Abstract
Background
Mosquitoes belonging to the Albitarsis Group (Anopheles: Nyssorhynchus) are of importance as malaria vectors across the Neotropics. The Group currently comprises six known species, and recent studies have indicated further hidden biodiversity within the Group. DNA barcoding has been proposed as a highly useful tool for species recognition, although its discriminatory utility has not been verified in closely related taxa across a wide geographic distribution.
Methods
DNA barcodes (658 bp of the mtDNA Cytochrome c Oxidase - COI) were generated for 565 An. albitarsis s.l. collected in Argentina, Brazil, Colombia, Paraguay, Trinidad and Venezuela over the past twenty years, including specimens from type series and type localities. Here we test the utility of currently advocated barcoding methodologies, including the Kimura-two-parameter distance model (K2P) and Neighbor-joining analysis (NJ), for determining species delineation within mosquitoes of the Neotropical Albitarsis Group of malaria vectors (Anopheles: Nyssorhynchus), and compare results with Bayesian analysis.
Results
Species delineation through barcoding analysis and Bayesian phylogenetic analysis, fully concur. Analysis of 565 sequences (302 unique haplotypes) resolved nine NJ tree clusters, with less than 2% intra-node variation. Mean intra-specific variation (K2P) was 0.009 (range 0.002 - 0.014), whereas mean inter-specific divergence were several-fold higher at 0.041 (0.020 - 0.056), supporting the reported "barcoding gap". These results show full support for separate species status of the six known species in the Albitarsis Group (An. albitarsis s.s., An. albitarsis F, An. deaneorum, An. janconnae, An. marajoara and An. oryzalimnetes), and also support species level status for two previously detected lineages - An. albitarsis G & An. albitarsis I (designated herein). In addition, we highlight the presence of a unique mitochondrial lineage close to An. deaneorum and An. marajoara (An. albitarsis H) from Rondônia and Mato Grosso in southwestern Brazil. Further integrated studies are required to confirm the status of this lineage.
Conclusions
DNA barcoding provides a reliable means of identifying both known and undiscovered biodiversity within the closely related taxa of the Albitarsis Group. We advocate its usage in future studies to elucidate the vector competence and respective distributions of all eight species in the Albitarsis Group and the novel mitochondrial lineage (An. albitarsis H) recovered in this study.
Similar content being viewed by others
Background
The need to understand the systematics of the Neotropical Albitarsis Group (Anopheles subgenus Nyssorhynchus) [1] is primarily driven by the operational requirement to reliably distinguish which component taxa are involved in malaria transmission. The group is of great epidemiological importance as three of the five formally described species are proven regional malaria vectors in Brazil: An. deaneorum Rosa-Freitas [2–4], An. janconnae Wilkerson and Sallum (= An. albitarsis E) [5] and An. marajoara Galvão and Damasceno [6–8]. The vector status of An. oryzalimnetes Wilkerson and Motoki (= An. albitarsis B), An. albitarsis Lynch-Arribálzaga and the informally named species An. albitarsis "F" [9], are unknown. The group, along with An. braziliensis, is easily recognized in the adult stage by a pair of white scale stripes on sternum I [10]. The taxonomy of this group is historically complex and a comprehensive review is given in Motoki et al.[11].
Recent studies using COI and white gene sequences [12], in addition to the complete mitochondrion DNA [13] for five species belonging to Albitarsis Group, suggested that An. marajoara from Manaus (Brazil) should be regarded as a separate taxon in the Group (= An. albitarsis G) [12, 13]. In the past year, two further lineages have been proposed suggesting that species discovery in the Albitarsis Group is far from complete. One, closely related to An. janconnae, was detected in the Caribbean region of Colombia based on COI, white gene and second internal transcribed spaces (ITS2) sequences [14]. The other, closely related to An. deaneorum, was found in Acrelândia, Acre, Brazil, using white and NADH dehydrogenase subunit 6 (ND6) gene sequences [15]. Neither of these proposed lineages was named.
Sequence data exist for mitochondrial COI[12, 13, 16, 17], NADH dehydrogenase subunit 4 (ND4) [17], ND6[17] and whole mitochondrial genomes [13], and the nuclear white[12, 14], ITS2 [9, 14, 18] and Dominant receptor (D2) [18] regions. However, a single marker is still to be identified, which can to separate all known species and recognized lineages in the Albitarsis Group to ensure accurate species identification for studies on vector competence, for accurate distribution mapping, and to facilitate vector control efforts. Early attempts to use the ITS2 as a species diagnostic marker in the group [19] were hampered by the inadvertent inclusion of undiscovered taxa [9, 13, 14] and extensive intragenomic variation, which rendered the proposed ITS2-PCR assay unusable [20].
Mitochondrial genes are considered better markers than nuclear genes because of their abundance (1000's copies per cell), lack of introns, limited exposure to recombination, and haploid mode of inheritance [21]. DNA barcoding - which relies on the genetic variation within a standardized region of the COI gene - has been promoted as a reliable method for the identification of species in a variety of both invertebrate and vertebrate taxa [22]. Krzywinski et al.[13] sequenced whole mitochondrial genomes of six confirmed and putative taxa in the Albitarsis Group and showed that the 5' half of the COI "barcode region" is clearly more variable than its 3' half. COI barcoding studies of the mosquito fauna of Canada [23] and India [24], and within the genus Anopheles[24–27] have shown exceptional promise for species-level determinations. However, there has been no rigorous testing of the utility of the barcoding region in large populations of geographically widespread and closely related species such as those in the Albitarsis Group.
