Abstract
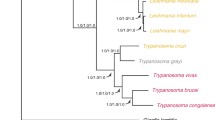
The genomes of several tsetse-transmitted African trypanosomes (Trypanosoma brucei brucei, T. b. gambiense, T. vivax, T. congolense) have been sequenced and are available to search online. The trypanosome strains chosen for the genome sequencing projects were selected because they had been well characterised in the laboratory, but all were isolated several decades ago. The purpose of this short review is to provide some background information on the origins and biological characterisation of these strains as a source of reference for future users of the genome data. With high throughput sequencing of many more trypanosome genomes in prospect, it is important to understand the phylogenetic relationships of the genome strains.
Similar content being viewed by others
Review
The genome sequence of Trypanosoma brucei brucei TREU 927/4 was published in 2005 [1] and that of T. b. gambiense Dal 972 clone 1 in 2010 [2]. Genome sequencing projects for T. vivax Y486 and T. congolense IL3000 are also complete [3]. The trypanosome strains chosen for sequencing were selected because they had been well characterised in the laboratory, but all were isolated several decades ago (Table 1). The purpose of this short review is to provide some background information on the origins and biological characterisation of these strains as a source of reference for future users of the genome data. The history of the discovery of these trypanosome species has been recently reviewed [4].
Trypanosoma brucei brucei TREU 927/4
Isolation and phenotype
Trypanosoma brucei brucei clone TREU 927/4 was chosen as the representative T. brucei for the genome project, because it displays the full range of known phenotypes for T. brucei, barring human infectivity. TREU 927/4 is capable of complete cyclical development within the tsetse fly, including mating [5] and produces short stumpy forms during bloodstream infection in the mammalian host [6]. TREU 927/4 is a clone derived from the isolate GPAL/KE/70/EATRO 1534 [5] that originates from Kiboko, Kenya, an area where human trypanosomiasis is unknown [7]. Nevertheless, there is some doubt about the status of TREU 927/4 with regard to human infectivity, since it has a degree of resistance to human serum [8], though it lacks the SRA gene that is characteristic of the human infective subspecies T. b. rhodesiense from East Africa [9].
The isolate GPAL/KE/70/EATRO 1534 was one of a collection of 15 T. brucei subgroup isolates obtained from wild caught tsetse flies of the species Glossina pallidipes from Kiboko, Kenya [7, 10, 11]. Each isolate was derived from the metacyclic population of a single infected fly by inoculation of macerated salivary glands into rodents; the bloodstream forms were subsequently used to study the antigenic types circulating in wild-caught flies [11]. Like other pleomorphic T. b. brucei isolates, TREU 927/4 is easily grown as bloodstream forms in rodents or procyclics in vitro; it has also been adapted to the bloodstream form in in vitro culture [6] and has been transmitted via G. morsitans morsitans and G. pallidipes in the laboratory.
Relationship to other T. brucei strains
TREU 927/4, together with other T. b. brucei isolates from Kiboko, has been characterised by various molecular methods in a number of different studies. The consensus from these analyses is that TREU 927/4 represents a subgroup of T. b. brucei that is widespread in East Africa.
Initial studies of isoenzyme variation showed that the Kiboko isolates had a high frequency of rare polymorphisms for several enzymes, notably threonine dehydrogenase (TDH) and malate dehydrogenase (MDH), and that these unusual isoenzyme patterns were shared by T. brucei subgroup isolates from other areas with abundant wild animals in Kenya (Kibwezi, Meru and Maasai Mara), Zambia (Luangwa valley) and Tanzania (Serengeti) [12–14]. As a consequence, these isolates appeared as a separate clade distinct from other T. brucei subgroup isolates in cluster analysis [12–14]. The clade was initially dubbed the kiboko group [12] and later split into 2 groups: kiboko and kakumbi[13]. Only a minority (estimated < 7%) of the many isolates of the T. brucei subgroup that have been characterised belong to the kiboko/kakumbi isoenzyme group [12, 13]. The association with areas of abundant wildlife in East Africa suggests transmission cycles primarily involving wild mammals and tsetse. However, some kiboko/kakumbi strains have been isolated from domestic animals and three came from humans in the Luangwa Valley, Zambia [13].
