed to be involved in Zn transport (Williams & Mills [2005]).
Starch accumulation in Zn-deficient roots and shoots
As shown in Table 2, genes encoding starch synthase, ADP-glucose pyrophosphorylase (AGPase) large subunit, AGPase small subunit, α-1,4-glucan branching enzyme, and phosphoglucomutase, which are thought to be involved in starch metabolism, were up-regulated by Zn deficiency. Sucrose synthase, which reversibly converts sucrose to ADP-glucose, was up-regulated in Zn-deficient shoots and slightly up-regulated in roots, while α-1,4-glucan phosphorylase, which catalyzes the phospholysis of a linear glucan chain to reversibly yield glucose 1-phosphate (Smith et al. [2003]), was also up-regulated. Although multiple pathways may exist for starch synthesis (Baroja-Fernández et al. [2004]), the above enzymes may be sufficient for starch synthesis. Moreover, several genes involved in sugar transporter were up-regulated especially in shoots. The expression of these genes increased more significantly, when plants were subjected to Zn deficiency and analysed by 22 K microarray analysis ( Additional file 1: Table S1).
The starch content was higher in both roots and shoots grown under Zn-deficient conditions (Figure 1a), corresponding to the gene expression pattern revealed by our microarray analysis. Iodine–starch reaction staining revealed starch accumulation in Zn-deficient roots, but not in control roots or Fe-deficient roots (Figure 1b–d). In Zn-deficient roots, several starch granules were detected in the cortex, especially in the cells close to the sclerenchyma and endodermis (Figure 1d, f, g), and additional granules were observed in the pericycle (Figure 1e). In shoots, starch granules were mainly found in the mesophyll cells of the control plants (Figure 1h); however, in Zn-deficient shoots, the location of the starch granules was different among the leaves within a plant. Starch accumulated in the mesophyll cells of the youngest mature leaves of Zn-deficient rice (Figure 1i). When the youngest leaf was immature, starch accumulated in the bundle sheath cells of older leaves (Figure 1j). At the large vein of old leaves, starch granules were observed both in the mesophyll cells and in the bundle sheath cells (Figure 1k).
Quantification of starch (a) and localization of starch granules in Zn-deficient and Zn-sufficient (control) plants by iodine staining (b–k). (b) Zn- and Fe-sufficient roots (control); Zn-deficient roots (−Zn) and Fe-deficient roots (−Fe), (c) control roots, (d–g) Zn-deficient roots, (h) medium vein of old control leaf, (i) medium vein of Zn-deficient newest mature leaf, (j) medium vein of Zn-deficient old leaf, (k) large vein of Zn-deficient old leaf. Bar = 100 μm. cor.; cortex, mes.; mesophyll cells, b.s.; bundle sheath cells.
Zn-deficient rice strongly expresses inactive RNase
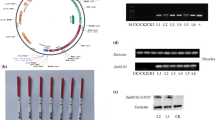
Among the 27,800 genes analyzed, Os09g0537700 (AK061438; OsRNS4), which is predicted to encode S-like RNase, showed the highest induction ratio in roots (Table 3). This gene was also up-regulated in Zn-deficient shoots. Other genes encoded putative RNases, were not significantly up-regulated by Zn deficiency. In rice eight genes for S-like RNase have been described (MacIntosh et al. [2010]) and among them only six were included in our microarray experiment. In addition, the transcript levels of AK061438 in Zn-deficient shoots were much higher than those of the other RNases. Northern blot analysis revealed that the expression of two S-like RNases increased in Zn-deficient shoots, but not in Fe-deficient or Cu-deficient shoots (Figure 2); Mn deficiency slightly induced expression. Although the expression ratio of AK061438 in Zn-deficient roots was very high, the transcript level in roots was much lower than in shoots. Northern blot analysis as well as 22 K microarray analysis of rice plants indicated that expression of OsRNS5 also increases by Zn deficiency (Figure 2 & Additional file 1: Table S2).