Herein we investigate the utility of the standard DNA COI barcode region for species identification using both the standard "simple" barcode methodology [22], NJ [28] and K2P model [29] to differentiate known and unknown species from 565 members of the Albitarsis Group, collected across a wide geographic range in South America in the last 20 years. For comparison, a subset of the full dataset is further investigated in a phylogenetic Bayesian framework [30].
Methods
Specimens and data access
Full specimen records (collection locality, coordinates, specimen identifiers, location of voucher specimens etc.) and all genetic data (edited chromatograms, consensus COI sequence files and corresponding GenBank numbers) are publicly accessible under the project code MBIK (Albitarsis Group Barcoding) on the BOLD website (http://www.boldsystems.org), as part of the Mosquito Barcoding Initiative (MBI). Collection data and distribution maps are also publicly available through MosquitoMap (http://www.mosquitomap.org). Specimens utilized in the molecular study were all morphologically verified as An. albitarsis s.l., using the original descriptions, and available keys [1, 31] and include topotypic material for An. albitarsis s.s., An. deaneorum, An. marajoara and An. oryzalimnetes, and type series material of An. janconnae. Voucher specimens and/or their DNA extracts for the majority of specimens used in this study are stored at -80°C in the archive collections of Walter Reed Biosystematics Unit, Smithsonian Institution, Museum Support Center, Suitland, Maryland, USA, or in the Culicid DNA Collection of the Molecular Systematics Laboratory, Natural History Museum, London.
Sequence generation
DNA barcodes (658 bp, excluding primers) were generated for 565 specimens collected across South America using the published high-throughput DNA extraction and PCR amplification protocols [27] using the LCO1490 & HCO2198 primers of Folmer et al.[32]. Sequencing reactions were carried out in both directions with the Big Dye® Terminator Kit on an ABI 3730 automated sequencer (PE Applied Biosystems). Sequences were edited in Sequencher™ v4.8 (Genes Codes Corporation, Ann Arbor, MI), and translated to amino acids in MacClade v.4.06 [33]. Similarities with publicly available sequences were assessed using BLAST (Basic Local Alignment Search Tool), available at http://blast.ncbi.nlm.nih.gov/Blast.cgi, and comparisons with unpublished barcode records checked through the IDS (Identification System) of the Barcode of Life database (BOLD, available at http://www.barcodinglife.org).
Data analysis
To test the resolution of customarily advocated barcoding methodologies [22], 302 unique haplotypes (n = 565) COI barcodes were first imported into PAUP* v.4.0 [34]. Following construction of a pairwise distance matrix using K2P [29], a bootstrapped [35] NJ tree [28] was generated using 1,000 replicates to produce an unrooted consensus tree. Sequence divergence was calculated in MEGA v.4.0 [36] using the K2P model. Anopheles braziliensis Chagas [GenBank: DQ076238] and An. darlingi Root [GenBank: DQ076236] were used as outgroup taxa to the Albitarsis Group, following previously published studies [14, 16].
A Bayesian phylogenetic analysis [30] was carried out using five specimens from each distinctive COI lineage found by NJ-K2P analysis (including respective type localities where possible). These sequences were added to the publicly available COI sequences of [14, 16] (trimmed to 658 bp). Recently published COI data [12, 13] overlapped those already included in our dataset and were therefore not included in the analysis. MrBayes [30] was performed online at: http://cbsuapps.tc.cornell.edu/mrbayes.aspx. The Bayesian analysis was run for 10 million generations with two parallel searches using three heated and one cold Markov chain, with the first 5 million generations discarded as burn-in. The best-fit model HKY + I + G was selected by the AIC criterion in MrModeltest 2.3 [37]. The trees generated were edited in Figtree v1.2.1 [38].
Results
Sequence statistics
MtDNA COI sequences (658 bp) were generated for 565 An. albitarsis s.l. collected across South America in the past twenty years: Argentina (n = 38), Brazil (n = 448), Colombia (n = 21), Paraguay (n = 23), Trinidad (n = 19) and Venezuela (n = 16). Among the 565 sequences were 302 unique haplotypes. Average nucleotide composition percentages for all 565 Albitarsis Group sequences were 38.5% (T), 15.8% (C), 29.4% (A) and 16.2% (G) (Table 1). Alignments were unambiguous: amino acid translations showed no stop codons, indicating that all sequences represented functional protein coding genes, not pseudogenes. The amino acid reading frame starts at the second base of the primer-edited sequences. The 302 unique nucleotide haplotypes, translated to 14 unique AA sequences (data not shown). All species in the Albitarsis Group were represented by the most common AA sequence, therefore AA derived phylogenies are not informative for species differentiation within this group.
