Abstract
Background
The digenean species of Echinostoma (Echinostomatidae) with 37 collar spines that comprise the so-called ‘revolutum’ species complex, qualify as cryptic due to the interspecific homogeneity of characters used to differentiate species. Only five species were considered valid in the most recent revision of the group but recent molecular studies have demonstrated a higher diversity within the group. In a study of the digeneans parasitising molluscs in central and northern Europe we found that Radix auricularia, R. peregra and Stagnicola palustris were infected with larval stages of two cryptic species of the ‘revolutum’ complex, one resembling E. revolutum and one undescribed species, Echinostoma sp. IG. This paper provides morphological and molecular evidence for their delimitation.
Methods
Totals of 2,030 R. auricularia, 357 R. peregra and 577 S. palustris were collected in seven reservoirs of the River Ruhr catchment area in Germany and a total of 573 R. peregra was collected in five lakes in Iceland. Cercariae were examined and identified live and fixed in molecular grade ethanol for DNA isolation and in hot/cold 4% formaldehyde solution for obtaining measurements from fixed materials. Partial fragments of the mitochondrial gene nicotinamide adenine dinucleotide dehydrogenase subunit 1 (nad 1) were amplified for 14 isolates.
Results
Detailed examination of cercarial morphology allowed us to differentiate the cercariae of the two Echinostoma spp. of the ‘revolutum’ species complex. A total of 14 partial nad 1 sequences was generated and aligned with selected published sequences for eight species of the ‘revolutum’ species complex. Both NJ and BI analyses resulted in consensus trees with similar topologies in which the isolates from Europe formed strongly supported reciprocally monophyletic lineages. The analyses also provided evidence that North American isolates identified as E. revolutum represent another cryptic species of the ‘revolutum’ species complex.
Conclusion
Our findings highlight the need for further analyses of patterns of interspecific variation based on molecular and morphological evidence to enhance the re-evaluation of the species and advance our understanding of the relationships within the ‘revolutum’ group of Echinostoma.
Similar content being viewed by others
Background
The digenean species of Echinostoma Rudolphi, 1809 (Echinostomatidae) with 37 collar spines that comprise the so-called Echinostoma ‘revolutum’ complex, qualify as cryptic (sensu Bickford et al.[1]; see also Pérez-Ponce de León and Nadler [2] for a recent review) due to the interspecific homogeneity of characters used to differentiate species. Only five species, the Eurasian Echinostoma revolutum (Frölich, 1802), E. echinatum (Zeder, 1803) and E. jurini (Skvortsov, 1924), the North American E. trivolvis (Cort, 1914) and the African E. caproni Richard, 1964, were considered valid in the most recent revision of the group using for species delimitation a single morphological feature of the larval stages (the number of pores of the para-oesophageal gland-cells in the cercaria), the specificity towards the first intermediate host (at the familial level), the ability to infect avian or mammalian hosts (or both) and geographical range on a global scale (continents) [3–5] (but see Kostadinova and Gibson [6] for a critical review). It is worth noting that E. echinatum has not been formally described and justified in a taxonomic publication and is not recognised as valid [see 6 for details]. However, recent molecular studies have demonstrated a higher diversity within the ‘revolutum’ species complex. Thus one African species, Echinostoma deserticum Kechemir et al., 2002, and a yet unidentified species from New Zealand were distinguished based on molecular data [7] (see also [8]), and E. trivolvis was found to represent a species complex [9]. Additional data on the geographical distribution of the Echinostoma spp. have also been obtained. E. revolutum was recorded in Australia [7] and North America [10, 11], Echinostoma paraensei Lie & Basch, 1967 in Australia and South America [7], and E. cf. robustum in North and South America [11].
The pioneer molecular studies, predominantly based on laboratory strains, have revealed that the mitochondrial nad 1 gene provides a better resolution for investigating relationships within the problematic Echinostoma ‘revolutum’ species complex in comparison with the nuclear rRNA spacers and the mitochondrial cox 1 gene [12, 13]. The subsequent DNA-based studies [7, 9–11, 14] have provided a framework for investigating genetic variation in natural Echinostoma spp. populations and revealed novel data on the cryptic variation, identification and geographical distribution of the species of the ‘revolutum’ complex.