An alignment of the amino acid sequences of the RNases is shown in Figure 3a. All of the RNases contain eight conserved cysteine residues involved in the three-dimensional formation of RNase (Rabijns et al. [2002]); however, the amino acid sequences encoded by AK061438 and AK109411 lack two histidine residues (replaced by Lys66 and Tyr125 in AK061438, and by Ala67 and Ser125 in AK109411) that are essential for RNA degradation (Kurihara et al. [1996]). In addition, the glutamate residue, which is also essential for RNA degradation, is not conserved in AK109411 (replaced by Ala121). These histidine and glutamate residues are conserved in RNases in other plant species (Figure 3b), excluding CalsepRRP, which is an inactive RNase because it lacks one histidine residue (replaced by Lys70) essential for RNA degradation (Van Damme et al. [2000]). Therefore, the proteins encoded by AK061438 and AK109411 may be inactive for RNA degradation (MacIntosh et al. [2010]).
Comparison of the amino acid sequences of RNase and RNase-related proteins in plants. (a) All of the amino acid sequences predicted from the rice full-length cDNA database. (b) Partial amino acid sequences from C. sepium (CalsepRRP), A. thaliana (RNS1, RNS2, and RNS3), M. charantia (RNase MC), and L. esculentum (RNase LE). Gaps were introduced to maximize the homologies. Identical or similar residues are boxed in black and grey, respectively. The three charged residues involved in the active site of RNase are indicated by asterisks, and the two aromatic residues, which presumably maintain the conformational stability of the site, are indicated by a black circle. The sequences of the eight conserved cysteine residues in rice are indicated by C1–C8.
In contrast, RNA degradation increased in the crude extract of Zn-deficient citrullus (Sharma et al. [1981]) and black gram (Pandey et al. [2002]). RNA degradation in a crude extract of Zn-deficient rice shoots was also higher than that in control shoots (Figure 4a), and it was inhibited by adding ZnSO4 but not FeSO4 (Figure 4b). These findings agree with a previous report showing that Zn ions inhibited RNA degradation in a crude extract of avena leaves (Wyen et al. [1971]).
RNAse activity in crude extracts of rice shoots. (a) 10 μg of total RNA from Zn-deficient rice shoots were incubated with 5 μg, 1 μg, and 0.2 μg of protein from crude extracts of Zn-deficient shoots (−Zn) and control shoots (C). 3 μg of incubated total RNA were loaded into each lane. (b) 10 μg of total RNA from Zn-deficient rice shoots and 2 μg of protein from crude extracts of Zn-deficient shoots were incubated together with 1 mM or 10 mM ZnSO4 or FeSO4. 3 μg of incubated total RNA were loaded into each lane.
Discussion
The expression of genes involved directly or indirectly in Zn transport increases under Zn deficiency. Moreover, the genes involved in starch synthesis and transport were up-regulated by Zn deficiency. Both Zn-deficient roots and shoots accumulated starch (Figure 1), in line with the gene expression pattern (Table 2). Starch may be used as a carbon source; therefore, it is assumed that Zn-deficient plants synthesize starch to withstand temporary abiotic stress. However, it is also possible that disruption of glycolysis by Zn deficiency causes an over-accumulation of soluble sugar, which ultimately results in starch accumulation. It is already reported that the activity of FBP aldolase in the glycolysis pathway decreased under Zn-deficiency in the leaves of oat and clover, and suggested that one of the reasons for a growth defect in plants grown under Zn-deficient conditions is the breakdown of normal carbohydrate metabolism (Quinlan-Watson [1951]). The concentration of soluble sugar and starch increase in Zn-deficient bean shoots (Marschner & Cakmak [1989]). An increase in soluble sugar would cause osmotic stress. We speculate that the starch might be synthesized to avoid osmotic stress due to increased soluble sugar in cells under Zn deficiency. Up-regulation of the genes that encode soluble sugar transporters due to Zn-deficiency suggests that sugar transporters help distribute sugar to avoid osmotic stress (Table 2).
Zn-deficient plants have increased RNase activity (Sharma et al. [1981]; Pandey et al. [2002]). We showed that Zn-deficient rice shoots also have increased RNA degradation (Figure 4a). Our microarray analysis revealed that the expression of one S-like RNase increased under Zn deficiency, and the transcript levels were much higher than those of other RNases (Table 3). However, unexpectedly, this RNase protein seemed to have no enzymatic activity as the histidine and glutamate residues essential for RNA degradation were changed (Figure 3a, b; (MacIntosh et al. [2010]; Kurihara et al. [1996]; Van Damme et al. [2000])). Our microarray analysis also showed that no gene for active RNases were up-regulated by Zn deficiency. In addition, Zn ions inhibited the RNase activity in a crude extract from Zn-deficient shoots (Figure 4b). These findings suggest that RNA degradation may be controlled by the Zn concentration at a cellular level rather than by the level of RNase mRNA.
