Abstract
Background
Species of the fungal genus Trichoderma are important industrial producers of cellulases and hemicellulases, but also widely used as biocontrol agents (BCAs) in agriculture. In the latter function Trichoderma species stimulate plant growth, induce plant defense and directly antagonize plant pathogenic fungi through their mycoparasitic capabilities. The recent release of the genome sequences of four mycoparasitic Trichoderma species now forms the basis for large-scale genetic manipulations of these important BCAs. Thus far, only a limited number of dominant selection markers, including Hygromycin B resistance (hph) and the acetamidase-encoding amdS gene, have been available for transformation of Trichoderma spp. For more extensive functional genomics studies the utilization of additional dominant markers will be essential.
Results
We established the Escherichia coli neomycin phosphotransferase II-encoding nptII gene as a novel selectable marker for the transformation of Trichoderma atroviride conferring geneticin resistance. The nptII marker cassette was stably integrated into the fungal genome and transformants exhibited unaltered phenotypes compared to the wild-type. Co-transformation of T. atroviride with nptII and a constitutively activated version of the Gα subunit-encoding tga3 gene (tga3Q207L) resulted in a high number of mitotically stable, geneticin-resistant transformants. Further analyses revealed a co-transformation frequency of 68% with 15 transformants having additionally integrated tga3Q207L into their genome. Constitutive activation of the Tga3-mediated signaling pathway resulted in increased vegetative growth and an enhanced ability to antagonize plant pathogenic host fungi.
Conclusion
The neomycin phosphotransferase II-encoding nptII gene from Escherichia coli proved to be a valuable tool for conferring geneticin resistance to the filamentous fungus T. atroviride thereby contributing to an enhanced genetic tractability of these important BCAs.
Similar content being viewed by others
Background
Trichoderma species are among the most frequently isolated soil fungi. While Trichoderma reesei is an efficient producer of cellulolytic and hemicellulolytic enzymes for industrial applications, other species such as the mycoparasites Trichoderma atroviride, Trichoderma harzianum, and Trichoderma virens represent important biocontrol agents (BCAs) applied in agriculture[1]. Trichoderma- based BCAs rely on the capability of these fungi to establish themselves in the plant rhizosphere, to stimulate plant growth and to induce plant defense against pathogens in addition to their ability to antagonize plant-pathogenic fungi by mycoparasitism[1–3]. Based on the fact that more than 60% of all registered biofungicides used for plant disease control are based on Trichoderma[4], there is an increasing interest to understand the modes of action of these fungi and the underlying molecular processes in greater detail. To gain a deeper understanding on how mycoparasitism in Trichoderma is induced by living host fungi intracellular signal transduction pathways have been investigated. Studies with both T. atroviride and T. virens revealed important roles of G protein-coupled receptors and heterotrimeric G proteins[5–10]. In T. atroviride, the Tga3 Gα protein was shown to regulate mycoparasitism-relevant processes, such as the attachment to the host fungus, and the production of cell wall-degrading enzymes and antifungal secondary metabolites. According to these essential functions, tga3 gene deletion mutants were avirulent, i.e. unable to attack and lyse host fungi[10].
For the genetic manipulation of T. atroviride various transformation techniques, inlcuding biolistic, Agrobacterium-mediated, and protoplast-based methods have been established[11]. These approaches basically rely on only two dominant selection markers, the Hygromycin B resistance-conferring hph gene and the acetamidase-encoding amdS gene enabling the fungus to grow on acetamide as sole nitrogen source[12]. While hph proved to be a reliable marker for gene deletion as well as the ectopic integration of expression and silencing constructs in T. atroviride, transformation with amdS may result in background growth especially when the integrating DNA rendered transformants less viable. In order to facilitate serial gene deletions and re-transformations in this important biocontrol fungus, the establishment of additional selection markers is crucial.
The neomycin phosphotransferase II-encoding nptII gene probably is the most widely used selectable marker for the transformation of plants[13]. In recent years, nptII has been applied for the transformation of filamentous fungi revealing its usefulness in these organisms[14–18].
Here we report the establishment of the nptII gene as a dominant selectable marker for the genetic transformation of T. atroviride using geneticin (G418) resistance. Subsequently, nptII was applied in co-transformation experiments to generate mutants expressing a constitutively active version of the tga3 Gα subunit-encoding gene. The consequences of the constitutive activation of Tga3-mediated signaling on T. atroviride growth, conidiation and mycoparasitism are presented.