The molecular karyotype of TREU 927/4 was deduced from chromosome-separation gels and hybridisation with a comprehensive set of gene probes [15]. The nuclear genotype has been analysed by restriction fragment length polymorphisms (RFLPs), mini- and microsatellites. Reinforcing the isoenzyme results, analysis of T. brucei subgroup isolates using RFLP data from repetitive DNA probes placed TREU 927/4 in a discrete cluster with 2 other Kiboko tsetse isolates [16, 17]. Minisatellite analysis of the original trypanosome population from which TREU 927/4 was cloned showed it to contain several genotypes [18]. Minisatellites are hypervariable and the Kiboko strains, like other collections of trypanosome isolates from particular locations, show population specific alleles [19]. A recent study using STRUCTURE to analyse microsatellite genotype data from a total of 142 T. brucei subgroup strains placed TREU 927/4 in a cluster designated Kiboko B, including 18 other T. b. brucei isolates derived from wild animals (mostly lions and hyenas) in Serengeti, Tanzania, and other tsetse and livestock isolates from Kiboko and Meru in Kenya [20]. In summary, there is agreement from all genotyping analyses to date that TREU 927/4 belongs to a particular group of T. b. brucei isolates that is found predominantly in wildlife areas of East Africa.
Complementary to these data from analyses of the nuclear genome, are data on variation in the sequence of kinetoplast DNA maxicircles, which constitute the mitochondrial genome of trypanosomes. Initial analysis of RFLPs showed that the maxicircles of kiboko/kakumbi strains could be distinguished from those of other T. brucei subgroup isolates by several polymorphisms [21]. Sequencing of the maxicircle COI (cytochrome c oxidase subunit I) gene allowed detailed analysis of mitochondrial haplotypes and revealed that isolates of the kiboko/kakumbi group, including TREU 927/4, share a limited range of haplotypes not found among other T. brucei subgroup isolates [20]. There was a particularly striking correlation between maxicircle haplotypes and the corresponding microsatellite data for the cluster of 19 T. b. brucei isolates designated Kiboko B, which included TREU 927/4 [20]. This reinforces the distinctive nature of this group of strains and also demonstrates that genetic exchange between this group and other T. brucei genotypes is not sufficiently frequent to break up this association.
Trypanosoma brucei gambiense DAL 972 clone 1
Isolation and phenotype
T. b. gambiense is the causative organism of human trypanosomiasis in West and Central Africa and has been divided into two groups or types based on phenotypic and genotypic characteristics [22]. Most isolates belong to Tbg1, which conforms to the classical description of T. b. gambiense as a pathogen that causes a chronic disease in human patients and manifests very low parasitaemia; it is typically slow growing in experimental rodents and is better adapted to transmission by palpalis than morsitans group tsetse flies [23]. Tbg1 is responsible for most cases of human trypanosomiasis in Africa. By contrast Tbg2 grows well in experimental rodents and is easily transmitted by morsitans group tsetse [23]; it has rarely been isolated from patients and has thus far been found only in Côte d'Ivoire (Ivory Coast) and Burkina Faso (Upper Volta) [16, 24]. Tbg2 isolates are genetically heterogeneous [16, 24, 25].
DAL 972 (MHOM/CI/86/DAL972) clone 1 was chosen as the representative Tbg1 for the genome project, because it was originally isolated from a patient in West Africa (Daloa focus in Côte d'Ivoire) [26], and has the phenotypic features typical of Tbg1. DAL 972 clone 1 has been very little passaged and has an extremely chronic phenotype in normal experimental rodents, but can be grown in immunosuppressed or immunodeficient mice such as SCID (severe combined immunodeficiency) mice. It has been transmitted via G. palpalis gambiensis, G. m. morsitans and G. pallidipes in the laboratory, and grows satisfactorily as procyclics in vitro in Cunningham's medium.
Relationship to other T. brucei strains
By biochemical characterisation, DAL 972 is a typical Tbg1 with specific isoenzyme patterns for alanine and aspartate aminotransferases [27], and has all three characteristic Tbg1 genetic markers, namely genes for the variant surface glycoproteins LiTat 1.3 and VSG AnTat 11.17, and the flagellar pocket glycoprotein, TgsGP [28, 29]. Note that TgsGP is not correctly annotated in the DAL 972 genome, because this gene has been subject to a rearrangement relative to TREU 927 resulting in lack of synteny for one homologue of chromosome 2 [30]. In addition, DAL 972 was shown to have the characteristic Tbg1 RFLP pattern for VSG 117 (AnTat 1.8) [31, 32].