However, in contrast with the wealth of sequences gathered recently from North America, which have revealed high diversity (six cryptic lineages) within the ‘revolutum’ complex of Echinostoma[9, 11], data from European natural populations are virtually lacking. Thus, of the eight species described and/or recorded from Europe, i.e. E. revolutum, E. paraulum Dietz, 1909, E. jurini (Skvortsov, 1924), E. miyagawai Ishii, 1932, E. robustum Yamaguti, 1935, E. bolschewense (Kotova, 1939), E. nordiana (Baschkirova, 1941), E. friedi Toledo et al., 2000 [3, 5, 15–22], sequence data are available only for E. revolutum[7, 12–14] and E. friedi (GenBank AJ564379).
In a study of the digeneans parasitising molluscs in central and northern Europe we found that Radix auricularia (Linnaeus, 1758), Radix peregra (Müller, 1774) and Stagnicola palustris (Müller, 1774) were infected with larval stages of two species of the Echinostoma ‘revolutum’ complex of cryptic species, one resembling E. revolutum sensu stricto (s.s.) and one undescribed species (see also [23]). Here we describe the cercariae of these two species and provide morphological and molecular evidence for their delimitation. Further, we extend the approaches of Morgan and Blair [7, 13], Kostadinova et al.[14] and Detwiler et al.[11] to the relationships within the ‘revolutum’ species complex inferred from the nad 1 gene with the newly-generated sequence data from natural infections in snails in Europe. Phylogenetic analyses revealed the presence of additional cryptic lineages of the Echinostoma ‘revolutum’ species complex.
Methods
Sample collection
Totals of 2,030 R. auricularia, 357 R. peregra and 577 S. palustris were collected during 2009–2012 in seven reservoirs of the River Ruhr catchment area (North Rhine-Westphalia, Germany): Baldeneysee (51°24′20.08"N, 7°2′22.47"E); Harkortsee (51°23′40.56"N, 7°24′8.27"E); Hengsteysee (51°24′52.17"N, 7°27′42.55"E); Hennetalsperre (51°19′50.97"N, 8°15′46.82"E); Kemnader See (51°25′19.05"N, 7°15′43.07"E); Sorpetalperre (51°20′ 15.01"N, 7°56′46.18"E); and Versetalsperre (51°10′55.71"N, 7°40′57.12"E). Seven distinct samples of R. peregra (a total of 573 snails) were collected in five localities in Iceland: Lakes Family Park (64°08′15"N, 21°52′03"W) and Nordic House (64°08′19"N, 21°56′45"W) in Reykjavik; Opnur (63°58′43"N, 21°10′37"W); Raudavatn (64°05′35"N, 21°47′14"W); and Helgavogur, Lake Myvatn (65°38′04"N, 16°55′28"W) in May and August 2012. Snails were collected randomly with a strainer or picked by hand from stones and floating vegetation along the shore at several sampling sites at each reservoir. In the laboratory, snails were labelled and placed individually into beakers with a small amount of lake water, and kept under a light source for up to 5 days to stimulate emergence of cercariae. Thereafter, snails were measured, dissected and examined for prepatent infections.
Morphological data
Cercariae were examined and identified live using the data from the keys of Faltýnková et al.[24, 25] and other relevant primary sources [3, 18–22]. Digital photographs of live cercariae (and rediae) were taken with a digital camera of an Olympus BX51 microscope. Vital stains (Neutral Red and Nile Blue sulphate) were used for visualisation of the para-oesophageal gland-cells of the cercariae. Measurements (in micrometres) were taken from the digital images with the aid of QuickPHOTO CAMERA 2.3 image analysis software or the program ImageJ [26]. Upon preliminary identification, two samples of cercariae (rediae) per isolate were fixed: (i) in molecular grade ethanol for DNA isolation and sequencing; and (ii) in hot/cold 4% formaldehyde solution for obtaining measurements from fixed materials. Snails were identified using Glöer [27]. Although R. peregra and R. ovata (Draparnaud, 1805) have recently been treated as junior synonyms of R. balthica (Linnaeus, 1758) we used the name R. peregra following the molecular studies of Bargues et al.[28] and Huňová et al.[29] which provide sequences for snails sampled in both central Europe and Iceland.