Results and discussion
Sensitivity of untransformed T. atroviride to geneticin
The sensitivity of T. atroviride strain P1 to geneticin was tested prior to transformation. Fungal growth from mycelial agar plug inocula placed on PDA plates supplemented with increasing amounts of geneticin were effectively impaired by concentrations of 10–60 μg/ml. Mycelia growth was completely inhibited by concentrations exceeding 60 μg/ml (Figure1A). Conidial germination was completely blocked by concentrations of ≥ 20 μg/ml (Figure1B). Based on these results, a geneticin concentration of 80 μg/ml was used for the selection of transformants.
Expression of the nptII gene confers geneticin resistance to T. atroviride
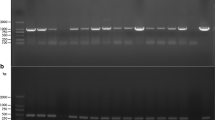
T. atroviride protoplasts were transformed with 10 μg of the circular plasmid pII99[19]. Within 3–4 days, colonies of various sizes emerged on selective medium (PDA supplemented with 80 μg/ml geneticin). 14 colonies were isolated and transferred individually on a fresh set of selection plates, on which 8 colonies survived. After purification of these geneticin-resistant transformants to mitotic stability by three rounds of single spore isolation, integration of the nptII gene was examined. PCR analysis with oligonucleotides pII99Fw and pII99Rev (Table1) resulted in the expected 888-bp amplicon in all tested transformants (Figure2). To confirm that the nptII gene product itself has no unwanted phenotypic effects, fungal growth rates, conidiation behavior and mycoparasitic abilities of the transformants were assessed in comparison to the wild-type. All tested transformants exhibited an unaltered phenotype (Table2, Figure3), demonstrating that the nptII gene is a suitable selectable marker for transformation of T. atroviride.
Screening of T. atroviridec transformants for the presence of the nptII gene. Genomic DNA was isolated from geneticin-resistant transformants and screened by PCR for the presence of the nptII gene using primers pII99Fw and pII99Rev (Table1) which yielded a specific amplicon of 888-bp. The ~600-bp band amplified from transformant 1 indicates the presence of an additional incomplete nptII gene copy. M, DNA size standard; -, wild-type; 1–8, transformants; +, plasmid pII99.
Co-transformation of T. atroviride with nptII and tga3Q207L
The tga3 gene encodes the class III adenylate cyclase-activating Gα subunit of T. atroviride[10]. To generate a constitutively activated version of Tga3, a point mutation resulting in the replacement of glutamine by leucine at position 207 of the gene (tga3Q207L) was introduced. This single amino acid exchange has previously been shown to result in an impairment of the intrinsic GTPase activity of the corresponding Gα subunits[20], leading to a constitutively active protein which stimulates downstream effectors even in the absence of receptor activation.
Mutants expressing the constitutively active tga3Q207L version were generated by co-transforming T. atroviride protoplasts with plasmids pKAtga3 (harboring tga3Q207L) and pII99. 30 colonies were isolated and transferred individually on PDA-geneticin selection plates of which 22 colonies developed. After purifying these transformants to mitotic stability, integration of the nptII gene into their genomes was examined by PCR as described above. Again, the expected 888-bp fragment could be amplified from all 22 transformants (Figure4), proving the suitabilty of this selection marker for T. atroviride once more.
Screening of T. atroviride co-transformants for the presence of the nptII gene. Genomic DNA was isolated from 22 geneticin-resistant transformants resulting from co-transformation of T. atroviride P1 with plasmids pKAtga3 and pII99 and analyzed by PCR for integration of the nptII gene using primers pII99Fw and pII99Rev. M, DNA size standard; +, plasmid pII99; 1–22, transformants; -, wild-type.