Tbg1 isolates typically have low DNA contents compared to other members of the T. brucei subgroup [33, 34]. The DNA content of DAL 972 was at the high end of the range for Tbg1, but lower than T. b. brucei or T. b. rhodesiense[34]. In addition, the molecular karyotypes of Tbg1 isolates typically have lower numbers of minichromosomes than T. b. brucei or T. b. rhodesiense[33] and this is also true for DAL 972 [34].
Trypanosoma vivax Y486
Isolation and phenotype
T. vivax is a major pathogen of ruminants both in Africa, where it is transmitted cyclically by tsetse flies, and in South America, where it is transmitted mechanically by biting flies such as tabanids. Despite its importance as a livestock pathogen, T. vivax has received little attention because it is difficult to cultivate in the laboratory. Unlike T. brucei, T. vivax is typically not infective to laboratory rodents, but can be gradually adapted to these hosts by for example co-injection with ruminant serum or immunosuppression by sublethal irradiation [35]. However, three strains of T. vivax (Y486, Y58, V953) from naturally infected cattle in Zaria, Nigeria were isolated directly into mice [36]. These spontaneously mouse infective strains grow to high parasitaemia in outbred mice and have been widely used in laboratory studies. The ability to clone antigenic variants of T. vivax Y486 [37] led to the purification and characterization of T. vivax variant surface glycoproteins [38]; T. vivax Y486 is ILRAD Duttonella antigen repertoire 1, ILDAR 1. As the most well characterized of the three Zaria T. vivax strains, Y486 was chosen for the genome project.
The Zaria T. vivax strains are infective to calves, sheep and goats, as well as mice, rats and rabbits, and are readily transmissible by a wide range of tsetse species (G. m. centralis, G. m. morsitans, G. pallidipes, G. austeni, G. brevipalpis, G. tachinoides, G. palpalis palpalis, G. p. gambiensis, G. fuscipes fuscipes) [37, 39–42]. Infection in these flies was typically found in the labrum and hypopharynx and sometimes also in the cibarium [41, 42]. Unlike T. brucei and T. congolense, T. vivax does not multiply in the tsetse midgut and hence does not grow in vitro as procyclics. Although culture systems to maintain T. vivax bloodstream forms in vitro were developed by several groups [43–45], none has come into general use. Likewise, there are several published methods from the same era for in vitro differentiation of bloodstream forms into epimastigotes and subsequent development to infective metacyclics [46–48]. Recently, a simple in vitro cultivation method for epimastigotes of IL 1392, a derivative of Y486, has been described that is robust enough to allow transfection and also gives rise to infective metacyclics [49].
Relationship to other T. vivax strains
The Nigerian reference isolate, T. vivax Y486, is a representative of the West African form of T. vivax. Before the advent of the Polymerase chain reaction (PCR), knowledge of genetic diversity in T. vivax was severely limited because of the difficulty of obtaining enough trypanosomes for analysis. However, differences in pathogenicity were recognised between East and West African T. vivax strains, the West African strains being generally regarded as more pathogenic to cattle than East African strains [50], although highly pathogenic haemorhaggic T. vivax strains are also known in East Africa [51, 52]. West African T. vivax strains can be identified by specific repetitive DNA sequences and are phylogenetically distinct from East African strains [53–56]. South American T. vivax strains have been shown to have close genetic similarity to West African strains [57], in agreement with the historical evidence that T. vivax was imported into the New World in cattle from West Africa [58]. Thus, T. vivax Y486 also represents the South American form of T. vivax.
Trypanosoma congolense IL3000
Isolation and phenotype
IL 3000 is a derivative of strain Transmara I, which was isolated from a bovine in the Transmara region of Kenya in 1966 [59, 60]. Antigenic variation has been studied in this strain (designated ILRAD Nannomonas antigen repertoire 2, ILNAR 2) and variant antigen genes have been characterized from both bloodstream and metacyclic IL 3000 trypanosomes [61, 62]. Presumably this was one of the deciding factors in choosing IL3000 for the genome project, although several other T. congolense strains such as TREU 1457 have also been widely used in experimental studies.