Molecular data
Total genomic DNA was isolated from ethanol-fixed single rediae and/or 10–50 pooled cercariae obtained from a single snail individual by placing the samples in 200 μL of a 5% suspension of deionised water and Chelex® containing 0.1 mg/mL proteinase K, followed by incubation at 56°C for 3 h, boiling at 90°C for 8 min, and centrifugation at 14,000 g for 10 min. Polymerase chain reaction (PCR) amplifications of partial fragments of the mitochondrial gene nicotinamide adenine dinucleotide dehydrogenase subunit 1 (nad 1) were performed in 25 μl reactions using Ready-To-Go-PCR Beads (GE Healthcare, UK) containing ~2.5 units of puReTaq DNA polymerase, 10 mM Tris–HCl (pH 9.0), 50 mM KCl, 1.5 mM MgCl2, 200 mM of each dNTP and stabilisers including BSA, 10 mM of each PCR primer, and 50 ng of template DNA. The following PCR primers were used: forward NDJ11 (equivalent to JB11 in [13]) 5′-AGA TTC GTA AGG GGC CTA ATA-3′ and reverse NDJ2a: 5′-CTT CAG CCT CAG CAT AAT-3′[14]. The thermocycling profile comprised initial denaturation at 95°C for 5 min, followed by 35 cycles with 30 s denaturation at 94°C, 20 s primer annealing at 48°C, and 45 s at 72°C for primer extension, with a final extension step of 4 min at 72°C.
PCR amplicons were purified using Qiagen QIAquick™ PCR Purification Kit (Qiagen Ltd, UK) and sequenced directly for both strands using the PCR primers. Sequencing was performed on an ABI Prism 3130xl automated sequencer using ABI Big Dye chemistry (ABI Perkin-Elmer, UK) according to the manufacturer’s protocol. Contiguous sequences were assembled and edited using MEGA v5 [30] and submitted to GenBank (accession numbers shown in Table 1).
Newly-generated and published nad 1 sequences for Echinostoma spp. (see Table 1 for details) were aligned using Clustal W implemented in MEGA v5 with reference to the amino acid translation, using the echinoderm and flatworm mitochondrial code [31]. Species boundaries were assessed via neighbour-joining (NJ) analyses of Kimura-2-parameter distances using MEGA v5 (nodal support estimated using 1,000 bootstrap resamplings) and Bayesian inference (BI) analysis using MrBayes 3.2 [32, 33]. The best-fitting model of nucleotide substitution estimated prior to BI analysis using jModelTest 2.1 [34] was the Hasegawa-Kishino-Yano model including estimates of invariant sites and among-site rate heterogeneity (HKY + I + G).
BI log-likelihoods were estimated with default prior probabilities and likelihood model settings (nst = 2; rates = invgamma; ngammacat = 4) over 106 generations via 4 simultaneous Markov Chain Monte Carlo chains (nchains = 4) with a sampling frequency of 100. The first 25% of the samples were discarded (sump burnin = 2500) as determined by the stationarity of lnL assessed with Tracer v. 1.4 [35]; the remaining trees were used to construct the 50% majority-rule consensus tree and to estimate the nodal support as posterior probability values [36]. Genetic distances (uncorrected p-distance) were calculated with MEGA v5.
Results
Morphological identification of infections in natural snail populations
We found larval stages of Echinostoma spp. in the snail populations sampled in three of the seven reservoirs in the River Ruhr drainage in Germany and in two of the five lakes in Iceland (see Table 2 for details on hosts and localities). Three lymnaeid snail species acted as first intermediate hosts of Echinostoma spp. of the ‘revolutum’ species complex in the areas studied: R. peregra in the lakes in Iceland and R. auricularia, R. peregra and S. palustris in the reservoirs in Germany. Prevalences were usually low (typically 1-3%) but occasionally higher values were registered (Table 2).
Detailed examination of cercarial morphology allowed us to identify two types of echinostomatid cercariae among the isolates sampled in Iceland and Germany (Figures 1, 2, 3, 4, 5). Both types belong to the ‘revolutum’ species complex of Echinostoma which is characterised by the following features of the cercariae: (i) 37 collar spines with an arrangement 5-6-15-6-5 (5 angle and 6 lateral spines on each side and 15 dorsal spines in a double row; Figures 1C, 2D,E, 3C, 4D,E); (ii) tail with a tip forming a highly contractile attenuated process and seven prominent tegumental fin-folds (2 dorsal, 3 ventral and 2 ventro-lateral; Figures 1B, 2C, 3B, 4C); and (iii) a flame-cell formula 2[(3 + 3 + 3) + (3 + 3 + 3)] = 36.
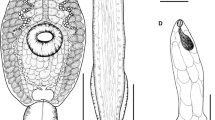
Echinostoma sp. IG, drawings of live cercaria. A. Body, ventral view. B. Tail, lateral view (note that only one of the two ventro-lateral fin-folds is illustrated). C. Head collar. D. Schematic illustration of the para-oesophageal gland-cells. Abbreviations: d, dorsal fin-fold; v, ventral fin-fold. Scale-bars: A, B, 100 μm; C, 50 μm.