Screening for tga3Q207L in geneticin-resistant transformants
Southern analysis was performed to confirm that the tga3Q207L gene had been successfully integrated into the genome of the geneticin-resistant transformants. Two bands (1703-bp and 2272-bp) resulting from the endogenous tga3 gene were observed in the wild-type after digestion of its genomic DNA with Sac I (Figure5A and B). While transformant 3/3 exhibited two additional bands indicative for an ectopic single-copy integration of the tga3Q207L gene, only one band in addition to those resulting from the endogenous tga3 gene locus was observed in transformant 4/5 (Figure5A and B). To discern whether two bands are overlying each other in the Southern blot or whether this banding pattern indicates integration of a truncated copy of tga3Q207L in transformant 4/5, the presence of the full-length tga3Q207L expression cassette was tested by PCR using primers tga3PromFw and M13Fw (Table1). For both transformants a specific amplicon of 3343-bp was obtained (Figure6), confirming integration of a complete tga3Q207L expression cassette into the genome of transformant 4/5. To show that the integrated tga3Q207L gene actually is expressed, tga3 transcripts were sequenced. Four and two out of nine sequenced transcripts represented the mutated tga3 allele in the ectopic co-transformants 3/3 and 4/5, respectively, proving expression of the tga3Q207L gene copy.
Analysis of T. atroviride co-transformants for the presence of the tga3Q207Lgene. (A) Schematic drawing of the endogenous tga3 gene locus and of the transforming plasmid pKAtga3 harboring the plasmid pKAtga3 with tga3Q207Lexpression cassette gene including 1030-bp of the 5′ and 783-bp of the 3′ non-coding regions. The point mutation resulting in the Q207L amino acid exchange is indicated by a bold type arrow. Grey arrows indicate primers used for the PCR-based approaches. (B) Southern blot of genomic DNA digested with Sac I and hybridized with a probe containing ~ 1118-bp of the tga3 gene. The two bands of 1703-bp and 2272-bp present in all lanes including the wild-type (−) correspond to the endogenous tga3 gene copy. The presence of additional bands in co-transformants 3/3 (lane 7) and 4/5 (lane 11) indicate ectopic integration of the tga3Q207L gene. (C) The tga3 gene locus was amplified by PCR as given in the Methods section and the resulting 3377-bp amplicon digested with Bpu10 I. As an additional Bpu10 I recognition site was generated in tga3Q207L by introducing the A to T mutation, Bpu10 I digestion resulted in the two indicative fragments (1004-bp, 1392-bp) in co-transformants with homologous integration of tga3Q207L whereas the 2396-bp fragment remained undigested in the wild-type and ectopic co-transformants 3/3 (lane 12) and 4/5 (lane 13). WT; wildtype; M, DNA ladder.
Confirmation of integration of a full-length tga3Q207Lgene in ectopic co-transformants 3/3 and 4/5. The presence of a complete copy of the tga3Q207L gene in co-transformants 3/3 (lane 2) and 4/5 (lane 3) was confirmed by PCR using primers tga3PromFw, which binds to the 5′ non-coding region of tga3, and M13Fw, which binds to the backbone of plasmid pKAtga3 (Table1). Both co-transformants yielded the expected 3343-bp amplicon. Lane 1, wild-type; lane 4, plasmid pKAtga3.
To detect putative homologous recombinants among the remaining 20 transformants that showed the wild-type banding pattern in the Southern analysis, the tga3 gene locus was amplified by PCR. To this end, a primer which anneals upstream of the 5′ flanking sequences present in the tga3Q207L expression cassette (Figure5A and Methods section) was used. The resulting amplicon was digested with Bpu10 I. An additional Bpu10 I recognition site is present in tga3Q207L which originated from introducing the A to T mutation (Figure5A). In 13 transformants Bpu10 I digestion resulted in the 1004-bp and 1392-bp fragments indicative for the introduced mutation indicating that these are co-transformants with tga3Q207L integrated at the homologous tga3 gene locus (Figure5C).
In summary, 15 out of 22 transformants (68%) with a stable integration of nptII had additionally integrated the tga3Q207L gene into their genomes. A comparative co-transformation of T. atroviride with pKAtga3 (harboring tga3Q207L) and pAN7-1 (harboring the hygromycin B phosphotransferase-encoding hph gene) resulted in a co-transformation rate of 43% (data not shown). The yield of co-transformants obtained in this study by using either nptII or hph as selection marker is in accordance with previous publications reporting co-transformation efficiencies of 30 - 50% for T. harzianum and 90% for T. longibrachiatum[21–24].