T. congolense generally grows well in laboratory rodents and bloodstream forms of IL3000 have been grown in mice as well as bloodstream form culture [60, 63]. T. congolense is transmissible by a range of tsetse species and the life cycle is similar to that of T. brucei with proliferation of procyclics in the midgut and transmission of metacyclics in the saliva; for T. congolense, production of metacyclics occurs in the proboscis [58, 64]. The tsetse developmental stages of T. congolense are more amenable to in vitro culture than those of T. brucei and methods for the in vitro production of attached epimastigotes and metacyclics pioneered by Gray and colleagues [65] have allowed the analysis of epimastigote and metacyclic as well as procyclic and bloodstream form populations. In this way epimastigote and metacyclic populations of sufficient purity for EST analysis were obtained from IL3000 [66] and recently the complete life cycle of IL3000 has been reproduced in vitro[63]. The ability to culture epimastigotes of IL3000 led to the characterisation of an epimastigote-specific, GPI-anchored surface glycoprotein called CESP [67], adding to the list of surface molecules specific to tsetse developmental forms of T. congolense, including GARP (glutamic acid/alanine rich protein) [68, 69], PRS (protease-resistant surface molecule) [70] and T. congolense procyclin [71]. It is possible to transfect T. congolense procyclics fairly easily by electroporation [63, 72, 73], but much more difficult to transfect bloodstream forms [63].
Relationship to other T. congolense strains
Three genetically distinct subgroups are currently recognized within T. congolense: savannah, forest and Kenya Coast or kilifi [74]. The most abundant and widespread is T. congolense savannah and IL3000 represents this subgroup. The savannah and forest subgroups were originally distinguished by different isoenzyme patterns [75, 76], and subsequently by unique repetitive DNA sequences which provide targets for species-specific PCR identification [77]. A further genetically distinct subgroup was isolated from livestock on the Kenya Coast [78] and appears to have a restricted distribution in East Africa. Although phylogenetic analysis places these three T. congolense subgroups in a single clade [79, 80], they are arguably sufficiently genetically divergent to warrant recognition as separate species [81].
Conclusions
This short review has brought together background information on the origins and biological characterisation of the four African tsetse-transmitted genome strains as a source of reference for future users of the genome data.
References
Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B: The genome of the African trypanosome Trypanosoma brucei. Science. 2005, 309: 416-422. 10.1126/science.1112642.
Jackson AP, Sanders M, Berry A, McQuillan J, Aslett MA, Quail MA, Chukualim B, Capewell P, MacLeod A, Melville SE: The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human African trypanosomiasis. PLoS Negl Trop Dis. 2010, 4 (4): e658-10.1371/journal.pntd.0000658.
Jackson AP, Berry A, Aslett M, Allison HC, Burton P, Vavrova-Anderson J, Brown R, Browne H, Corton N, Hauser H: Antigenic diversity is generated by distinct evolutionary mechanisms in African trypanosome species. PNAS. 2012, 109: 3416-3421. 10.1073/pnas.1117313109.
Steverding D: The history of African trypanosomiasis. Parasites and Vectors. 2008, 1: 3-10.1186/1756-3305-1-3.
Turner CMR, Sternberg J, Buchanan N, Smith E, Hide G, Tait A: Evidence that the mechanism of gene exchange in Trypanosoma brucei involves meiosis and syngamy. Parasitology. 1990, 101: 377-386. 10.1017/S0031182000060571.
van Deursen FJ, Shahi SK, Turner CMR, Hartmann C, Guerra-Giraldez C, Matthews KR, Clayton CE: Characterisation of the growth and differentiation in vivo and in vitro of bloodstream form Trypanosoma brucei strain TREU 927. Mol Biochem Parasitol. 2001, 112: 163-171. 10.1016/S0166-6851(00)00359-5.
Goedbloed E, Ligthart G, Minter D: A comparison of Trypanosoma brucei subgroup isolates from Glossina pallidipes in two areas of Kenya. Trans Roy Soc Trop Med Hyg. 1971, 65: 261-263. 10.1016/0035-9203(71)90272-0.
Turner CMR, McLellan S, Lindergard LAG, Bisoni L, Tait A, MacLeod A: Human infectivity trait in Trypanosoma brucei: stability, heritability and relationship to sra expression. Parasitology. 2004, 129: 445-454. 10.1017/S0031182004005906.
Radwanska M, Chamekh M, Vanhamme L, Claes F, Magez S, Magnus E, De Baetselier P, Buscher P, Pays E: The serum resistance-associated gene as a diagnostic tool for the detection of Trypanosoma brucei rhodesiense. Am J Trop Med Hyg. 2002, 67: 684-690.
Dar FK, Wilson AJ, Goedbloed E, Ligthart GS, Minter D: Serological studies on trypanosomiasis in East Africa. I: introduction and techniques. Ann Trop Med Parasit. 1973, 67: 21-29.