Echinostoma sp. IG, microphotographs of live cercaria. A. Body, ventral view. B. Dorsal view showing para-oesophageal gland-cells and outlets (staining with Neutral Red) C. Tail, lateral view. D. Head collar, ventral view showing angle and lateral spines. E. Head collar, dorsal view, showing dorsal collar spines. Scale-bars: A, C, 100 μm; B, D, E, 50 μm.
Echinostoma revolutum , drawings of live cercaria. A. Body, ventral view. B. Tail, lateral view (note that only one of the two ventro-lateral fin-folds is illustrated). C. Head collar. D. Schematic illustration of the para-oesophageal gland-cells. Abbreviations: d, dorsal fin-fold; v, ventral fin-fold; vl, ventro-lateral fin-fold. Scale-bars: A, B, 100 μm; C, 50 μm.
Echinostoma revolutum , microphotographs of live cercaria. A. Body, ventral view. B. Ventral view showing outlets of para-oesophageal gland-cells (staining with Neutral Red). C. Tail, lateral view. D. Head collar, ventral view showing angle and lateral spines. E. Head collar, dorsal view showing dorsal collar spines. Scale-bars: A, C, 100 μm; B, D, E, 50 μm.
Eleven isolates (three ex R. peregra from Iceland, plus two ex R. auricularia, five ex R. peregra and one ex S. palustris from Germany) were identified as E. revolutum based on cercarial morphology and especially the presence of 12 small para-oesophageal gland-cells with long ducts, located between pharynx and ventral sucker [24] (Figures 2B, 4B). However, seven isolates of cercariae, one ex R. peregra from Iceland and six ex R. auricularia from Germany, further referred to as Echinostoma sp. IG (indicating the origin of the isolates i.e. Iceland and Germany) exhibited slight differences from the isolates identified as E. revolutum as follows: (i) collar spines with blunt (Figures 1C, 2D,E) vs sharp (Figures 3C, 4D,E) tips; (ii) para-oesophageal gland-cell outlets opening at the margin of the oral sucker only (one dorsal pair, four dorsolateral pairs, and one ventro-lateral pair; see Figures 1D, 2B) vs openings present on the ventral surface of the body (one pair at the level of pharynx; the remaining i.e. one dorsal pair, one dorsolateral pair, and three ventro-lateral pairs opening at the margin of the oral sucker, see Figures 3D, 4B); and (iii) distal dorsal tail fin-fold large vs less prominent (length 40-60% of tail length vs 20-38%; width c.70% of tail width vs 20-30%; compare Figures 1B, 2C and 3B, 4C; Table 3). Comparison of the metrical data obtained for live cercariae revealed that Echinostoma sp. IG had a shorter tail, with distinctly larger distal dorsal fin-fold and shorter distal ventral fin-fold (Table 3). Furthermore, although it was difficult to observe the fin-folds in fixed material thus rendering differentiation difficult, the cercariae of Echinostoma sp. IG were characterised by a distinctly more elongate, narrower body and a shorter tail (Figure 5; Table 3); this represents another distinguishing feature for the two European species studied by us.
Molecular analysis
A total of 14 partial nad 1 sequences was generated (11 for E. revolutum and 3 for Echinostoma sp. IG; Table 1). These sequences were aligned with selected published sequences representing the data available for eight species of the ‘revolutum’ species complex of Echinostoma generated from both laboratory strains [13] and natural isolates [9, 11, 14]; two otherwise unpublished sequences were also retrieved from GenBank (see Table 1 for details). The aligned dataset included 39 sequences and was comprised of 472 nt positions after trimming the ends to match the shortest aligned sequences. Sequences for Echinoparyphium spp. of Kostadinova et al.[14] were used as outgroups (Table 1).
Both NJ and BI analyses resulted in consensus trees with similar topologies (see Figure 6 for a phylogeny inferred from genetic distances and BI). The newly-generated sequences for E. revolutum formed a strongly supported clade which included a sequence for E. revolutum (s.s.) of Kostadinova et al.[14] (see also [6]). On the other hand, the sequences for the isolates identified as Echinostoma sp. IG formed a strongly supported reciprocally monophyletic lineage, basal to Echinostoma spp., which also incorporated the sequence for an isolate from Wales (UK) provisionally identified as Echinostoma cf. friedi by Kostadinova et al.[14]. The isolates comprising this lineage also exhibited the highest levels of divergence from the isolates of Echinostoma spp. analysed (p-distance range 17.2-21.6%; divergence from E. friedi (AJ564379) (p-distance range 18.9-19.1%).