Determination of barcode clusters
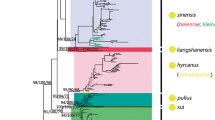
Following NJ-K2P analysis, nine distinct clusters were recovered (Figure 1), six of which corresponded to the previously determined species (An. albitarsis s.s. (n = 109), An. albitarsis F (n = 44), An. deaneorum (n = 36), An. marajoara (n = 70), An. janconnae (n = 96), and An. oryzalimnetes (n = 51), with a further three clusters corresponding to the previously described lineages An. albitarsis G (n = 105) [12, 13] and sp. nr. An. janconnae[14] (herein called An. albitarsis I) (n = 12), and a newly detected lineage designated herein as An. albitarsis H (n = 42). Anopheles albitarsis s.s., An. janconnae, An. marajoara and An. oryzalimnetes were supported by high bootstrap values (BSV) (88%, 73%, 78% and 100%, respectively), as was An. albitarsis G (BSV 96%) and the additional lineage An. albitarsis I (BSV 92%). However, An. deaneorum, An. albitarsis F and the new lineage An. albitarsis H were poorly supported with BSV's of < 70%.
Bootstrapped NJ tree, based on 1000 replicates of K2P data matrices. Bootstrapped NJ tree of 302 unique COI haplotypes generated from 565 individuals belonging to Albitarsis Group, based on K2P distance algorithm. Bootstrap values less than 70% are not shown. *Highlights taxa regarded as species in this study. **Novel lineage. Outgroup taxa include An. darlingi [GenBank: DQ076236] and An. braziliensis [GenBank: DQ076238]. The bold lines indicate the nine phylogroups considered here.
Genetic divergence was estimated based on the nine clusters determined with NJ-K2P analysis (Figure 2, Figure 3). The overall mean distance (K2P) was 0.037. The average intra-specific genetic divergence was 0.009 (0.002 - 0.014) and the average inter-specific divergence was 0.04 (0.020 to 0.056). The most genetically divergent clusters were An. marajoara with An. janconnae (0.056) and An. janconnae with An. albitarsis G (0.054); while the most similar pairs were An. albitarsis H - An. marajoara (0.020) and An. albitarsis H - An. deaneorum (0.024) (Figure 2, Figure 3). Anopheles albitarsis I, with a mean genetic divergence of 0.045 (range 0.027 - 0.053), showed consistently higher genetic divergence with respect to all other taxa in the Albitarsis Group (Figure 2, Figure 3).
Mean inter- and intra- specific of K2P distances in nine species clusters. The species clusters were defined using the NJ-K2P distance method commonly utilized in barcoding. Numbers in boldface indicate intra-specific genetic divergence. Numbers underlined indicate the lowest (An. marajoara with An. albitarsis H) and highest (An. janconnae with An. marajoara) values between the species and lineages. Specimen numbers per species are indicated in parentheses.
Plot of K2P distance matrices of the nine clusters determined using NJ-K2P distances. Y-axis: genetic divergence and X-axis clusters. mar: An. marajoara; albG: An. albitarsis G; dea: An. deaneorum; alb: An. albitarsis s.s.; albH: An. albitarsis H; ory: An. oryzalimnetes; jan: An. janconnae; albF: An. albitarsis F; albI: An. albitarsis I.
Bayesian Analysis
Bayesian analysis supported all formally described species and previously indicated lineages detected with NJ-K2P (posterior probability P = 0.8 - 1), however, the newly recognized lineage, An. albitarsis H, is unresolved (P = 0.4, not shown) (Figure 4). The monophyly of the Albitarsis Group is confirmed and four species groups are recognized as follow: Group 1, An. janconnae, An. albitarsis F and An. albitarsis I; Group 2, An. albitarsis G, An. albitarsis H, An. deaneorum and An. marajoara; Group 3, An. albitarsis s.s.; Group 4, An. oryzalimnetes. Groups 2, 3 and 4 are more closely related, and they are paraphyletic with respect to Group 1. The lineage proposed by Gutierrez et al.[14] from the Colombian Caribbean coast showed strong correlation (P = 0.99) with An. albitarsis I. Lehr et al.[16] undetermined COI sequences grouped here with An. albitarsis G and An. albitarsis H (Figure 4) (see Figure 5), [16].
MrBayes tree generated using GenBank sequences and topotypic specimens and lineages found with NJ-K2P distances. Bold lines indicate the nine phylogroups considered here: albH: An. albitarsis H; mar: An. marajoara; dea: An. deaneorum; albG: An. albitarsis G; alb: An. albitarsis s.s.; ory: An. oryzalimnetes; albI: An. albitarsis I; albF: An. albitarsis F; jan: An. janconnae. Outgroups: An. darlingi and An. braziliensis. P values less than 70% are not shown. *Newly determined species. **Novel lineage. COI sequences from previously published works are indicated in blue [16] and red [14].