Goedbloed E, Ligthart GS, Minter DM, Wilson AJ, Dar FK, Paris J: Serological studies on trypanosomiasis in East Africa. II: Comparisons of antigenic types of Trypanosoma brucei subgroup organisms isolated from wild tsetse flies. Ann Trop Med Parasit. 1973, 67: 31-43.
Gibson WC, Marshall TFdC, Godfrey DG: Numerical analysis of enzyme polymorphism: a new approach to the epidemiology and taxonomy of trypanosomes of the subgenus Trypanozoon. Adv Parasitol. 1980, 18: 175-246.
Godfrey DG, Baker RD, Rickman LR, Mehlitz D: The distribution, relationships and identification of enzymic variants within the subgenus Trypanozoon. Adv Parasitol. 1990, 29: 1-74.
Mathieu-Daude F, Tibayrenc M: Isozyme variability of Trypanosoma brucei s.l.: genetic, taxonomic, and epidemiological significance. Expt Para. 1994, 78: 1-19. 10.1006/expr.1994.1001.
Melville SE, Leech V, Gerrard CS, Tait A, Blackwell JM: The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Mol Biochem Parasitol. 1998, 94: 155-173. 10.1016/S0166-6851(98)00054-1.
Hide G, Cattand P, Le Ray D, Barry DJ, Tait A: The identification of Trypanosoma brucei subspecies using repetitive DNA sequences. Mol Biochem Parasitol. 1990, 39: 213-226. 10.1016/0166-6851(90)90060-Y.
Hide G, Buchanan N, Welburn S, Maudlin I, Barry JD, Tait A: Trypanosoma brucei rhodesiense: characterisation of stocks from Zambia, Kenya, and Uganda using repetitive DNA probes. Expt Para. 1991, 72: 430-439. 10.1016/0014-4894(91)90089-F.
MacLeod A, Turner CMR, Tait A: A high level of mixed Trypanosoma brucei infections in tsetse flies detected by three hypervariable minisatellites. Mol Biochem Parasitol. 1999, 102: 237-248. 10.1016/S0166-6851(99)00101-2.
MacLeod A, Turner CMR, Tait A: The detection of geographical substructuring of Trypanosoma brucei populations by the analysis of minisatellite polymorphisms. Parasitology. 2001, 123: 475-482.
Balmer O, Beadell JS, Gibson W, Caccone A: Phylogeography and taxonomy of Trypanosoma brucei. PLoS NTD. 2011, 5: e961-
Gibson WC, Fase-Fowler F, Borst P: Further analysis of intraspecific variation in Trypanosoma brucei using restriction site polymorphisms in the maxi-circle of kinetoplast DNA. Mol Biochem Parasitol. 1985, 15: 21-36. 10.1016/0166-6851(85)90026-X.
Gibson WC: Will the real Trypanosoma brucei gambiense please stand up?. Parasitol Today. 1986, 2: 255-257. 10.1016/0169-4758(86)90011-6.
Richner D, Brun R, Jenni L: Production of metacyclic forms by cyclical transmission of West African Trypanosoma (Trypanozoon) brucei isolates from man and animals. Acta Trop. 1988, 45: 309-319.
Mehlitz D, Zillmann U, Scott CM, Godfrey DG: Epidemiological studies on the animal reservoir of gambiense sleeping sickness. III. Characterisation of Trypanozoon stocks by isoenzymes and sensitivity to human serum. Tropenmed Parasit. 1982, 33: 113-118.
Biteau N, Bringaud F, Gibson W, Truc P, Baltz T: Characterization of Trypanozoon isolates using a repeated coding sequence and microsatellite markers. Mol Biochem Parasitol. 2000, 105: 185-201.
Dukes P, Kaukus A, Hudson KM, Asonganyi T, Gashumba JK: A new method for isolating Trypanosoma brucei gambiense from sleeping sickness patients. Trans Roy Soc Trop Med Hyg. 1989, 83: 636-639. 10.1016/0035-9203(89)90379-9.
Stevens JR, Lanham SM, Allingham R, Gashumba JK: A simplified method for identifying subspecies and strain groups in Trypanozoon by isoenzymes. Ann Trop Med Parasit. 1992, 86: 9-28.
Bromidge T, Gibson W, Hudson KM, Dukes P: Identification of Trypanosoma brucei gambiense by PCR amplification of variant surface glycoprotein genes. Acta Trop. 1993, 53: 107-119. 10.1016/0001-706X(93)90023-5.