Neighbour-joining (NJ) phylogram reconstructed using the newly-generated and retrieved from GenBank nad1 sequences (472 nt positions) for Echinostoma spp. of the ‘ revolutum ’ species complex. Outgroup: Echinoparyphium spp. Nodal support (bootstrap values > 70% shown only) inferred from 1,000 replicates; these are followed by posterior probabilities from BI analysis. The scale-bar indicates expected number of substitutions per site. Sequence identification as in GenBank followed by a letter: D, Detwiler et al. [9, 11]; K, Kostadinova et al.[14]; M, Marcilla et al. (GenBank); M & B, Morgan & Blair [7, 13].
Unexpectedly, the European isolates of E. revolutum and those obtained from natural infections in Lymnaea elodes and Ondatra zibethicus (L.) in North America by Detwiler et al.[11] formed two strongly supported sister lineages. This solution (both NJ and BI analyses) was consistent with the distinctly higher inter-lineage divergence (p-distance; 4.9–6.8%) compared with intra-lineage divergence (p-distance range, European isolates: 0–2.1%, North American isolates: 0.4–1.1%). These data indicate that the North American isolates represent another cryptic species of the ‘revolutum’ species complex.
Another unexpected result was that the sequence for Echinostoma revolutum of Morgan and Blair [7, 13] (AF025832; isolate from Europe) exhibited a strong association with the sequence for Echinostoma friedi of Marcilla et al. (unpublished, GenBank AJ564379) based on an isolate of this species recently described by these authors [22] from Spain (p-distance 0.8%; divergence from nearest neighbours, i.e. Echinostoma robustum sensu Detwiler et al.[11], of 4.9-9.1%. The clade comprising the former two European isolates and those of E. robustum from North America exhibited a complex structure suggesting the existence of at least three species (subclade support indicated in Figure 6).
Discussion
The combined morphological and DNA-based approaches in this first intensive screening of Radix spp. for infections with Echinostoma spp. allowed us to delineate two cryptic species of the ‘revolutum’ complex in central and northern Europe. Furthermore, comparative sequence analyses depicted three additional cryptic lineages in North America.
Both distance- and model-based phylogenies provided high support for reciprocal monophyly of Echinostoma sp. IG. The isolates of this lineage, that evidently represents a new species, awaiting further formal description after a discovery of the adult parasite stage, were found to be clearly distinguishable among the European isolates by using both morphological and molecular evidence. Although the identification of the European isolates of Echinostoma spp. followed the standard taxonomic practice, the detection of the new cryptic species required substantial taxonomic expertise. This involved detailed knowledge on the variation of the features used for species delimitation based on thorough morphological examination of a large number of cercariae from each isolate. The corroboration of our hypothesis for the distinct species status of the two species of Echinostoma parasitising snail populations in Germany and Iceland on the basis of molecular data thus may appear secondary.
However, the distinguishing features are difficult to detect and/or subject to variation (reviewed in Kostadinova and Gibson [6]). For example, Kanev [3] described 16 ducts and pores of para-oesophageal gland-cells in the cercariae of E. revolutum ex Lymnaea stagnalis; of these, 12 were located on the oral sucker and four on the ventral surface. On the other hand, we detected only 12 small para-oesophageal gland-cells in the cercariae of E. revolutum ex Radix spp.; Faltýnková et al.[24] also provided this number for E. revolutum ex L. stagnalis. It is worth noting that recent field studies indicate that E. revolutum most commonly occurs in L. stagnalis in Europe [23, 24], infections with this species have occasionally been reported in the past from R. auricularia, R. peregra and R. ovata (Draparnaud, 1805) [22, 37–45]. Further molecular study would reveal whether Echinostoma spp. of the ‘revolutum’ species complex parasitising L. stagnalis and Radix spp. are conspecific or represent as yet undiscovered cryptic species. We believe that ‘reciprocal illumination’ sensu Hennig [46] of morphological characters upon a molecular-based species delimitation has a strong potential for delineating species boundaries within the ‘revolutum’ complex of cryptic species.