Phenotypic characterization of co-transformants
The Gα subunit Tga3 was previously shown to play crucial roles for vegetative growth and mycoparasitism-relevant signal transduction processes in T. atroviride. Δtga3 mutants exhibited drastically reduced growth rates compared to the parental strain and were avirulent, i.e. unable to parasitize known susceptible host fungi[10].
For further analyses, co-transformants 3/3 and 4/5, which harbor the constitutively active Tga3 allele integrated ectopically, and co-transformant 2/1, which has integrated tga3Q207L at the homologous locus, were used. All three co-transformants showed a ~20% increased growth rate compared to the wild-type and a transformant only harboring nptII (Table2). While Δtga3 mutants continuously produced spores also in the dark[10], tga3Q207L co-transformants showed a conidiation behavior similar to the wild-type (Figure7), with the characteristic feature being production of concentric rings of conidiophores in response to the light/dark cycle as previously described for T. atroviride T. atroviride e.g.[25]. Furthermore, the number of produced conidia was similar to that of the wild-type and the nptII transformants (Table2).
Phenotype of nptII/tga3Q207Lco-transformants. Colony morphology of co-transformants 3/3 and 4/5 (with ectopic integration of the tga3Q207L gene) and co-transformant 2/1 (with homologous integration of the tga3Q207L gene) in comparison to the T. atroviride wild-type and a transformant having integrated nptII only upon growth on PDA for 8 days at 28°C either in the dark or under alternating light/dark (8/16 h) conditions. Co-transformants with either ectopic or homologous integration of tga3Q207L showed normal light-induced conidiation behavior. Pictures were taken after 4 and 8 days.
Previous studies furthermore showed that T. atroviride Δtga3 mutants were unable to attack and parasitize host fungi[10], while T. reesei mutants expressing the constitutively active Gα protein gna3 QL exhibited a higher efficiency of antagonism against Pythium ultimum[26]. The effect of the constitutively active Tga3Q207L protein on the mycoparasitic interaction between T. atroviride and the host fungus R. solani was assessed in plate confrontation assays. At the onset of mycoparasitism, tga3Q207L-expressing co-transformants behaved similar to the wild-type, whereas after eight and 14 days of incubation the co-transformants showed faster and more advanced mycoparasitic overgrowth and host lysis (Figure8). These results reveal that constitutive signaling via Tga3 leads to enhanced mycoparasitism in T. atroviride.
Mycoparasitic abilities of co-transformants. The antagonistic activity of co-transformants 3/3 and 4/5 (with ectopic integration of the tga3Q207L gene) and co-transformant 2/1 (with homologous integration of the tga3Q207L gene) in comparison to the T. atroviride wild-type was assessed in plate confrontation assays using R. solani as host fungus. While after four days of incubation at 28°C all strains showed a similar onset of mycoparasitism, after eight and 14 days the co-transformants exhibited more advanced overgrowth and host lysis compared to the wild-type.
Conclusions
In this study we present the successful establishment of the neomycin phosphotransferase-encoding nptII gene from E. coli as a robust selection marker for the genetic transformation of the biocontrol fungus T. atroviride. Respective transformants showed stable geneticin resistance, had no unwanted phenotypic alterations, and their phenotypes were similar both in the presence and absence of geneticin (Additional file1). The fact that no spontaneous geneticin-resistant T. atroviride colonies were detected makes the nptII selection marker a valuable alternative to the commonly used Hygromycin B phosphotransferase-encoding hph gene.
Co-transformation of T. atroviride with nptII and the tga3Q207L gene on two different plasmids resulted in a high number of colonies with stable geneticin resistance. 15 out of 22 (68%) tested transformants were real co-transformants also having integrated tga3Q207L. Interestingly, the majority of the co-transformants showed an integration of tga3Q207L at the homologous tga3 gene locus. Despite the fact that both plasmids were used in their circular forms for transformation, this high number of homologous recombinants may have resulted from the long flanking regions present in the tga3Q207L expression cassette which may have favored homologous double crossover events.
Constitutive activation of Tga3-mediated signaling led to an enhanced ability of T. atroviride to antagonize the phytopathogenic fungus R. solani thereby confirming the important role of G protein signaling in mycoparasitism.