Gibson W, Nemetschke L, Ndung'u J: Conserved sequence of the TgsGP gene in Group 1 Trypanosoma brucei gambiense. Infect Genet Evol. 2010, 10: 453-458. 10.1016/j.meegid.2010.03.005.
Felu C, Pasture J, Pays E, Perez-Morga D: Diagnostic potential of a conserved genomic rearrangement in the Trypanosoma brucei gambiense-specific TGSGP locus. Am J Trop Med Hyg. 2007, 76: 922-929.
Dukes P, Gibson WC, Gashumba JK, Hudson KM, Bromidge TJ, Kaukus A, Assonganyi T, Magnus E: Absence of the LiTat 1.3 (CATT antigen) gene in Trypanosoma brucei gambiense stocks from Cameroon. Acta Trop. 1992, 51: 123-134. 10.1016/0001-706X(92)90054-2.
Kanmogne GD, Stevens JR, Asonganyi T, Gibson WC: Characterisation of Trypanosoma brucei gambiense isolates using restriction fragment length polymorphisms in 5 variant surface glycoprotein genes. Acta Trop. 1996, 61: 239-254. 10.1016/0001-706X(96)00006-X.
Dero B, Zampetti-Bosseler F, Pays E, Steinert M: The genome and the antigen gene repertoire of Trypanosoma brucei gambiense are smaller than those of T. b. brucei. Mol Biochem Parasitol. 1987, 26: 247-256. 10.1016/0166-6851(87)90077-6.
Kanmogne GD, Bailey M, Gibson W: Wide variation in DNA content among isolates of Trypanosoma brucei ssp. Acta Trop. 1997, 63: 75-87. 10.1016/S0001-706X(96)00600-6.
Gathuo HKW, Nantulya VM, Gardiner PR: Trypanosoma vivax: adaptation of two East African stocks to laboratory rodents. J Protozoo. 1987, 34: 48-53.
Leeflang P, Buys J, Blotkamp C: Studies on Trypanosoma vivax - Infectivity and serial maintenance of natural bovine isolates in mice. Int J Parasitol. 1976, 6: 413-417. 10.1016/0020-7519(76)90027-8.
Barry JD: Antigenic variation during Trypanosoma vivax infections of different host species. Parasitology. 1986, 92: 51-65. 10.1017/S0031182000063447.
Gardiner PR, Pearson TW, Clarke MW, Mutharia LM: Identification and isolation of a variant surface glycoprotein from Trypanosoma vivax. Science. 1987, 235: 774-777. 10.1126/science.3810164.
De Gee ALW, Ige K, Leeflang P: Studies on Trypanosoma vivax: Transmission of mouse infective T. vivax by tsetse flies. Int J Parasitol. 1976, 6: 419-421. 10.1016/0020-7519(76)90028-X.
Moloo SK, Sabwa CL, Kabata JM: Vector competence of Glossina pallidipes and Glossina morsitans centralis for Trypanosoma vivax, Trypanosoma congolense and T. b. brucei. Acta Trop. 1992, 51: 271-280. 10.1016/0001-706X(92)90045-Y.
Moloo SK, Kutuza SB, Desai J: Comparative study on the infection rates of different Glossina species for East African and West African Trypanosoma vivax stocks. Parasitology. 1987, 95: 537-542. 10.1017/S0031182000057966.
Jefferies D, Helfrich MP, Molyneux DH: Cibarial infections of Trypanosoma vivax and T. congolense in Glossina. Parasitol Res. 1987, 73: 289-292. 10.1007/BF00531079.
Zweygarth E, Gray MA, Kaminsky R: Axenic in vitro cultivation of Trypanosoma vivax trypomastigote forms. Trop Med Parasitol. 1991, 42: 45-48.
Brun R, Moloo SK: In vitro cultivation of animal infective forms of a West African Trypanosoma vivax stock. Acta Trop. 1982, 39: 135-141.
Hirumi H, Hirumi K, Moloo SK, Shaw MK: In vitro cultivation of bloodstream trypomastigotes of Trypanosoma vivax without feeder layer cells. J Protozoo Res. 1991, 1: 1-12.
Hirumi H, Nelson RT, Hirumi K: Complete cyclic development of Trypanosoma vivax in vitro. J Protozoo. 1983, 30: A6-
Gumm ID: The axenic cultivation of insect forms of Trypanosoma (Duttonella) vivax and development to the infective metacyclic stage. J Protozoo. 1991, 38: 163-171.