The function of the inactive RNase induced by Zn deficiency is not clear. An inactive S-like RNase accumulate in the rhizomes of Calystegia sepium and is suggested to play a role in vegetative storage proteins (Van Damme et al. [2000]). Thus, Zn-deficient rice might accumulate inactive RNase for storage of amino acids. The inactive RNase, also, accumulate in response to drought stress (Salekdeh et al. [2002]), one of the most fatal stresses for plants. Therefore, the induction of the inactive RNase could be due to fatal stress for plants caused subsequently by Zn-deficiency stress. However, in our experiment, Fe-deficient stress decreased the expression of AK061438 and AK109411 (Figure 4), although Fe-deficient stress is more detrimental to plant growth than other micronutrient-deficient stresses. Moreover, the expression of AK061438 decreased under sulfur-deficient stress (Ohkama-Ohtsu et al. [2004]). These data suggest that the induction of putative inactive RNases is specifically caused by Zn-or water-deficient stress rather than by general stress conditions in plants.
Taken together, Zn-deficient rice plants accumulate starch that can be used as carbon and nitrogen sources. This might be involved in the tolerance to Zn deficiency, damage by Zn-deficient stress, or Zn homeostasis. To better understand these mechanisms, it is important to elucidate the differences between Zn-efficient and Zn-inefficient cultivars, and the molecular functions of each gene involved in tolerance to Zn deficiency should be revealed.
Conclusions
Microarray analysis of Zn deficiency rice revealed the up-regulation of several genes involved in starch synthesis in Zn deficient rice roots and shoots. The accumulation of starch granules in the cortical cells of these tissues further supported these results. A gene encoding inactive RNase was much more highly transcribed than those encoding active RNases. Moreover, the level of RNA degradation in a crude extract of Zn-deficient shoots reduced after addition of Zn to a crude extract of Zn-deficient shoots. These results suggest that the tolerance of rice plants to low levels of Zn may be promoted by the accumulation of starch and inactive RNase proteins.
Methods
Plant materials and growth conditions
Rice seeds (Oryza sativa L.) were germinated for 1 week at room temperature on paper towels soaked with distilled water. After germination, the seedlings were transferred to a Saran net floating on nutrient solution in a glasshouse for 2 weeks. Two week old plants were transferred to a 20-L plastic container containing a nutrient solution as described previously (Suzuki et al. [2006]) with or without ZnSO4, The pH of the nutrient solution was adjusted daily to 5.5, and was renewed weekly. To compare the response of rice to Zn deficiency with that to deficiencies in other micronutrients, rice seeds were germinated for 1 week and then transferred to the nutrient solution described above. After 1 week, the seedlings were transferred to fresh nutrient solution without Zn, Fe, Cu or Mn and grown for 2 additional weeks.
Microarray analysis
Roots and shoots of four week old plants subjected to Zn deficiency or grown under control conditions were collected, frozen in liquid nitrogen, and stored at −80°C until use. RNA was extracted from the roots and shoots of three plants, and total RNA (200 ng) from the Zn deficient and Zn sufficient plants were labelled with Cy3 or Cy5 using an Agilent Low RNA Input Fluorescent Linear Amplification Kit (Agilent Technologies), and rice 44 K oligo-DNA microarray analysis was performed in duplicate using color swaps, according to the manufacturer’s instructions, as described previously (Bashir et al. [2011]; Ishimaru et al. [2007]). The induction ratios shown in the tables were calculated as the relative increases in expression under Zn deficiency compared to the level of expression under control conditions.
