Abstract
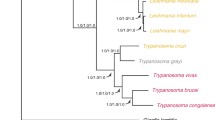
Trypanosoma evansi is the causative agent of the animal trypanosomiasis surra, a disease with serious economic burden worldwide. The availability of the genome of its closely related parasite Trypanosoma brucei allows us to compare their genetic and evolutionarily shared and distinct biological features. The complete genomic sequence of the T. evansi YNB strain was obtained using a combination of genomic and transcriptomic sequencing, de novo assembly, and bioinformatic analysis. The genome size of the T. evansi YNB strain was 35.2 Mb, showing 96.59% similarity in sequence and 88.97% in scaffold alignment with T. brucei. A total of 8,617 protein-coding genes, accounting for 31% of the genome, were predicted. Approximately 1,641 alternative splicing events of 820 genes were identified, with a majority mediated by intron retention, which represented a major difference in post-transcriptional regulation between T. evansi and T. brucei. Disparities in gene copy number of the variant surface glycoprotein, expression site-associated genes, microRNAs, and RNA-binding protein were clearly observed between the two parasites. The results revealed the genomic determinants of T. evansi, which encoded specific biological characteristics that distinguished them from other related trypanosome species.
Similar content being viewed by others
References
Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J Mol Biol 215, 403–410.
Altschul, S., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402.
Archer, S.K., Luu, V.D., de Queiroz, R.A., Brems, S., and Clayton, C. (2009). Trypanosoma brucei PUF9 regulates mRNAs for proteins involved in replicative processes over the cell cycle. PLoS Pathog 5, e1000565.
Berriman, M., Ghedin, E., Hertz-Fowler, C., Blandin, G., Renauld, H., Bartholomeu, D.C., Lennard, N.J., Caler, E., Hamlin, N.E., Haas, B., et al. (2005). The genome of the African trypanosome Trypanosoma brucei. Science 309, 416–422.
Birhanu, H., Fikru, R., Said, M., Kidane, W., Gebrehiwot, T., Hagos, A., Alemu, T., Dawit, T., Berkvens, D., Goddeeris, B.M., et al. (2015). Epidemiology of Trypanosoma evansi and Trypanosoma vivax in domestic animals from selected districts of Tigray and Afar regions, Northern Ethiopia. Parasit Vectors 8, 212.
Birney, E., Clamp, M., and Durbin, R. (2004). GeneWise and Genomewise. Genome Res 14, 988–995.
Blencowe, B.J. (2006). Alternative splicing: new insights from global analyses. Cell 126, 37–47.
Borst, P. (2002). Antigenic variation and allelic exclusion. Cell 109, 5–8.
Brenndörfer, M., and Boshart, M. (2010). Selection of reference genes for mRNA quantification in Trypanosoma brucei. Mol Biochem Parasitol 172, 52–55.
Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J Mol Biol 268, 78–94.
Carnes, J., Anupama, A., Balmer, O., Jackson, A., Lewis, M., Brown, R., Cestari, I., Desquesnes, M., Gendrin, C., Hertz-Fowler, C., et al. (2015). Genome and phylogenetic analyses of Trypanosoma evansi reveal extensive similarity to T. brucei and multiple independent origins for dyskinetoplasty. PLoS Negl Trop Dis 9, e3404.
Cech, T.R., and Steitz, J.A. (2014). The noncoding RNA revolution—trashing old rules to forge new ones. Cell 157, 77–94.
Conesa, A., Götz, S., García-Gómez, J.M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676.
Desquesnes, M., Kamyingkird, K., Pruvot, M., Kengradomkij, C., Bossard, G., Sarataphan, N., and Jittapalapong, S. (2009). Antibody-ELISA for Trypanosoma evansi: application in a serological survey of dairy cattle, Thailand, and validation of a locally produced antigen. Prevent Vet Med 90, 233–241.
El-Sayed, N.M., Myler, P.J., Bartholomeu, D.C., Nilsson, D., Aggarwal, G., Tran, A.N., Ghedin, E., Worthey, E.A., Delcher, A.L., Blandin, G., et al. (2005). The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309, 409–415.