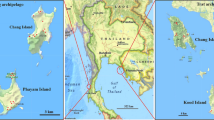
Map of South America showing the verified distributions of members of the Albitarsis Group based on COI DNA sequences. Locality data for 565 An. albitarsis s.l. specimens were plotted alongside those GenBank sequences whose identity has been molecularly verified within [14, 16]. alb: An. albitarsis s.s.; albF: An. albitarsis F; albG: An. albitarsis G; albH: An. albitarsis H; albI: An. albitarsis I; dea: An. deaneorum; jan: An. janconnae; mar: An. marajoara; ory: An. oryzalimnetes. Localities determined from GenBank COI sequences are indicated as follow: An. albitarsis F [GenBank: DQ076234] [16], fuchsia diamond with heavy blue edging, and An. albitarsis I [GenBank: GQ153597-610] [14] black circles with bright green edging.
Discussion
The analysis of COI sequences from members of the Albitarsis Group using NJ-K2P distances revealed nine distinct clusters: six of these correspond to the formerly defined species An. albitarsis s.s., An. deaneorum, An. janconnae, An. marajoara, An. oryzalimnetes and An. albitarsis F, and another to An. albitarsis G [13], which we believe to comprise a separate species in the Albitarsis Group along with "sp. nr. An. janconnae"[14], named An. albitarsis I. The remaining cluster, which we believe to represent a separate lineage, is named An. albitarsis H. Currently available synonyms for the Albitarsis Group as discussed in Motoki et al.[11] (An. albitarsis (Anopheles albitarsis var. limai Galvão and Lane, Anopheles albitarsis imperfectus Corrêa and Ramos)), An. marajoara (An. albitarsis domesticus Galvão and Damasceno) and the status An. allopha as a nomen dubium[1] are in our opinion sustained, with none referring to the newly recognized species/lineages presented here.
The monophyly of the Albitarsis Group is strongly supported with a high P value (P = 1), which echoes earlier studies [14–17]. BLAST searches of COI sequences revealed 99-100% homology with sequences reported for An. albitarsis s.s. [GenBank: DQ076204-DQ076208], An. deaneorum [GenBank: DQ076226, DQ076230], An. janconnae [GenBank: DQ076233], An. marajoara [GenBank: DQ076216] and An. oryzalimnetes [GenBank: DQ076210-DQ076215] [16]. This further reinforces our confidence in the identities of these taxa. The respective distributions and taxonomic positions of the informally designated taxa (An. albitarsis F, G, and I) and new lineage An. albitarsis H within the Albitaris Group are discussed in relation to earlier works below.
Anopheles albitarsis F
Anopheles albitarsis F was originally proposed as a putative new species in the Albitarsis Group based on ITS2 and white gene sequences [9] from specimens collected in Vichada, Colombia. One of the individuals assumed by Lehr et al.[16] to be An. albitarsis E (= An. janconnae), was unresolved in their Bayesian phylogeny (sample C14, Portuguesa, Venezuela [GenBank: DQ076234]), but this was confirmed as An. albitarsis F in our Bayesian analysis (Figure 4). Correlation of our data with these earlier studies confirms the wider distribution of the species outside of Colombia. As well as Vichada, Colombia [9] and Portuguesa, Venezuela [16], we further report it from the states of Cojedes and Zulia in Venezuela and in St George East and St Andrew/St David in Trinidad (Table 2, Figure 5).
Morphologically, An. albitarsis F is similar to the proven malaria vector An. marajoara[9], thus the wider distribution of this cryptic taxon in Venezuela and Trinidad is of epidemiological significance. Based on morphology and RAPD profiling [18], An. marajoara was believed to be the only member of the Albitarsis Group present in western Venezuela [8], where it is a proven secondary vector of Plasmodium vivax 210 [39]. However, these studies predate the discovery of An. albitarsis F and it is not known if the species-diagnostic RAPD assay (developed for the four known members of the Albitarsis Group at that time) [40] could distinguish An. marajoara from An. albitarsis F. Given the data presented herein, it seems likely that "An. marajoara" reported in Venezuela [8] corresponds to An. albitarsis F. It is also probable that "population C" from Venezuela and Colombia [41, 42] identified on the basis of chromosomal analysis, may also be An. albitarsis F. Further sampling is needed to assess the true identity of An. albitarsis s.l. in Venezuela and assess whether An. albitarsis F is the only member of the group present there, or whether An. marajoara and An. albitarsis F are sympatric in that country. This is also true for the population of An. marajoara in Trinidad, which is regarded as a secondary vector in the region [43]. An ecological study stated that An. marajoara was identified from the island based on RAPD profiles and ITS2 sequences [43], yet our data clearly show all samples tested from Trinidad to be An. albitarsis F. Concurrent species determination, distribution mapping and vector incrimination studies are needed to assess the true impact of An. marajoara and An. albitarsis F on malaria transmission across their range.