Quantification of starch and iodine staining
Rice seeds were germinated for 1 week and transferred to nutrient solution containing Zn. After 2 weeks, the seedlings were transferred to a Zn-deficient nutrient solution for 3 weeks and harvested for analysis. The samples were collected 2 h before sunset. To determine the level of starch, each plant was ground using a mortar and pestle with liquid nitrogen and dried overnight at 65°C. Around 50 mg of dried plant tissue was used for starch quantification, employing a total starch assay procedure kit (Megazyme, Bray, Ireland). To determine the localization of the starch granules, leaf blades and roots were cut with a scalpel into approximately 1-cm sections. These sections were embedded in 5% agar and then cut into 80- to 130-μm sections using a DTK-100 microslicer (Dosaka EM Co. Ltd., Kyoto, Japan) and stained with iodine. To remove the pigment in the leaf, the plant sections were soaked in 80% ethanol for 1 month.
Measuring RNA degradation
RNA degradation was detected by electrophoresis (Van Damme et al. [2000]). Total RNA from rice shoots grown under Zn-deficient treatment for 3 weeks was used as a substrate. A 10-μg sample of total RNA was incubated in 20 μL of 25 M Tris–HCl (pH 7.4) containing 25 mM KCl and 5 mM MgCl2 in the presence of crude extracts from Zn-deficient and control shoots at 30°C for 15 min. To investigate the effects of metal on RNA degradation, 10 μg of RNA were incubated with 1 mM and 10 mM ZnSO4 and FeSO4 under the conditions described above. A 3-μg aliquot of incubated RNA diluted with distilled water was loaded onto a 1.2% agarose gel containing ethidium bromide. The extracts were prepared from 0.1 g of ground tissue mixed with 1 mL of Tris buffer. The amount of protein in the crude extract was quantified by the Bradford method.
Northern blot analysis
Total RNA was extracted from roots and shoots, and 10 μg per lane were electrophoresed in 1.2% (w/v) agarose gels containing 0.66 M formaldehyde, transferred to Hybond-N+ membrane (Amersham Biosciences UK Ltd., Buckinghamshire, UK), and hybridized with probes at 65°C. Northern blots were analyzed using BAS 3000 (FujiFilm, Tokyo, Japan). The full-length rice cDNAs (Rice Genome Resource Center, Tsukuba, Japan) were used for labelling the probes.
Authors’ information
M.S.; Present address; Aichi Steel Corporation, 1, Wanowari, Arao-Machi, Tokai-Shi, Aichi 476–8666, Japan H.I.; Present address: Division of Plant Sciences, National Institute of Agrobiological Sciences, 2-1-2 Kannondai, Tsukuba, Ibaraki 305–8602, Japan. M.T.; Present address: Faculty of Agriculture, Utsunomiya University - 350 Mine, Utsunomioya, Tochigi 321–8505, Japan.
Abbreviations
- AGPase:
-
ADP-glucose pyrophosphorylase
- FBP aldolase:
-
Fructose-1,6-bisphosphate aldolase.
References
Baroja-Fernández E, Muñoz FJ, Zandueta-Criado A, Morán-Zorzano MT, Viale AM, Alonso-Casajús N, Pozueta-Romero J: Most of ADP·glucose linked to starch biosynthesis occurs outside the chloroplast in source leaves. Proc Natl Acad Sci USA 2004,101(35):13080–13085. 10.1073/pnas.0402883101
Bashir K, Nishizawa NK: Deoxymugineic acid synthase: A gene important for Fe-acquisition and homeostasis. Plant Signal Behav 2006,1(6):292.
Bashir K, Inoue H, Nagasaka S, Takahashi M, Nakanishi H, Mori S, Nishizawa N: Cloning and characterization of deoxymugineic acid synthase genes from graminaceous plants. J Biol Chem 2006,281(43):32395–32402. 10.1074/jbc.M604133200
Bashir K, Ishimaru Y, Nishizawa NK: Iron uptake and loading into rice grains. Rice 2010,3(2):122–130. 10.1007/s12284-010-9042-y
Bashir K, Ishimaru Y, Shimo H, Nagasaka S, Fujimoto M, Takanashi H, Tsutsumi N, An G, Nakanishi H, Nishizawa NK: The rice mitochondrial iron transporter is essential for plant growth. Nat Commun 2011, 2: 322.