Stiles JK, Wallbanks KR, Molyneux DH: Metacyclogenesis of Trypanosoma vivax in vitro - attachment to chitosan gel. Ann Trop Med Parasit. 1990, 84: 197-200.
D'Archivio S, Medina M, Cosson A, Chamond N, Rotureau B, Minoprio P, Goyard S: Genetic engineering of Trypanosoma (Duttonella) vivax and in vitro differentiation under axenic conditions. PLoS NTD. 2011, 5: e1461-
Stephen LE: Trypanosomiasis, a veterinary perspective. 1986, Oxford: Pergamon Press
Gardiner PR, Assoku RKG, Whitelaw DD, Murray M: Hemorrhagic lesions resulting from Trypanosoma vivax infection in Ayrshire cattle. Vet Parasitol. 1989, 31: 187-197. 10.1016/0304-4017(89)90069-1.
Magona JW, Walubengo J, Odimin JT: Acute haemorrhagic syndrome of bovine trypanosomosis in Uganda. Acta Trop. 2008, 107: 186-191. 10.1016/j.actatropica.2008.05.019.
Dickin SK, Gibson WC: Hybridisation with a repetitive DNA probe reveals the presence of small chromosomes in Trypanosoma vivax. Mol Biochem Parasitol. 1989, 33: 135-142. 10.1016/0166-6851(89)90027-3.
Masake RA, Majiwa PAO, Moloo SK, Makau JM, Njuguna JT, Maina M, Kabata J, OleMoiYoi OK, Nantulya VM: Sensitive and specific detection of Trypanosoma vivax using the polymerase chain reaction. Expt Para. 1997, 85: 193-205. 10.1006/expr.1996.4124.
Morlais I, Ravel S, Grebaut P, Dumas V, Cuny G: New molecular marker for Trypanosoma (Duttonella) vivax identification. Acta Trop. 2001, 80: 207-213. 10.1016/S0001-706X(01)00160-7.
Adams ER, Hamilton PB, Rodrigues AC, Malele II, Delespaux V, Teixeira MMG, Gibson WC: New Trypanosoma (Duttonella) vivax genotypes from tsetse flies in East Africa. Parasitology. 2010, 137: 641-650. 10.1017/S0031182009991508.
Cortez AP, Ventura RM, Rodrigues AC, Batista JS, Paiva F, Anez N, Machado RZ, Gibson WC, Teixeira MMG: The taxonomic and phylogenetic relationships of Trypanosoma vivax from South America and Africa. Parasitology. 2006, 133: 159-169. 10.1017/S0031182006000254.
Hoare CA: The Trypanosomes of Mammals. 1972, Oxford: Blackwell Scientific Publications
Wellde B, Lotzsch R, Deindl G, Sadun E, Williams J, Warui G: Trypanosoma congolense. I. Clinical observations of experimentally infected cattle. Expt Para. 1974, 36: 6-19. 10.1016/0014-4894(74)90107-6.
Hirumi H, Hirumi K: In vitro cultivation of Trypanosoma congolense bloodstream forms in the absence of feeder cell layers. Parasitology. 1991, 102: 225-236. 10.1017/S0031182000062533.
Majiwa PAO, Matthyssens G, Williams RO, Hamers R: Cloning and analysis of Trypanosoma (Nannomonas) congolense ILNAT 2.1 VSG gene. Mol Biochem Parasitol. 1985, 6: 97-108.
Eshita Y, Urakawa T, Hirumi H, Fish WR, Majiwa PA: Metacyclic form-specific variable surface glycoprotein-encoding genes of Trypanosoma (Nannomonas) congolense. Gene. 1992, 113: 139-148. 10.1016/0378-1119(92)90389-7.
Coustou V, Guegan F, Plazolles N, Baltz T: Complete in vitro life cycle of Trypanosoma congolense: Development of genetic tools. PLoS Negl Trop Dis. 2010, 4 (4): e618-
Peacock L, Cook S, Ferris V, Bailey M, Gibson W: The life cycle of Trypanosoma (Nannomonas) congolense in the tsetse fly. Parasites and Vectors. 2012,
Gray MA, Ross CA, Taylor AM, Luckins AG: In vitro cultivation of Trypanosoma congolense: the production of infective metacyclic trypanosomes in cultures initiated from cloned stocks. Acta Trop. 1984, 41: 343-353.