Anopheles albitarsis G
Mean COI genetic intra-specific distance within An. albitarsis G was 0.010, whereas mean inter-specific divergence within the Albitarsis Group were four-fold higher (0.041) (Figure 2, Figure 3), and well above accepted species delimitation divergence thresholds routinely employed in DNA barcoding [22–27]. Based on consistent differences in DNA sequence in one individual (C10 [GenBank: DQ076225]) from four different DNA markers [40], Wilkerson et al. first suggested the presence of hidden genetic diversity within An. marajoara from Manaus, Brazil. Bayesian analysis of the entire COI gene showed that sample C10 grouped closest with An. deaneorum, but with low support (P = 0.69) [16]. Because of this, these authors questioned the validity of An. deaneorum arguing: "if An. deaneorum is a separate species from An. marajoara, then An. marajoara may consist of two or more species in Amazonian Brazil". The presence of An. albitarsis G as a cryptic species in the Albitarsis Group was later confirmed in further DNA studies [12, 13]. BLAST of our An. albitarsis G sequences returned 100% homology with GenBank: DQ076225 (C10, from Manaus, Brazil, as An. marajoara) and GenBank: DQ076217 (C2, from Itaituba, Brazil, as An. marajoara) [16]. GenBank: DQ076221 (Itaituba, Brazil) [16] still shows deviation in the Bayesian phylogeny. However, at this time we are unable to assess whether this reflects further hidden genetic diversity, whether these differences are due to poor sequence quality in the first instance, or are indeed real and reflective of geographic distance as all other specimens sequenced are from Manaus, Brazil (Figure 4). Further sampling from Itaituba and other localities is needed.
Anopheles albitarsis H
This novel mitochondrial lineage was detected in 42 specimens collected in Rondônia and Mato Grosso, Brazil. COI sequences showed highest similarity with An. marajoara and An. deaneorum (0.020 and 0.024, respectively), both values being significantly higher than its intra-specific divergence (0.010) (Figure 2, Figure 3). BLAST of our sequences revealed 99% homology with GenBank: DQ076222 from Matupá and GenBank: DQ076223 from Peixoto de Azevedo (both Mato Grosso, Brazil) and GenBank: DQ076224 from Ariquemes, Rondônia, Brazil (as An. marajoara samples C7, C8 and C9) [16]. These three samples were shown to form a cluster distinct from An. marajoara, which is further supported in our Bayesian analysis (Figure 4).
Based on sequence variation, the barcoding community generally accept species delineation if the intraspecific variation is less than 1%, while the inter-specific variation is at least 2% different from its nearest congener [22–27, 44]. Average inter-specific divergences between known taxa in the Albitarsis Group is 0.036, however, inter-specific values between An. deanorum, An. marajoara and Anopheles albitarsis H are much lower, ranging from 0.020 - 0.025. Our COI data supports the suggestion of Lehr et al.[16], exposing this grouping as cryptic complex in its own right within the Albitarsis Group. The status of the mitochondrial lineage An. albitarsis H is far from resolved. Further detailed ecological, genetic and morphological studies are necessary before we can speculate whether this lineage is reflective of a new species within the Albitarsis Group.
The presence of a cryptic species near to An. deaneorum (using ND6 and white gene sequences [15]) and near to An. marajoara (by allozymes and mtDNA RFLPs [45]), were detected in populations of An. albitarsis s.l. from the same and neighbouring localities to where An. albitarsis H was detected in this study.
Anopheles albitarsis I
Brochero et al.[9] first reported An. albitarsis F in Vichada, Colombia (east of the Cordillera Oriental) in sympatry with another taxon determined as An. marajoara based on morphological keys [10]. Microsatellite analysis of five populations of An. marajoara in Colombia revealed two incompletely isolated gene pools separated by the eastern Andean cordillera [46]. Using the entire COI, white gene and ITS2 sequences, Gutierrez et al.[14] reported a new lineage in the Albitarsis Group closely related to An. janconnae from northwestern Colombia. Sequences generated in our study share 99% homology with these COI sequences [GenBank: GQ153597-GQ153610] [14], and this cluster was highly supported in our Bayesian analysis (P = 0.99) (Figure 5). We clearly show that the lineage previously reported by Gutierrez et al.[14] as sp. nr. An. janconnae in the Caribbean region of Colombia and by Brochero et al.[46] as An. marajoara, is a separate species in the Albitarsis Group, which we call An. albitarsis I.
Despite the small sample size analysed, this species is clearly distinct from other members of the Albitarsis Group based on COI sequence data, with an intra-specific divergence of 0.008 and mean inter-specific divergence of 0.044 (0.027 with An. albitarsis F - 0.053 with An. marajoara) (Figure 2). That the sister taxon An. albitarsis F and An. albitarsis I were detected in sympatry in the geographically separate localities of Tibú, Norte de Santander, Colombia and Rio Socuavo, Zulia, Venezuela (Figure 5), provides further evidence to support their separate species status within the Albitarsis Group.
Distribution of Albitarsis Group in South America based on COI sequences
Locality data for all molecularly confirmed Neotropical Albitarsis Group specimens used in this study (n = 565) and correlated COI data from previous studies [14, 16] were plotted using the mapping methodology developed by Foley [47] (Figure 5). This figure shows the distributions of An. albitarsis F and An. albitarsis I to be restricted to northern South America (Colombia, Venezuela and Trinidad), whereas An. albitarsis s.s. is found in southern Brazil, northern Argentina and Paraguay. Anopheles marajoara is present in the central east and west region of Brazil and An. deaneorum is in southwestern Brazil. Anopheles oryzalimnetes has a wide distribution in Brazil, mainly in the central region (Figure 5). Three species in the Albitarsis Group and the new lineage An. albitarsis H appear endemic only to Brazil to date: An. marajoara (Amapá, Mato Grosso, Pará, Rondônia), An. janconnae is present in northern Brazil (Roraima and Pará states), An. albitarsis G in Amazonian Brazil (Amazonas and Pará states) and An. albitarsis H in Rondônia and Mato Grosso, Brazil. Some species and lineages were found to be sympatric: for example, An. marajoara with An. oryzalimnetes and An. janconnae in Pará, Brazil; An. albitarsis H, An. deaneorum, An. marajaora and An. oryzalimnetes in Mato Grosso, Brazil; An. oryzalimnetes and An. albitarsis in São Paulo, Brazil, amongst others (Figure 5).