Broadley MR, White PJ, Hammond JP, Zelko I, Lux A: Zinc in plants. New Phytol 2007,173(4):677–702. 10.1111/j.1469-8137.2007.01996.x
Cakmak I, Gülüt KY, Marschner H, Graham RD: Efect of zinc and iron deficiency on phytosiderophore release in wheat genotypes differing in zinc efficiency. J Plant Nutr 1994,17(1):1–17. 10.1080/01904169409364706
Cakmak I, Sari N, Marschner H, Ekiz H, Kalayci M, Yilmaz A, Braun HJ: Phytosiderophore release in bread and durum wheat genotypes differing in zinc efficiency. Plant Soil 1996,180(2):183–189. 10.1007/BF00015301
Cakmak I, Ekiz H, Yilmaz A, Torun B, Köleli N, Gültekin I, Alkan A, Eker S: Differential response of rye, triticale, bread and durum wheats to zinc deficiency in calcareous soils. Plant Soil 1997,188(1):1–10. 10.1023/A:1004247911381
Gao X, Zou C, Zhang F, van der Zee S, Hoffland E: Tolerance to zinc deficiency in rice correlates with zinc uptake and translocation. Plant Soil 2005,278(1):253–261. 10.1007/s11104-005-8674-y
Hacisalihoglu G, Hart JJ, Wang Y-H, Cakmak I, Kochian LV: Zinc efficiency is correlated with enhanced expression and activity of zinc-requiring enzymes in wheat. Plant Physiol 2003,131(2):595–602. 10.1104/pp.011825
Hajiboland R, Yang XE, Römheld V, Neumann G: Effect of bicarbonate on elongation and distribution of organic acids in root and root zone of Zn-efficient and Zn-inefficient rice (Oryza sativa L.) genotypes. Environ Exp Bot 2005,54(2):163–173. 10.1016/j.envexpbot.2004.07.001
Hoffland E, Wei C, Wissuwa M: Organic anion exudation by lowland rice (Oryza sativa L.) at zinc and phosphorus deficiency. Plant Soil 2006,283(1):155–162. 10.1007/s11104-005-3937-1
Inoue H, Higuchi K, Takahashi M, Nakanishi H, Mori S, Nishizawa NK: Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. Plant J 2003,36(3):366–381. 10.1046/j.1365-313X.2003.01878.x
Ishimaru Y, Suzuki M, Kobayashi T, Takahashi M, Nakanishi H, Mori S, Nishizawa NK: OsZIP4, a novel zinc-regulated zinc transporter in rice. J Exp Bot 2005,56(422):3207–3214. 10.1093/jxb/eri317
Ishimaru Y, Masuda H, Suzuki M, Bashir K, Takahashi M, Nakanishi H, Mori S, Nishizawa NK: Overexpression of the OsZIP4 zinc transporter confers disarrangement of zinc distribution in rice plants. J Exp Bot 2007,58(11):2909–2915. 10.1093/jxb/erm147
Ishimaru Y, Bashir K, Fujimoto M, An G, Itai R, Tsutsumi N, Nakanishi H, Nishizawa NK: Rice-specific mitochondrial iron-regulated gene (MIR) plays an important role in iron homeostasis. Mol Plant 2009,2(5):1059–1066. 10.1093/mp/ssp051
Ishimaru Y, Bashir K, Nishizawa NK: Zn uptake and translocation in rice plants. Rice 2011,4(1):21–27. 10.1007/s12284-011-9061-3
Kitagishi K, Obata H: Effects of zinc on the nitrogen metabolism of meristematic tissues of rice plants with reference to protein synthesis. Soil Sci Plant Nutr 1986,32(3):397–405.
Kurihara H, Nonaka T, Mitsui Y, Ohgi K, Irie M, Nakamura KT: The crystal structure of ribonuclease Rh from Rhizopus niveus at 2.0 Å resolution. J Mol Biol 1996,255(2):310–320. 10.1006/jmbi.1996.0025
Lee S, Jeong H, Kim S, Lee J, Guerinot M, An G: OsZIP5 is a plasma membrane zinc transporter in rice. Plant Mol Biol 2010,73(4):507–517. 10.1007/s11103-010-9637-0
Ma JF, Shinada T, Matsuda C, Nomoto K: Biosynthesis of phytosiderophores, mugineic acids, associated with methionine cycling. J Biol Chem 1995,270(28):16549–16554. 10.1074/jbc.270.28.16549
Ma JF, Taketa S, Chang Y-C, Iwashita T, Matsumoto H, Takeda K, Nomoto K: Genes controlling hydroxylations of phytosiderophores are located on different chromosomes in barley (Hordeum vulgare L.). Planta 1999,207(4):590–596. 10.1007/s004250050522
MacIntosh G, Hillwig M, Meyer A, Flagel L: RNase T2 genes from rice and the evolution of secretory ribonucleases in plants. Mol Genet Genomics 2010,283(4):381–396. 10.1007/s00438-010-0524-9
Marschner H: Mineral nutrition of higher plants. 2nd edition. Academic, London; 1995:347–364.