Echinostoma sp. IG was found to be conspecific with an isolate from Wales (UK) provisionally identified as Echinostoma cf. friedi by Kostadinova et al.[14]. The lineage comprising this and the newly-sequenced isolates occupied a basal position (as in Kostadinova et al.[14]) and this is in sharp contrast with the phylogenetic solution based on nad 1 gene of Detwiler et al.[11]. These authors wrote that “A comparison of samples identified as E. robustum (U58102) and E. friedi (AY168937) reveals that they are found within the same monophyletic clade and thus do not qualify as distinct species according to a phylogenetic definition. Additionally, they are genetically similar (0.009 genetic divergence, ND1 …” and concluded that “the sample tentatively identified as E. friedi in Kostadinova et al. (2003) is genetically very similar to E. robustum”. Our results clearly indicate that the sequence for E. friedi from its type-locality in Spain (AJ564379; Marcilla et al. unpublished sequence in GenBank) and for the European isolate labelled as E. revolutum (AF025832) of Morgan and Blair [7, 12, 13] represent conspecific isolates; the genetic divergence between these two isolates was 0.8%, i.e. substantially lower than that (i.e. 18.9-19.1%) between the lineage containing E. cf. friedi (AY168937) of Kostadinova et al.[14] and the European isolate labelled as E. revolutum (nad 1 sequence AF025832; ITS sequence U58102) by Morgan and Blair [7, 12, 13]. We believe, therefore, that Detwiler et al.[11] have in fact used the otherwise unpublished sequence for E. friedi of Marcilla et al. (AJ564379) but have mislabelled it (as AY168937).
Kostadinova et al.[14] indicated a tentative affiliation to E. robustum of the isolates of the ‘Australian-German’ clade of Echinostoma spp. of Morgan and Blair [7], but suggested that this specific identification is pending a redescription of both larval and adult stages. The present results indicate that suggesting synonymy for the European isolate studied by Morgan and Blair [7, 12, 13] and E. friedi should await examination of a larger number of molecularly characterised natural isolates of the European species of the ‘revolutum’ complex since our knowledge on cryptic diversity in this group is still limited. This suggestion is supported by the discovery of two genetically distinct, geographically separated lineages of E. revolutum: E. revolutum s.s. from Europe and E. revolutum of Detwiter et al.[11] from North America, thus demonstrating that the suggestion for the cosmopolitan distribution of this species [11] appears to be a result of cryptic variation. Indeed, these authors noted that their results of network analyses indicate gene flow and population expansion within North America but not on a global scale. The taxonomy of the North American species can be further scrutinised using the morphological data available for cercariae and/or experimentally developed adults [11, 47].
Conclusion
The results of our study suggest that further analyses of patterns of interspecific variation based on a combination of molecular and well-documented morphological data would enhance the re-evaluation of the species and advance our understanding of the relationships within the ‘revolutum’ group of Echinostoma.
References
Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I: Cryptic species as a window on diversity and conservation. Trends Ecol Evol. 2007, 22: 148-155. 10.1016/j.tree.2006.11.004.
Pérez-Ponce de León G, Nadler SA: Critical Comment: What we don’t recognize can hurt us: A plea for awareness about cryptic species. J Parasitol. 2010, 96: 453-464. 10.1645/GE-2260.1.
Kanev I: Life-cycle, delimitation and redescription of Echinostoma revolutum (Frölich, 1802) (Trematoda: Echinostomatidae). Syst Parasitol. 1994, 28: 125-144. 10.1007/BF00009591.
Kanev I, Dimitrov V, Radev V, Fried B: Redescription of Echinostoma trivolvis (Cort, 1914) with a discussion of its identity. Syst Parasitol. 1995, 32: 61-70. 10.1007/BF00009468.
Kanev I, Fried B, Dimitrov V, Radev V: Redescription of Echinostoma jurini (Skvortzov, 1924) with a discussion of its identity and characteristics. Ann Naturhist Mus Wien. 1995, 97B: 37-53.
Kostadinova A, Gibson DI: The systematics of the echinostomes. Echinostomes as Experimental Models for Biological Research. Edited by: Fried B, Graczyk TK. 2000, Dordrecht: Kluwer Academic Publishers, 31-57.
Morgan JAT, Blair D: Mitochondrial ND1 gene sequences used to identify echinostome isolates from Australia and New Zealand. Int J Parasitol. 1998, 28: 493-502. 10.1016/S0020-7519(97)00204-X.
Kechemir N, Jourdane J, Mas-Coma S: Life cycle of a new African echinostome species reproducing by parthenogenesis. J Nat Hist. 2002, 36: 1777-1784. 10.1080/00222930110062633.
Detwiler JT, Zajac AM, Minchella DJ, Belden LK: Revealing cryptic parasite diversity in a definitive host: echinostomes in muskrats. J Parasitol. 2012, 98: 1148-1155. 10.1645/GE-3117.1.
Sorensen RE, Curtis J, Minchella DJ: Intraspecific variation in the rDNA its loci of 37-collar-spined echinostomes from North America: implications for sequence-based diagnoses and phylogenetics. J Parasitol. 1998, 84: 992-997. 10.2307/3284633.