Whilst this study significantly contributes to the verified distribution of members of the Albitarsis Group, it is important to note that samples were not available from Bolivia, Costa Rica, French Guiana, Guatemala, Guyana, Panama, Peru, Suriname and Uruguay, where species of the Group have previously been reported [48]. We advocate the utility of the COI barcoding region for the correct species identification of An. albitarsis s.l. specimens in these regions, allowing complete species level distribution maps to be constructed for all eight species and the new lineage recognised in this study.
Conclusions
The results reported here using COI barcoding of specimens collected over 20 years from a wide geographic range in South America (Argentina, Brazil, Colombia, Paraguay, Trinidad and Venezuela), including topotypic specimens of the formally described taxa, helped resolve the taxonomic status and the distribution of the Albitarsis Group in South America. Contrary to intra-specific variation detected in previous studies using ITS2 [20], we consider the COI barcode region to be a robust marker of choice for species delimitation in the Albitarsis Group, not least because amplification of a short region (658 bp) yielded similar results to studies based on the entire mitochondria [13] or full COI gene [12, 14, 16]. We now consider the following species to comprise the Albitarsis Group: An. albitarsis s.s., An. deaneorum, An. janconnae, An. marajoara, An. oryzalimnetes, An. albitarsis F, An. albitarsis G and An. albitarsis I. In addition, we uncovered a new mitochondrial lineage, An. albitarsis H, which requires further sampling and sequencing of additional markers before its true species status can be resolved. This first comprehensive study of species in the Albitarsis Group across a wide geographic range in South America and provides a firm systematic basis for future studies that ideally should include ecology, biogeography, population genetics analysis, and vector incrimination of these species, particularly in malaria endemic regions where some of these new taxa may be acting as vectors.
References
Linthicum KJ: A revision of the Argyritarsis Section of the subgenus Nyssorhynchus of Anopheles (Diptera: Culicidae). Mosq Syst. 1988, 20: 98-270.
Klein TA, Lima JB, Tada MS: Comparative susceptibility of anopheline mosquitoes to Plasmodium falciparum in Rondonia, Brazil. Am J Trop Med Hyg. 1991, 44: 598-603.
Klein TA, Lima JB, Tada MS, Miller R: Comparative susceptibility of anopheline mosquitoes in Rondonia, Brazil to infection by Plasmodium vivax. Am J Trop Med Hyg. 1991, 45: 463-470.
Rubio-Palis Y, Zimmerman RH: Ecoregional classification of malaria vectors in the Neotropics. J Med Entomol. 1997, 34: 499-510.
Povoa MM, de Souza RT, Lacerda RN, Rosa ES, Galiza D, de Souza JR, Wirtz RA, Schlichting CD, Conn JE: The importance of Anopheles albitarsis E and An. darlingi in human malaria transmission in Boa Vista, state of Roraima, Brazil. Mem Inst Oswaldo Cruz. 2006, 101: 163-168.
Conn JE, Wilkerson RC, Segura MNO, De Souza RTL, Schlichting CD, Wirtz RA, Povoa MM: Emergence of a new Neotropical malaria vector facilitated by human migration and changes in land use. Am J Trop Med Hyg. 2002, 66: 18-22.
Moreno JE, Rubio-Palis Y, Paez E, Perez E, Sanchez V, Vaccari E: Anopheles (Anopheles) neomaculipalpus: a new malaria vector in the Amazon basin?. Med Vet Entomol. 2005, 19: 329-332.
Rubio-Palis Y, Wilkerson R, Guzman H: Morphological characters of adult Anopheles (Nyssorhynchus) marajoara in Venezuela. J Am Mosq Control Assoc. 2003, 19: 107-114.
Brochero HH, Li C, Wilkerson RC: A newly recognized species in the Anopheles (Nyssorhynchus) albitarsis complex (Diptera: Culicidae) from Puerto Carreno, Colombia. Am J Trop Med Hyg. 2007, 76: 1113-1117.
Faran ME, Linthicum KJ: A handbook of the Amazonian species of Anopheles (Nyssorhynchus) (Diptera: Culicidae). Mosq Syst. 1981, 13: 1-81.
Motoki MT, Wilkerson RC, Sallum MA: The Anopheles albitarsis complex with the recognition of Anopheles oryzalimnetes Wilkerson and Motoki, n. sp. and Anopheles janconnae Wilkerson and Sallum, n. sp. (Diptera: Culicidae). Mem Inst Oswaldo Cruz. 2009, 104: 823-850.