Fadda, A., Ryten, M., Droll, D., Rojas, F., Färber, V., Haanstra, J.R., Merce, C., Bakker, B.M., Matthews, K., and Clayton, C. (2014). Transcriptome-wide analysis of trypanosome mRNA decay reveals complex degradation kinetics and suggests a role for co-transcriptional degradation in determining mRNA levels. Mol Microbiol 94, 307–326.
Gazestani, V.H., Lu, Z., and Salavati, R. (2014). Deciphering RNA regulatory elements in trypanosomatids: one piece at a time or genomewide? Trends Parasitol 30, 234–240.
Glover, L., Hutchinson, S., Alsford, S., McCulloch, R., Field, M.C., and Horn, D. (2013). Antigenic variation in African trypanosomes: the importance of chromosomal and nuclear context in VSG expression control. Cell Microbiol 15, 1984–1993.
Grab, D.J., Webster, P., Ito, S., Fish, W.R., Verjee, Y., and Lonsdale-Eccles, J.D. (1987). Subcellular localization of a variable surface glycoprotein phosphatidylinositol-specific phospholipase-C in African trypanosomes. J Cell Biol 105, 737–746.
Hahn, S., and Young, E.T. (2011). Transcriptional regulation in Saccharomyces cerevisiae: Transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189, 705–736.
Iseli, C., Jongeneel, C.V. and Bucher, P. (1999). ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol 1999, 138–148.
Ivens, A.C., Peacock, C.S., Worthey, E.A., Murphy, L., Aggarwal, G., Berriman, M., Sisk, E., Rajandream, M.A., Adlem, E., Aert, R., et al. (2005). The genome of the kinetoplastid parasite, Leishmania major. Science 309, 436–442.
Jensen, R.E., Simpson, L., and Englund, P.T. (2008). What happens when Trypanosoma brucei leaves Africa. Trends Parasitol 24, 428–431.
Jurka, J., Kapitonov, V.V., Pavlicek, A., Klonowski, P., Kohany, O., and Walichiewicz, J. (2005). Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110, 462–467.
Kamena, F., Tamborrini, M., Liu, X., Kwon, Y.U., Thompson, F., Pluschke, G., and Seeberger, P.H. (2008). Synthetic GPI array to study antitoxic malaria response. Nat Chem Biol 4, 238–240.
Kim, D., Langmead, B., and Salzberg, S.L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360.
Lanham, S.M., and Godfrey, D.G. (1970). Isolation of salivarian trypanosomes from man and other mammals using DEAE-cellulose. Exp Parasitol 28, 521–534.
Li, B. (2015). DNA double-strand breaks and telomeres play important roles in trypanosoma brucei antigenic variation. Eukaryot Cell 14, 196–205.
Liu, S., Zhou, X., Hao, L., Piao, X., Hou, N., and Chen, Q. (2017). Genome-wide transcriptome analysis reveals extensive alternative splicing events in the protoscoleces of Echinococcus granulosus and Echinococcus multilocularis. Front Microbiol 8, 929.
Lunde, B.M., Moore, C., and Varani, G. (2007). RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 8, 479–490.
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., He, G., Chen, Y., Pan, Q., Liu, Y., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1, 18.
Mallick, B., Ghosh, Z., and Chakrabarti, J. (2008). MicroRNA switches in Trypanosoma brucei. Biochem BioPhys Res Commun 372, 459–463.
Manna, P.T., Boehm, C., Leung, K.F., Natesan, S.K., and Field, M.C. (2014). Life and times: synthesis, trafficking, and evolution of VSG. Trends Parasitol 30, 251–258.
McCulloch, R., and Field, M.C. (2015). Quantitative sequencing confirms VSG diversity as central to immune evasion by Trypanosoma brucei. Trends Parasitol 31, 346–349.
Mcdowell, M.A., Ransom, D.M., and Bangs, J.D. (1998). Glycosylphosphatidylinositol-dependent secretory transport in Trypanosoma brucei. Biochem J 335, 681–689.
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., Garimella, K., Altshuler, D., Gabriel, S., Daly, M., et al. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303.
Mestdagh, P., Van Vlierberghe, P., De Weer, A., Muth, D., Westermann, F., Speleman, F., and Vandesompele, J. (2009). A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol 10, R64.
Mistry, J., Finn, R.D., Eddy, S.R., Bateman, A., and Punta, M. (2013). Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res 41, e121.