Detwiler JT, Bos DH, Minchella DJ: Revealing the secret lives of cryptic species: Examining the phylogenetic relationships of echinostome parasites in North America. Mol Phylogenet Evol. 2010, 55: 611-620. 10.1016/j.ympev.2010.01.004.
Morgan JAT, Blair D: Nuclear rDNA ITS sequence variation in the trematode genus Echinostoma: an aid to establishing relationships within the 37-collar-spine group. Parasitology. 1995, 111: 609-615. 10.1017/S003118200007709X.
Morgan JAT, Blair D: Relative merits of nuclear ribosomal internal transcribed spacers and mitochondrial CO1 and ND1 genes for distinguishing among Echinostoma species (Trematoda). Parasitology. 1998, 116: 289-297. 10.1017/S0031182097002217.
Kostadinova A, Herniou EA, Barrett J, Littlewood DTJ: Phylogenetic relationships of Echinostoma Rudolphi, 1809 (Digenea: Echinostomatidae) and related genera re-assessed via DNA and morphological analyses. Syst Parasitol. 2003, 54: 159-176. 10.1023/A:1022681123340.
Dietz E: Die Echinostomiden der Vögel. Zool Anz. 1909, 34: 180-192.
Dietz E: Die Echinostomiden der Vögel. Zool Jahrb. 1910, 12: S256-S512.
Iskova NI: Trematoda, Part 4. Echinostomatata. Fauna Ukrainy. Vol. 34. 1985, Kiev: Naukova Dumka, (In Russian)
Našincová V: Contribution to the distribution and the life history of Echinostoma revolutum in Central Europe. Věst Českoslov Společ Zool. 1986, 50: 70-80.
Našincová V: The life cycle of Echinostoma bolschewense (Kotova, 1939) (Trematoda: Echinostomatidae). Folia Parasitol. 1991, 38: 143-154.
Kostadinova A, Gibson DI, Biserkov V, Chipev N: Re-validation of Echinostoma miyagawai Ishii, 1932 (Digenea: Echinostomatidae) on the basis of experimental completion of its life-cycle. Syst Parasitol. 2000, 45: 81-108. 10.1023/A:1006241610689.
Kostadinova A, Gibson DI, Biserkov V, Ivanova R: A quantitative approach to the evaluation of the morphological variability of two echinostomes, Echinostoma miyagawai Ishii, 1932 and E. revolutum (Frölich, 1802) from Europe. Syst Parasitol. 2000, 45: 1-15. 10.1023/A:1006232612469.
Toledo R, Muñoz-Antolí C, Esteban JG: The life-cycle of Echinostoma friedi n. sp. (Trematoda: Echinostomatidae) in Spain and a discussion on the relationships within the ‘revolutum’ group based on cercarial chaetotaxy. Syst Parasitol. 2000, 45: 199-217. 10.1023/A:1006385902664.
Soldánová M, Selbach C, Sures B, Kostadinova A, Pérez-del-Olmo A: Larval trematode communities in Radix auricularia and Lymnaea stagnalis in a reservoir system of the Ruhr River. Parasit Vectors. 2010, 3: 56-10.1186/1756-3305-3-56.
Faltýnková A, Našincová V, Kablásková L: Larval trematodes (Digenea) of the great pond snail, Lymnaea stagnalis (L.), (Gastropoda, Pulmonata) in Central Europe: a survey of species and key to their identification. Parasite. 2007, 14: 39-51.
Faltýnková A, Našincová V, Kablásková L: Larval trematodes (Digenea) of planorbid snails (Gastropoda: Pulmonata) in Central Europe: a survey of species and key to their identification. Syst Parasitol. 2008, 69: 155-178. 10.1007/s11230-007-9127-1.
Abramoff MD, Magalhaes PJ, Ram SJ: Image processing with ImageJ. Biophotonics Internat. 2004, 11: 36-42.
Glöer P: Die Süßwassergastropoden Nord- und Mitteleuropas. 2, neubearbeitete Auflage. 2002, Hackenheim: ConchBooks
Bargues MD, Vigo M, Horák P, Dvořák J, Patzner RA, Pointier JP, Jackiewicz M, Meier–Brook C, Mas-Coma S: European Lymnaeidae (Mollusca: Gastropoda), intermediate hosts of trematodiases, based on nuclear ribosomal DNA ITS2 sequences. Infect Genet Evol. 2001, 1: 85-107. 10.1016/S1567-1348(01)00019-3.