McKeon SN, Lehr MA, Wilkerson RC, Ruiz JF, Sallum MA, Lima JB, Povoa MM, Conn JE: Lineage divergence detected in the malaria vector Anopheles marajoara (Diptera: Culicidae) in Amazonian Brazil. Malar J. 2010, 9: 271-
Krzywinski J, Li C, Morris M, Conn JE, Lima JB, Povoa MM, Wilkerson RC: Analysis of the evolutionary forces shaping mitochondrial genomes of a Neotropical malaria vector complex. Mol Phylogenet Evol. 2011, 58: 469-477.
Gutierrez LA, Orrego LM, Gomez GF, Lopez A, Luckhart S, Conn JE, Correa MM: A new mtDNA COI gene lineage closely related to Anopheles janconnae of the Albitarsis complex in the Caribbean region of Colombia. Mem Inst Oswaldo Cruz. 2010, 105: 1019-1025.
Bourke BP, Foster PG, Bergo ES, Calado DC, Sallum MA: Phylogenetic relationships among species of Anopheles (Nyssorhynchus) (Diptera, Culicidae) based on nuclear and mitochondrial gene sequences. Acta Trop. 2010, 114: 88-96.
Lehr MA, Kilpatrick CW, Wilkerson RC, Conn JE: Cryptic species in the Anopheles (Nyssorhynchus) albitarsis (Diptera: Culicidae) complex: Incongruence between Random Amplified Polymorphic DNA-Polymerase Chain Reaction identification and analysis of mitochondrial DNA COI gene Sequences. Ann Entomol Soc Am. 2005, 98: 908-917.
Wilkerson RC, Foster PG, Li C, Sallum MA: Molecular phylogeny of Neotropical Anopheles (Nyssorhynchus) albitarsis species complex (Diptera: Culicidae). Ann Entomol Soc Am. 2005, 98: 918-925.
Wilkerson RC, Parsons TJ, Klein TA, Gaffigan TV, Bergo E, Consolim J: Diagnosis by random amplified polymorphic DNA polymerase chain reaction of four cryptic species related to Anopheles (Nyssorhynchus) albitarsis (Diptera: Culicidae) from Paraguay, Argentina, and Brazil. J Med Entomol. 1995, 32: 697-704.
Li C, Wilkerson RC: Identification of Anopheles (Nyssorhynchus) albitarsis complex species (Diptera: Culicidae) using rDNA internal transcribed spacer 2-based polymerase chain reaction primes. Mem Inst Oswaldo Cruz. 2005, 100: 495-500.
Li C, Wilkerson RC: Intragenomic rDNA ITS2 variation in the neotropical Anopheles (Nyssorhynchus) albitarsis complex (Diptera: Culicidae). J Hered. 2007, 98: 51-59.
Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A: Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene. 1999, 238: 195-209.
Hebert PD, Cywinska A, Ball SL, deWaard JR: Biological identifications through DNA barcodes. Proc R Soc Lond B. 2003, 270: 313-321.
Cywinska A, Hunter FF, Hebert PD: Identifying Canadian mosquito species through DNA barcodes. Med Vet Entomol. 2006, 20: 413-424.
Kumar NP, Rajavel AR, Natarajan R, Jambulingam P: DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae). J Med Entomol. 2007, 44: 1-7.
Gonzalez R, Carrejo N, Wilkerson RC, Alarcon J, Alarcon-Ormasa J, Ruiz F, Bhatia R, Loaiza J, Linton YM: Confirmation of Anopheles (Anopheles) calderoni Wilkerson, 1991 (Diptera: Culicidae) in Colombia and Ecuador through molecular and morphological correlation with topotypic material. Mem Inst Oswaldo Cruz. 2010, 105: 1001-1009.
Laboudi M, Faraj C, Sadak A, Harrat Z, Boubidi SC, Harbach RE, El Aouad R, Linton YM: DNA barcodes confirm the presence of a single member of the Anopheles maculipennis group in Morocco and Algeria: An. sicaulti is conspecific with An. labranchiae. Acta Trop. 2011, 118: 6-13.
Ruiz F, Linton YM, Ponsonby DJ, Conn JE, Herrera M, Quinones ML, Velez ID, Wilkerson RC: Molecular comparison of topotypic specimens confirms Anopheles (Nyssorhynchus) dunhami Causey (Diptera: Culicidae) in the Colombian Amazon. Mem Inst Oswaldo Cruz. 2010, 105: 899-903.
Saitou N, Nei M: The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987, 4: 406-425.
Kimura M: A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980, 16: 111-120.
Ronquist F, Huelsenbeck JP: MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003, 19: 1572-1574.
Ranulfo G, Carrejo Nacy: Introducción al estudio taxónomico de Anopheles de Colombia: claves y notas de distribución. 2009, Colombia, Programa Editorial Universidad del Valle
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R: DNA primers for amplification of mitochondrial Cytochrome c Oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994, 3: 294-299.
Maddison DR, Maddison WR: Analysis of phylogeny and character evolution. 2003, Sunderland: Sinauer Associates
Swofford DL: Phylogenetic analysis using parsimony. 2002, Sinauer Associates, Inc. Publishers, 4
Felsenstein J: Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985, 39: 783-791.