Methods
Strains and media
Trichoderma atroviride strain P1 (ATCC 74058; teleomorph Hypocrea atroviridis) was used throughout this study. Fungal cultures were maintained on potato dextrose agar (PDA; Merck, Germany) at 28°C until intense sporulation. Rhizoctonia solani strain 1450 (strain collection of the Institute of Plant Pathology, University of Naples Federico II, Italy) was used as a plant pathogenic host. Escherichia coli JM109 served as a host for plasmid amplification and was grown as described by[27].
Sensitivity of T. atroviride mycelium and conidia to geneticin (G418)
Plasmid pII99[19] harbors the neomycin phosphotransferase-encoding nptII gene under control of the regulatory sequences of A. nidulans trpC (encoding a trifunctional protein involved in tryptophan biosynthesis,[28]). nptII confers resistance to geneticin (G418) and therefore allows the selection of transformed colonies on media containing this antibiotic. To determine the concentration of geneticin lethal for untransformed T. atroviride mycelia and conidia, either agar plugs covered with fungal mycelium or 103 conidia were placed on increasing concentrations (0–100 μg/ml) of geneticin (Roth, Karlsruhe, Germany) on PDA. A geneticin concentration of 80 μg/ml not allowing any growth of T. atroviride was used for transformant selection.
DNA procedures
Standard molecular techniques were performed according to[27]. Isolation of fungal DNA was carried out as described previously[29]. For standard PCR amplifications, recombinant Taq Polymerase (Fermentas, Vilnius, Lithuania) was used.
Expression plasmid construction and fungal transformation
The Phusion Site-Directed Mutagenesis Kit (Finnzymes, Vantaa, Finland) was used to construct a mutated version of tga3 [GenBank: AF452097], tga3Q207L, encoding a constitutively activated Gα protein with a single amino acid modification (Q207L). An equivalent mutation of the Tga3 orthologue in mammals resulted in the impairment of its intrinsic GTPase activity[30]. Corresponding mutations were introduced into other fungi in order to evaluate the function of Gα subunits e.g.[20, 31, 32]. Two oligonucleotides (tga3PhusionFw and tga3PhusionRev) (Table1) were designed based on the T. atroviride genomic tga3 sequence with tga3PhusionFw harboring the single-point mutation. In a PCR approach these oligonucleotides were used to amplify the tga3 gene including approximately 1000-bp of 3′ and 5′ non-coding sequences in plasmid pGEM-T (Promega, Madison, WI). The linear amplified target plasmid with the mutated tga3Q207L gene was re-circularized by ligation resulting in pKAtga3. The tga3Q207L-harboring insert was verified by sequencing to confirm that only the desired mutation had been inserted.
T. atroviride protoplasts were transformed with 10 μg of plasmid pII99 only or 10 μg of pKAtga3 in co-transformation with 2 μg of pII99 according to[33].
Recovery of transformants and molecular analysis
Putative transformants were selected on PDA plates containing 80 μg/ml geneticin, a concentration which prevented the growth of T. atroviride strain P1. Transformants were transferred individually onto PDA supplemented with 80 μg/ml geneticin. Geneticin-resistant strains were individually transferred to PDA for sporulation and colonies obtained from three subsequent single spore isolation steps were used for further analysis. The presence of nptII was tested by PCR using oligonucleotides pII99Fw and pII99Rev (Table1). For the purpose of comparison, hygromycin-resistant transformants were obtained by transforming T. atroviride with 10 μg of plasmid pAN7-1[34] only or 10 μg of pKAtga3 in co-transformation with 2 μg of pAN7-1. Transformants were selected on 200 μg/ml hygromycinB (Calbiochem, San Diego, USA).
Integration of tga3Q207L into the genome was analyzed by Southern blotting in transformants exhibiting a positive result in nptII screening. Standard molecular methods[27] were used for DNA electrophoresis and blotting. DNA labeling, hybridization and detection were performed according to the DIG High Primer DNA Labeling and Detection Starter Kit I protocols (Roche Applied Science). The DIG-labeled probe was amplified from pKAtga3 with oligonucleotides tga3ProbeFw and tga3ProbeRev (Table1) producing an amplicon of 1118-bp. For detecting homologous recombinants among the transformants, the tga3 gene locus was amplified by PCR using Phusion High-Fidelity DNA Polymerase (Thermo Fischer Scientific, Waltham, USA) and primers tga3-locus-check (which anneals upstream of the tga3 5′ flanking sequences present in the tga3Q207L expression cassette) and tga3TermRev. The resulting amplicon was digested with Bpu10 I and the banding pattern analyzed by agarose gel electrophoresis.