Huňová K, Kašný M, Hampl V, Leontovyč R, Kuběna A, Mikeš L, Horák P: Radix spp.: Identification of trematode intermediate hosts in the Czech Republic. Acta Parasitol. 2012, 57: 273-284. 10.2478/s11686-012-0040-7.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S: MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011, 28: 2731-2739. 10.1093/molbev/msr121.
Telford MJ, Herniou EA, Russell RB, Littlewood DTJ: Changes in mitochondrial genetic codes as phylogenetic characters: Two examples from the flatworms. Proc Nat Acad Sci USA. 2000, 97: 11359-11364. 10.1073/pnas.97.21.11359.
Huelsenbeck JP, Ronquist F: MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001, 17: 754-755. 10.1093/bioinformatics/17.8.754.
Ronquist F, Huelsenbeck JP: MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003, 19: 1572-1574. 10.1093/bioinformatics/btg180.
Posada D: jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008, 25: 1253-1256. 10.1093/molbev/msn083.
Rambaut A, Drummond AJ: Tracer v1.4. 2007, Available from http://beast.bio.ed.ac.uk/Tracer
Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP: Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001, 294: 2310-2314. 10.1126/science.1065889.
Golikova MN: An ecological and parasitological study of the biocoenoses of some lakes in the Kaliningrad region. IV. The trematode fauna of invertebrates. Vestn Lenin Univ Biol. 1960, 15: 80-94. (In Russian)
Žďárská Z: Larvální stádia motolic z vodních plžů na území ČSSR. Českoslov Parasitol. 1963, 10: 207-262. (In Czech)
Nezvalová J: Příspěvek k poznání cerkarií jižní Moravy. Folia Fac Sci Nat Univ Purkynianae Brunensis Biol. 1970, 515: 217-252. (In Czech)
Balůsek J, Vojtek J: Příspěvek k poznání našich cerkárií. Folia Fac Sci Nat Univ Purkynianae Brunensis Biol. 1973, 40: 3-43. (In Czech)
Stenko RP: On the trematode fauna of some Crimean freshwater molluscs and its changes induced by anthropogenic factors. Vestn Zool. 1976, 5: 90-91. (In Russian)
Stenko RP: On studying fauna of trematoda larvae of fresh-water Crimean mollusks. Vestn Zool. 1976, 5: 42-46. (In Russian)
Kanev I, Vassilev I, Bayssade-Dufour C, Albaret J-L, Cassone J: Chétotaxie cercarienne d’Echinostoma revolutum (Froelich, 1802) et E. echinatum (Zeder, 1803) (Trematoda, Echinostomatidae). Ann Parasit Hum Comp. 1987, 62: 222-234.
Väyrynen T, Siddall R, Valtonen ET, Taskinen J: Patterns of trematode parasitism in lymnaeid snails from northern and central Finland. Ann Zool Fenn. 2000, 37: 189-199.
Faltýnková A: Larval trematodes (Digenea) in molluscs from small water bodies near České Budějovice, Czech Republic. Acta Parasitol. 2005, 50: 49-55.
Hennig W: Phylogenetic systematics. 1966, Urbana: University of Illinois Press
Detwiler JT, Minchella DJ: Intermediate host availability masks the strength of experimentally-derived colonisation patterns in echinostome trematodes. Int J Parasitol. 2009, 39: 585-590. 10.1016/j.ijpara.2008.10.008.
Acknowledgements
We thank Blanka Škoríková for her kind help with the figures and Jana Köchling, Verena Altmann, Jessica Schwelm and Dr Ana Pérez-del-Olmo, for their assistance in sampling. This study was supported by the Czech Science Foundation (AF, AK, MS, SG, grant P505/10/1562); the ‘Sichere Ruhr’ project as part of the Bundesministerium für Bildung und Forschung (BMBF) program ‘Sustainable Water Management’ (BS, grant 02WRS1283); and the Research Fund of the University of Iceland (KS). CS benefited from a Deutsche Bundesstiftung Umwelt (DBU) PhD fellowship.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CS, MS and KS obtained the samples. CS, AF, MS and SG undertook the morphological study. SG carried out the sequencing and phylogenetic analysis. CS, SG, AF and MS prepared the first draft of the MS. KS, BS and AK conceived and coordinated the study and helped to draft the MS. All authors read and approved the final manuscript.
Simona Georgieva, Christian Selbach contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Georgieva, S., Selbach, C., Faltýnková, A. et al. New cryptic species of the ‘revolutum’ group of Echinostoma (Digenea: Echinostomatidae) revealed by molecular and morphological data. Parasites Vectors 6, 64 (2013). https://doi.org/10.1186/1756-3305-6-64
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-3305-6-64