Marschner H, Cakmak I: High light intensity enhances chlorosis and necrosis in leaves of zinc, potassium, and magnesium deficient bean (Phaseolus vulgaris) plants. J Plant Physiol 1989, 134: 308–315. 10.1016/S0176-1617(89)80248-2
Mori S, Nishizawa NK: Methionine as a dominant precursor of phytosiderophores in graminaceae plants. Plant Cell Physiol 1987,28(6):1081–1092.
Nishiyama R, Kato M, Nagata S, Yanagisawa S, Yoneyama T: Identification of Zn-nicotianamine and Fe-2’-deoxymugineic acid in the phloem saps from rice plants (Oryza sativa L.). Plant and Cell Physiology 2012, 53: 381–390. 10.1093/pcp/pcr188
Ohkama-Ohtsu N, KIshimoto N, Yazaki J, Fujii F, Shinbo K, Shimatani Z, Nagata Y, Hashimoto A, Ohta T, Sato Y, et al.: Up-regulation of genes for ferritin, RNase, and DnaJ in leaves of rice plants in response to sulfur deficiency. Soil Sci Plant Nutr 2004, 50: 771–775. 10.1080/00380768.2004.10408534
Pandey N, Pathak GC, Singh AK, Sharma CP: Enzymic changes in response to zinc nutrition. J Plant Physiol 2002,159(10):1151–1153. 10.1078/0176-1617-00674
Pearson JN, Rengel Z: Genotypic differences in the production and partitioning of carbohydrates between roots and shoots of wheat grown under zinc or manganese deficiency. Ann Bot 1997,80(6):803–808. 10.1006/anbo.1997.0523
Ptashnyk M, Roose T, Jones DL, Kirk GJD: Enhanced zinc uptake by rice through phytosiderophore secretion: a modelling study. Plant Cell Environ 2011,34(12):2038–2046. 10.1111/j.1365-3040.2011.02401.x
Quinlan-Watson TAF: Aldolase activity in zinc-deficient plants. Nature 1951,167(4260):1033–1034.
Rabijns A, Verboven C, Rouge P, Barre A, Van Damme EJM, Peumans WJ, De Ranter CJ: Structure of an RNase-related protein from Calystegia sepium. Acta Crystallogr Sect D 2002,58(4):627–633.
Rengel Z, Graham RD: Uptake of zinc from chelate-buffered nutrient solutions by wheat genotypes differing in zinc efficiency. J Exp Bot 1996,47(2):217–226. 10.1093/jxb/47.2.217
Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al.: Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 2000,290(5499):2105–2110. 10.1126/science.290.5499.2105
Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J: Proteomic analysis of rice leaves during drought stress and recovery. Proteomics 2002,2(9):1131–1145. 10.1002/1615-9861(200209)2:9<1131::AID-PROT1131>3.0.CO;2-1
Sharma CP, Gupta JP, Agarwala SC: Metabolic changes in citrullus subjected to zinc stress. J Plant Nutr 1981,3(1–4):337–344.