To prove that the tga3Q207L gene copy was indeed expressed, co-transformants 3/3 and 4/5 were grown on PDA plates for 3 days, RNA isolated and reverse-transcribed to cDNA, and fragments of the tga3 gene were amplified using primers Mut-tga3Fw and Mut-tga3Rev (Table1). After cloning the resulting 604-bp fragments into pGEM-T (Promega, Madison, WI), the inserts of nine colonies representing tga3 transcripts were sequenced to determine the ratio of wild-type and mutated alleles.
Phenotype analysis of transformants
For assessing growth of T. atroviride transformants and wild-type, 5 mm diameter mycelial agar plugs of the respective strain were placed in the center of a PDA plate and incubated at 28°C. Radial growth was measured every 24 h until the plate was fully covered. Conidia were quantified by plating a freshly harvested and filtered spore suspension of the respective strain containing 1 × 106 conidia onto PDA plates. Conidia produced on three independent plates after three days of incubation at 28°C were harvested and counted.
For plate confrontation assays, 5 mm mycelial agar plugs of the respective T. atroviride strains and R. solani as host were placed on PDA plates at a distance from each other of 4 cm and incubated in the dark at 28°C for 14 days. Pictures were taken at days four, eight, and 14.
Abbreviations
- BCA:
-
Biocontrol agent
- PDA:
-
Potato dextrose agar
- Bp:
-
Base-pair
- h:
-
Hour.
References
Druzhinina IS, Seidl-Seiboth V, Herrera-Estrella A, Horwitz BA, Kenerley CM, Monte E, Mukherjee PK, Zeilinger S, Grigoriev IV, Kubicek CP: Trichoderma: the genomics of opportunistic success. Nat Rev Microbiol. 2011, 9 (10): 749-759. 10.1038/nrmicro2637.
Harman GE, Howell CR, Viterbo A, Chet I, Lorito M: Trichoderma species–opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004, 2 (1): 43-56. 10.1038/nrmicro797.
Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, Thon M, Zeilinger S, Casas-Flores S, Horwitz BA, Mukherjee PK, Mukherjee M, Kredics L, Alcaraz LD, Aerts A, Antal Z, Atanasova L, Cervantes-Badillo MG, Challacombe J, Chertkov O, McCluskey K, Coulpier F, Deshpande N, von Döhren H, Ebbole DJ, Esquivel-Naranjo EU, Fekete E, Flipphi M, Glaser F, Gómez-Rodríguez EY: Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 2011, 12 (4): R40-10.1186/gb-2011-12-4-r40.
Verma M, Brar SK, Tyagi RD, Surampalli RY, Valero JR: Antagonistic fungi, Trichodrema spp.: panoply of biological control. Biochem Eng. 2007, 37: 1-20. 10.1016/j.bej.2007.05.012.
Brunner K, Omann M, Pucher ME, Delic M, Lehner SM, Domnanich P, Kratochwill K, Druzhinina I, Denk D, Zeilinger S: Trichoderma G protein-coupled receptors: functional characterisation of a cAMP receptor-like protein from Trichoderma atroviride. Curr Genet. 2008, 54 (6): 283-299. 10.1007/s00294-008-0217-7.
Mukherjee PK, Latha J, Hadar R, Horwitz BA: Role of two G-protein alpha subunits, TgaA and TgaB, in the antagonism of plant pathogens by Trichoderma virens. Appl Environ Microbiol. 2004, 70: 542-549. 10.1128/AEM.70.1.542-549.2004.
Omann M, Lehner S, Escobar C, Brunner K, Zeilinger S: The 7-transmembrane receptor Gpr1 governs processes relevant for the antagonistic interaction of Trichoderma atroviride with its host. Microbiology. 2012, 158: 107-118. 10.1099/mic.0.052035-0.
Rocha-Ramirez V, Omero C, Chet I, Horwitz BA, Herrera-Estrella A: Trichoderma atroviride G-protein alpha-subunit gene tga1 is involved in mycoparasitic coiling and conidiation. Eukaryot Cell. 2002, 1: 594-605. 10.1128/EC.1.4.594-605.2002.