Tamura K, Dudley J, Nei M, Kumar S: MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007, 24: 1596-1599.
Nylander J: MrModeltest v2. 2004, Evolutionary Biology Center, Uppsala University: Program distributed by the author, v2
Rambaut A: FigTree. [http://tree.bio.ed.ac.uk/software/figtree]v1.1.3
Rubio-Palis Y: Variation of the vectorial capacity of some anophelines in western Venezuela. Am J Trop Med Hyg. 1994, 50: 420-424.
Wilkerson RC, Gaffigan TV, Bento Lima J: Identification of species related to Anopheles (Nyssorhynchus) albitarsis by random amplified polymorphic DNA-polymerase chain reaction (Diptera: Culicidae). Mem Inst Oswaldo Cruz. 1995, 90: 721-732.
Kitzmiller J: Chromosomal differences among species of Anopheles mosquitoes. Mosq Syst. 1977, 9: 112-122.
Kreutzer RD, Kitzmiller JB, Rabbani MG: Cytogenetically distinguishable sympatric and allopatric populations of the mosquito Anopheles albitarsis. Acta Amazon. 1976, 3: 473-482.
Chadee DD, Wilkerson RC: Ecology of the malaria vector, Anopheles (Nyssorhynchus) marajoara Galvao and Damasceno in Trinidad, West Indies. J Am Mosq Control Assoc. 2006, 22: 22-28.
Ratnasingham S, Hebert PD: BOLD: The Barcode of Life Data System http://www.barcodinglife.org. Mol Ecol Notes. 2007, 7: 355-364.
Narang SK, Klein TA, Perera OP, Lima JB, Tang AT: Genetic evidence for the existence of cryptic species in the Anopheles albitarsis complex in Brazil: allozymes and mitochondrial DNA restriction fragment length polymorphisms. Biochem Genet. 1993, 31: 97-112.
Brochero H, Li C, Wilkerson R, Conn JE, Ruiz-Garcia M: Genetic structure of Anopheles (Nyssorhynchus) marajoara (Diptera: Culicidae) in Colombia. Am J Trop Med Hyg. 2010, 83: 585-595.
Foley D: A spreadsheet mapping approach for error checking and sharing collection point data. Biodiversity Informatics. 2011, 7: 137-142.
Walter Reed Biosystematics Unit (WRBU): Online Mosquito Catalogue. [http://www.mosquitocatalog.org]
Acknowledgements
This study forms part of the larger objectives of the Mosquito Barcoding Initiative (MBI), led by the senior authors YML & RCW. This investigation received financial support from: supporters of the Natural History Museum, London to further the activities of the Mosquito Barcoding Initiative (to YML); the UNICEF/UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (TDR) (grant A50252 to YML), Consortium for the Barcode of Life (CBOL) (to YML & RCW) and The Sloane Foundation (to YML & RCW); from the National Institute of Health (NIH), USA, (grant 2R01AI054139 to JEC) and COLCIENCIAS (grant 110134319196 to MLQ). We gratefully acknowledge Dr. Marta Moreno (Griffin Laboratory, Wadsworth Center, New York State Department of Health, Albany, NY, USA), Izis M. Carvalho and Galiza Primo (Instituto Evandro Chagas, Ananindeua, PA, Brazil) for their invaluable field assistance; Andrea Ormos for molecular laboratory support (Laboratories of Analytical Biology, Smithsonian Institution, Washington DC, USA), Dr. Desmond Foley (WRBU, Smithsonian Institution, Washington DC, USA) for elaboration of the map; Dr. Shazia Mahamdallie for helpful suggestions and discussions and Dr. Helena Brochero (Universidad Nacional de Colombia, Bogotá D.C., Colombia) for donation of An. albitarsis F samples.
This research was performed under a Memorandum of Understanding between the Walter Reed Army Institute of Research and the Smithsonian Institution, with institutional support provided by both organizations. The material to be published reflects the views of the authors and should not be construed to represent those of the Department of the Army or the Department of Defense. This manuscript was prepared while YML held a National Research Council Research Associateship Award at the Walter Reed Army Institute of Research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
FRL, RCW & YML conceived the ideas; YML, RCW, MLQ & JEC obtained funding; FRL, SNM, JEC, MLQ & RCW undertook fieldwork and/or donated samples; FRL, YML & DML carried out the molecular laboratory work; FRL, YML, SNM & RCW carried out the data analysis and interpretation; FRL wrote the draft manuscript; YML, RCW, JEC, MMP, MLQ & SNM revised the draft manuscript; FRL, RCW & YML carried out the final revisions and submitted the manuscript. All authors read and approved the final version of the manuscript.
Freddy Ruiz-Lopez, Richard C Wilkerson and Yvonne-Marie Linton contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ruiz-Lopez, F., Wilkerson, R.C., Conn, J.E. et al. DNA barcoding reveals both known and novel taxa in the Albitarsis Group (Anopheles: Nyssorhynchus) of Neotropical malaria vectors. Parasites Vectors 5, 44 (2012). https://doi.org/10.1186/1756-3305-5-44
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-3305-5-44