Shojima S, Nishizawa N-K, Fushiya S, Nozoe S, Irifune T, Mori S: Biosynthesis of phytosiderophores : In vitro biosynthesis of 2′-deoxymugineic acid from L-methionine and nicotianamine. Plant Physiol 1990,93(4):1497–1503. 10.1104/pp.93.4.1497
Smith AM, Zeeman SC, Thorneycroft D, Smith SM: Starch mobilization in leaves. J Exp Bot 2003,54(382):577–583. 10.1093/jxb/erg036
Suzuki M, Takahashi M, Tsukamoto T, Watanabe S, Matsuhashi S, Yazaki J, Kishimoto N, Kikuchi S, Nakanishi H, Mori S, et al.: Biosynthesis and secretion of mugineic acid family phytosiderophores in zinc-deficient barley. Plant J 2006,48(1):85–97. 10.1111/j.1365-313X.2006.02853.x
Suzuki M, Tsukamoto T, Inoue H, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK: Deoxymugineic acid increases Zn translocation in Zn-deficient rice plants. Plant Mol Biol 2008,66(6):609–617. 10.1007/s11103-008-9292-x
Van Damme EJM, Hao Q, Barre A, Rougé P, Van Leuven F, Peumans WJ: Major protein of resting rhizomes of Calystegia sepium (Hedge Bindweed) closely resembles plant RNases but has no enzymatic activity. Plant Physiol 2000,122(2):433–446. 10.1104/pp.122.2.433
Walter A, Römheld V, Marschner H, Mori S: Is the release of phytosiderophores in zinc-deficient wheat plants a response to impaired iron utilization? Physiol Plant 1994,92(3):493–500. 10.1111/j.1399-3054.1994.tb08841.x
Widodo B, Broadley MR, Rose T, Frei M, Pariasca-Tanaka J, Yoshihashi T, Thomson M, Hammond JP, Aprile A, Close TJ, et al.: Response to zinc deficiency of two rice lines with contrasting tolerance is determined by root growth maintenance and organic acid exudation rates, and not by zinc-transporter activity. New Phytol 2010,186(2):400–414. 10.1111/j.1469-8137.2009.03177.x
Williams LE, Mills RF: P1B-ATPases – an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci 2005,10(10):491–502. 10.1016/j.tplants.2005.08.008
Wissuwa M, Ismail AM, Yanagihara S: Effects of zinc deficiency on rice growth and genetic factors contributing to tolerance. Plant Physiol 2006,142(2):731–741. 10.1104/pp.106.085225
Wyen NV, Erdei S, Farkas GL: Isolation from avena leaf tissues of a nuclease with the same type of specificity towards RNA and DNA: Accumulation of the enzyme during leaf senescence. Biochim Biophys Acta 1971, 232: 472–483. 10.1016/0005-2787(71)90601-0
Yang X, Huang J, Jiang Y, Zhang H-S: Cloning and functional identification of two members of the ZIP (Zrt, Irt-like protein) gene family in rice (Oryza sativa L.). Mol Biol Rep 2009,36(2):281–287. 10.1007/s11033-007-9177-0
Zhang F, Römheld V, Marschner H: Effect of zinc deficiency in wheat on the release of zinc and iron mobilizing root exudates. Z für Pflanzenernährung Bodenkunde 1989,152(2):205–210. 10.1002/jpln.19891520211
Acknowledgments
We thank Dr. Yoshiaki Nagamura (National Institute of Agrobiological Sciences, Tsukuba, Japan) for the microarray analysis and for providing full-length cDNA clones. We also thank Dr. Hitoshi Obata (Mie University, Tsu, Japan) for helpful advice and Dr. Takanori Kobayashi (The University of Tokyo, Tokyo, Japan) for careful reading of the manuscript. This work was supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Green Technology Project IP-5003).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare no competing financial interests.
Authors’ contributions
MS, MK, HN, and NN designed the research, MS performed the research and MS, KB, HI, HN, and NN discussed the data and wrote the paper. All authors read and approved the final manuscript.
Electronic supplementary material
12284_2012_11_MOESM1_ESM.doc
Additional file 1: Table-S1.Microarray analysis of genes involved in carbohydrate metabolism and transport in rice. The values present the average ± SE (n = 4). Two weeks old rice plants were subjected to Zn deficiency for two more weeks. Table S2. Microarray profile of genes encoding RNase in rice. The values present the average ± SE (n = 4). Two weeks old rice plants were subjected to Zn deficiency for two more weeks. (DOC 62 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Suzuki, M., Bashir, K., Inoue, H. et al. Accumulation of starch in Zn-deficient rice. Rice 5, 9 (2012). https://doi.org/10.1186/1939-8433-5-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1939-8433-5-9