Reithner B, Brunner K, Schuhmacher R, Peissl I, Seidl V, Krska R, Zeilinger S: The G protein alpha subunit Tga1 of Trichoderma atroviride is involved in chitinase formation and differential production of antifungal metabolites. Fungal Genet Biol. 2005, 42 (9): 749-760. 10.1016/j.fgb.2005.04.009.
Zeilinger S, Reithner B, Scala V, Peissl I, Lorito M, Mach RL: Signal transduction by Tga3, a novel G protein alpha subunit of Trichoderma atroviride. Appl Environ Microbiol. 2005, 71 (3): 1591-1597. 10.1128/AEM.71.3.1591-1597.2005.
Schuster A, Schmoll M: Biology and Biotechnology of Trichoderma. Appl Microbiol Biotechnol. 2010, 87 (3): 787-799. 10.1007/s00253-010-2632-1.
Ruis-Diez B: Strategies for the transformation of filamentous fungi. J Appl Microbiol. 2002, 92 (2): 189-195. 10.1046/j.1365-2672.2002.01516.x.
Padilla IMG, Burgos L: Aminoglycoside antibiotics: structure, functions and effects on in vitro plant culture and genetic transformation protocols. Plant Cell Rep. 2010, 29: 1203-1213. 10.1007/s00299-010-0900-2.
Son H, Seo YS, Min K, Park AR, Lee J, Jin JM, Lin Y, Cao P, Hong SY, Kim EK, Lee SH, Cho A, Lee S, Kim MG, Kim Y, Kim JE, Kim JC, Choi GJ, Yun SH, Lim JY, Kim M, Lee YH, Choi YD, Lee YW: A phenome-based functional analysis of transcription factors in the cereal head blight fungus Fusarium graminearum. PLoS Pathog. 2011, 7 (10): e1002310-10.1371/journal.ppat.1002310.
Shimizu T, Ito T, Kanematsu S: Transient and multivariate system for transformation of a fungal plant pathogen, Rosellinia necatrix, using autonomously replicating vectors. Curr Genet. 2012, 58 (3): 129-138. 10.1007/s00294-012-0370-x.
Weiland JJ: Transformation of Pythium aphanidermatum to geneticin resistance. Current Genet. 2003, 42: 344-352. 10.1007/s00294-002-0359-y.
Rodríguez-Sáiz M, Lembo M, Bertetti L, Muraca R, Velasco J, Malcangi A, de la Fuente JL, Barredo JL: Strain improvement for cephalosporin production by Acremonium chrysogenum using geneticin as a suitable transformation marker. FEMS Microbiol Lett. 2004, 235 (1): 43-49.
Vijn I, Govers F: Agrobacterium tumefaciens-mediated transformation of the oomycete plant pathogen Phytophtora infestans. Mol Plant Pathol. 2003, 4 (6): 459-467. 10.1046/j.1364-3703.2003.00191.x.
Namiki F, Matsunaga M, Okuda M, Inoue I, Nishi K, Fujita Y, Tsuge T: Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis. Mol Plant Microbe Interact. 2001, 14: 580-584.
Masters SB, Miller RT, Chi MH, Chang FH, Beiderman B, Lopez NG, Bourne HR: Mutations in the GTP-binding site of GS alpha alter stimulation of adenylyl cyclase. J Biol Chem. 1989, 264: 15467-
Bae YS, Knudsen GR: Cotransformation of Trichoderma harzianum with beta-glucuronidase and green fluorescent protein genes provides a useful tool for monitoring fungal growth and activity in natural soils. Appl Environ Microbiol. 2000, 66: 810-10.1128/AEM.66.2.810-815.2000.
Bowen J, Crowhurst R, Templeton M, Stewart A: Molecular markers for a Trichoderma harzianum biological control agent: Introduction of the hygromycin B resistance gene and the β‐glucuronidase gene by transformation. New Zeal J Crop Hort Sci. 1996, 24: 219-228. 10.1080/01140671.1996.9513956.
Inglis PW, Queiroz PR, Valadares-Inglis MC: Transformation with green fluorescent protein of Trichoderma harzianum 1051, a strain with biocontrol activity against Crinipellis perniciosa, the agent of witches'-broom disease of cocoa. J Gen Appl Microbiol. 1999, 45: 63-67. 10.2323/jgam.45.63.