Helm JR, Hertz-Fowler C, Aslett M, Berriman M, Sanders M, Quail MA, Soares MB, Bonaldo MF, Sakurai T, Inoue N, Donelson JE: Analysis of expressed sequence tags from the four main developmental stages of Trypanosoma congolense. Mol Biochem Parasitol. 2009, 168: 34-42. 10.1016/j.molbiopara.2009.06.004.
Sakurai T, Sugimoto C, Inoue N: Identification and molecular characterization of a novel stage-specific surface protein of Trypanosoma congolense epimastigotes. Mol Biochem Parasitol. 2008, 161: 1-11. 10.1016/j.molbiopara.2008.05.003.
Bayne RAL, Kilbride EA, Lainson A, Tetley L, Barry D: A major surface antigen of procyclic stage Trypanosoma congolense. Mol Biochem Parasitol. 1993, 61: 295-310. 10.1016/0166-6851(93)90075-9.
Beecroft RP, Roditi I, Pearson TW: Identification and characterization of an acidic major surface glycoprotein from procyclic stage Trypanosoma congolense. Mol Biochem Parasitol. 1993, 61: 285-294. 10.1016/0166-6851(93)90074-8.
Butikofer P, Vassella E, Boschung M, Renggli CK, Brun R, Pearson TW, Roditi I: Glycosylphosphatidylinositol-anchored surface molecules of Trypanosoma congolense insect forms are developmentally regulated in the tsetse fly. Mol Biochem Parasitol. 2002, 119: 7-16. 10.1016/S0166-6851(01)00382-6.
Utz S, Roditi I, Renggli CK, Almeida IC, Acosta-Serrano A, Butikofer P: Trypanosoma congolense procyclins: Unmasking cryptic major surface glycoproteins in procyclic forms. Euk Cell. 2006, 5: 1430-1440. 10.1128/EC.00067-06.
Downey N, Donelson JE: Expression of foreign proteins in Trypanosoma congolense. Mol Biochem Parasitol. 1999, 104: 39-53. 10.1016/S0166-6851(99)00136-X.
Downey N, Donelson J: Search for promotors for the GARP and rRNA genes of Trypanosoma congolense. Mol Biochem Parasitol. 1999, 104: 25-38. 10.1016/S0166-6851(99)00135-8.
Gibson W: Species-specific probes for the identification of the African tsetse-transmitted trypanosomes. Parasitology. 2009, 136: 1501-1507. 10.1017/S0031182009006179.
Young CJ, Godfrey DG: Enzyme polymorphism and the distribution of Trypanosoma congolense isolates. Ann Trop Med Parasit. 1983, 77: 467-481.
Gashumba JK, Baker RD, Godfrey DG: Trypanosoma congolense: the distribution of enzymic variants in East and West Africa. Parasitology. 1988, 96: 475-486. 10.1017/S0031182000080112.
Masiga DK, Smyth AJ, Hayes PJ, Bromidge TJ, Gibson WC: Sensitive detection of trypanosomes in tsetse flies by DNA amplification. Int J Parasitol. 1992, 22: 909-918. 10.1016/0020-7519(92)90047-O.
Majiwa PAO, Masake RA, Nantulya VN, Hamers R, Matthyssons G: Trypanosoma congolense: identification of two karyotypic groups. EMBO J. 1985, 4: 3307-3313.
Stevens JR, Noyes H, Dover GA, Gibson WC: The ancient and divergent origins of the human pathogenic trypanosomes, Trypanosoma brucei and T. cruzi. Parasitology. 1999, 118: 107-116. 10.1017/S0031182098003473.
Hamilton PB, Stevens JR, Gaunt MW, Gidley J, Gibson WC: Trypanosomes are monophyletic: evidence from genes for glyceraldehyde phosphate dehydrogenase and small subunit ribosomal RNA. Int J Parasitol. 2004, 34: 1393-1404. 10.1016/j.ijpara.2004.08.011.
Gibson W: Resolution of the species problem in African trypanosomes. Int J Parasitol. 2007, 37: 829-838. 10.1016/j.ijpara.2007.03.002.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author declares that she has no competing interests.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Gibson, W. The origins of the trypanosome genome strains Trypanosoma brucei brucei TREU 927, T. b. gambiense DAL 972, T. vivax Y486 and T. congolense IL3000. Parasites Vectors 5, 71 (2012). https://doi.org/10.1186/1756-3305-5-71
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-3305-5-71