Mugnier, M.R., Cross, G.A.M., and Papavasiliou, F.N. (2015). The in vivo dynamics of antigenic variation in Trypanosoma brucei. Science 347, 1470–1473.
Nagamune, K., Nozaki, T., Maeda, Y., Ohishi, K., Fukuma, T., Hara, T., Schwarz, R.T., Sutterlin, C., Brun, R., Riezman, H., et al. (2000). Critical roles of glycosylphosphatidylinositol for Trypanosoma brucei. Proc Natl Acad Sci USA 97, 10336–10341.
Nilsson, D., Gunasekera, K., Mani, J., Osteras, M., Farinelli, L., Baerlocher, L., Roditi, I., and Ochsenreiter, T. (2010). Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog 6, e1001037.
Pertea, M., Pertea, G.M., Antonescu, C.M., Chang, T.C., Mendell, J.T., and Salzberg, S.L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33, 290–295.
Pinger, J., Chowdhury, S., and Papavasiliou, F.N. (2017). Variant surface glycoprotein density defines an immune evasion threshold for African trypanosomes undergoing antigenic variation. Nat Commun 8, 828.
Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., and Lopez, R. (2005). InterProScan: protein domains identifier. Nucleic Acids Res 33, W116–W120.
Rettig, J., Wang, Y., Schneider, A., and Ochsenreiter, T. (2012). Dual targeting of isoleucyl-tRNA synthetase in Trypanosoma brucei is mediated through alternative trans-splicing. Nucleic Acids Res 40, 1299–1306.
Richardson, J.B., Lee, K.Y., Mireji, P., Enyaru, J., Sistrom, M., Aksoy, S., Zhao, H., and Caccone, A. (2017). Genomic analyses of African Trypanozoon strains to assess evolutionary relationships and identify markers for strain identification. PLoS Negl Trop Dis 11, e0005949.
Siegel, T.N., Hekstra, D.R., Wang, X., Dewell, S., and Cross, G.A.M. (2010). Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res 38, 4946–4957.
Spitz, F., and Furlong, E.E.M. (2012). Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 13, 613–626.
Stanke, M., Keller, O., Gunduz, I., Hayes, A., Waack, S., and Morgenstern, B. (2006). AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res 34, W435–W439.
Stijlemans, B., Baral, T.N., Guilliams, M., Brys, L., Korf, J., Drennan, M., Van Den Abbeele, J., De Baetselier, P., and Magez, S. (2007). A glycosylphosphatidylinositol-based treatment alleviates trypanosomiasis-associated immunopathology. J Immunol 179, 4003–4014.
Tarailo-Graovac, M., and Chen, N. (2009). Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics Chapter 4: Unit 4.10.
Taylor, D.R. and Hooper, N.M. (2011). GPI-Anchored Proteins in Health and Disease (Springer New York).
Taylor, J.E. and Rudenko, G. (2006). Switching trypanosome coats: what’s in the wardrobe? Trends Genet 22, 614–620.
Thiel, T., Michalek, W., Varshney, R.K., and Graner, A. (2003). Exploiting EST databases for the development and characterization of genederived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106, 411–422.
Trapnell, C., Pachter, L., and Salzberg, S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111.
Yang, C., Suo, X., Huang, X., Zhang, G., Jia, Y., Wang, Q., and Shen, J. (2007). Protection of mice against homologous or heterologous infections with antiserum mixture to the predominant variable antigen type repertoire of Trypanosoma evansi YNB stock. Exp Parasitol 116, 53–58.
Xu, Z., and Wang, H. (2007). LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 35, W265–W268.
Acknowledgements
We thank Professor Lun Zhaorong from Sun Yat-sen University for providing the Trypanosoma brucei strain. We sincerely appreciate the technical assistance of the technicians at the Key Laboratory of Zoonosis, Shenyang Agriculture University. This study was supported by grants of the National Key Research and Development Program of China (2017YFD0500400 and 2017YFD0501200), the National Natural Science Foundation of China (81420108023 and 81772219), and distinguished scientist grant from Shenyang Agricultural University.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Zheng, L., Jiang, N., Sang, X. et al. In-depth analysis of the genome of Trypanosoma evansi, an etiologic agent of surra. Sci. China Life Sci. 62, 406–419 (2019). https://doi.org/10.1007/s11427-018-9473-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-018-9473-8