Sánchez-Torres P, González R, Pérez-González JA, González-Candelas L, Ramón D: Development of a transformation system for Trichoderma longibrachiatum and its use for constructing multicopy transformants for the egl1 gene. Appl Microbiol Biotechnol. 1994, 41: 440-446.
Steyaert JM, Weld RJ, Stewart A: Isolate-specific conidiation in Trichoderma in response to different nitrogen sources. Fungal Biol. 2010, 114 (2–3): 179-188.
Do Nascimento Silva R, Steindorff AS, Ulhoa CJ, Felix CR: Involvement of G-alpha protein GNA3 in production of cell wall-degrading enzymes by Trichoderma reesei (Hypocrea jecorina) during mycoparasitism againstPythium ultimum. Biotechnol Lett. 2009, 31: 531-536. 10.1007/s10529-008-9900-5.
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A laboratory Manual. 1989, New York: Cold Spring Harbor Laboratory Press, 2
Mullaney ER, Hamer JE, Roberti KA, Yelton MM, Timberlake WE: Primary structure of the trpC gene of Aspergillus nidulans. Mol Gen Genet. 1985, 199: 37-45. 10.1007/BF00327506.
Peterbauer CK, Lorito M, Hayes CK, Harman GE, Kubicek CP: Molecular cloning and expression of the nag1 gene (N-acetyl-β-D-glucosaminidase-encoding gene) from Trichoderma harzianum P1. Current Genet. 1996, 30: 325-331. 10.1007/s002940050140.
Berman DM, Wilkie TM, Gilman AG: GAIP and RGS4 Are GTPase-activating proteins for the Gi Subfamily of G protein α subunits. Cell. 1996, 86: 445-452. 10.1016/S0092-8674(00)80117-8.
Regenfelder E, Spellig T, Hartmann A, Lauenstein S, Bölker M, Kahmann R: G proteins in Ustilago maydis: transmission of multiple signals? . EMBO J. 1997, 16: 1934-1942. 10.1093/emboj/16.8.1934.
Segers GC, Nuss DL: Constitutively activated G [alpha] negatively regulates virulence, reproduction and hydrophobin gene expression in the chestnut blight fungus Cryphonectria parasitica. Fungal Genet Biol. 2003, 38: 198-208. 10.1016/S1087-1845(02)00534-0.
Peterbauer C, Litscher D, Kubicek C: The Trichoderma atroviride seb1 (stress response element binding) gene encodes an AGGGG-binding protein which is involved in the response to high osmolarity stress. Mol Genet Genomics. 2002, 268: 223-231. 10.1007/s00438-002-0732-z.
Punt PJ, Oliver RP, Dingemanse MA, Pouwels PH, van den Hondel CAMJJ: Transformation of Aspergillus nidulans based on the hygromycin B marker from Escherichia coli. Gene. 1987, 56: 117-124. 10.1016/0378-1119(87)90164-8.
Acknowledgements
This work was supported by the Austrian Science Fund FWF (grants P18109-B12 and V139-B20) and the Vienna Science and Technology Fund WWTF (grant LS09-036). We acknowledge Alexander Lichius for copyediting the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SZ conceived and designed the study and wrote parts of the manuscript. SG and MO participated in the design of the experiments, generated the transformants and contributed to manuscript writing. SG, CER, and TR performed phenotypic analyses. All authors read and approved the final manuscript.
Electronic supplementary material
13104_2012_2009_MOESM1_ESM.pdf
Additional file 1:Phenotype of nptII / tga3Q207Lco-transformants on PDA and PDA + 80μg/ml geneticin. The figure shows the colony morphology of co-transformants 3/3 and 4/5 (with ectopic integration of the tga3Q207L gene) and co-transformant 2/1 (with homologous integration of the tga3Q207L gene) grown on PDA and PDA + 80μg/ml geneticin for 4 days at 28°C in the dark. (PDF 271 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Gruber, S., Omann, M., Rodrìguez, C.E. et al. Generation of Trichoderma atroviride mutants with constitutively activated G protein signaling by using geneticin resistance as selection marker. BMC Res Notes 5, 641 (2012). https://doi.org/10.1186/1756-0500-5-641
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1756-0500-5-641